ABSTRACT
Breast cancer (BC) is a leading cause of cancer deaths in Libyan women. BRCA1 variants differ globally due to the diversity of genetic makeup and populations history. Their distribution, prevalence, and significance in Libyans remain largely unexplored. This study investigated the characteristics and distribution of BRCA1 variants in exons 5, 11, and 20 in Libyan families with BC. Thirty-six BC patients at ≤ 45 years, between 46–50 years and with a family history of breast, ovarian, pancreatic or prostate cancer in close relatives, or with triple-negative BC, were selected from 33 unrelated families during 2018–2020 at the National Cancer Institute, Sabratha, Libya. From these 33 families, 20 women (18 BC patients and two unaffected) were screened for BRCA1 exons 5, 11 and 20 using Sanger sequencing. All families completed an epidemiology and family history questionnaire. Twenty-seven variants (26 in exon 11 and 1 in exon 20, minor allele frequency of < 0.01) were detected in 10 of 18 unrelated families (55.6%.) Among the 27 variants, 26 (96%) were heterozygous. A frameshift pathogenic variant, c.2643del, and one novel variant c.1366A>G were identified. Furthermore, seven variants with unknown clinical significance were detected: c.1158T>A, c.1346C>G, c.1174C>G, c.3630 G>T, c.3599A>T, and c.3400 G>C in exon 11, and c.5244T>A in exon 20. Six variants with conflicting pathogenicity interpretations, c. 3460T>A, c. 3572 G>A, c. 3700 G>C, c. 1246C>G, c. 1344C>G, and c. 1054 G>A, were also identified. Twelve benign/likely benign variants were identified. Rare BRCA1 variants that have not been reported in North Africa were found in Libyan patients. These findings provide preliminary insights into the BRCA1 variants that could contribute to hereditary BC risk in Libyans. Further functional, computational, and population analyses are essential to determine their significance and potential impact on BC risk, which could ultimately lead to more personalized management strategies.
1. Introduction
Breast cancer (BC) remains a significant global health concern. Recent estimates from the Global Cancer Observatory (2022) reveal that it is the second most common cancer worldwide, with roughly 2.3 million new cases diagnosed annually, representing 11.6% of all new cancers. Tragically, it is also the fourth leading cause of cancer death among women, with an estimated 1.6 million deaths in 2022 (6.9% of all cancer deaths globally) [Citation1]. Incidence rates of BC in transitioned countries are higher, but rates in transitioning countries have been rapidly increasing, and mortality rates remain high [Citation1,Citation2]. Some of the most rapid increases are occurring in North Africa, which had a 37.5 and 15.2 per 100,000 age-standardized incidence and death rate in females in 2019, up 90.9 and 24.0% since 1990, respectively [Citation3]. The young age of onset and high histological grade of BC in the region point to the involvement of genetic factors [Citation4].
Numerous studies have demonstrated that variants in BC susceptibility genes, such as BRCA1 and BRCA2, increase the risk of developing BC [Citation5,Citation6]. These genes are hereditary risk factors for BC and are responsible for 25% of familial BC and 5–10% of all BC cases [Citation7]. The frequency of BRCA1 and BRCA2 gene germline alterations varies among different geographical and ethnic populations, with some variants being specific to certain ethnic groups while others are shared across all populations [Citation5–7].
Consanguineous marriage is common in North Africa, creating a genetic environment conducive to the emergence and persistence of specific, recurrent, and founder BRCA1/2 pathogenic variants [Citation4]. Beyond established BRCA1/2 variants, a growing body of research reveals the presence and impact of novel mutations in geographically and ethnically diverse populations within North Africa [Citation8]. These findings highlight the unique genetic landscapes, largely focused on European populations, and emphasize the unique genetic landscapes of these regions and their contribution to early-onset, hereditary, and male BC [Citation4,Citation9–11]. Whereas BRCA1 and BRCA2 are the most commonly investigated genes for their association with BC, the presence of variants of uncertain significance (VUS) adds another dimension. Among women of non-Ashkenazi ancestry, those of African origin had the highest rate of BRCA1/2 variants of uncertain significance (16.5%) compared to Western European women (5.7%) [Citation12]. Existing BRCA1 databases contain mostly European-specific variants, while African variants are underreported [Citation13].
Libya is a country with a diverse population, made up of various ethnic groups, with Arabs being the largest group [Citation14]. However, intermarriage has resulted in a mixed race of Imazighen and Arab ancestry that accounts for about 93% of the current population. The remaining 7% consists of Imazighen, Tuareg, Tebou, Black Africans, and small groups of Greeks, Armenians, Cretans, and Maltese [Citation15]. Unlike other North African countries, BRCA1/2 research in Libya is lacking [Citation9]. This gap is due to limited access to clinical genetic services, fueled by both high costs and scarce availability of genetic testing. Discovering BRCA1/2 variants in an understudied population like Libyans will provide new information on the genetic landscape of hereditary BC in North Africa, improve the understanding of how ethnic diversity impacts variant actionability, and inform the development of more personalized approaches to cancer treatment. The purpose of this study was to investigate the distribution and characteristics of BRCA1 variants in exons 5, 11 and 20 in Libyan families These exons were chosen because they represent hotspots for variants within their respective domains: exon 5 in the RING (Really Interesting New Gene) domain, exon 11 in the region encoded by exons 11–13, and exon 20 in the BRCT (BRCA1 C-terminal) domain [Citation16,Citation17]. These exons exhibit high rates of clinically significant variations [Citation16]. Notably, exon 11 encompasses 60% of the coding sequence, making it a particularly relevant target for variant analysis [Citation17].
2. Materials and methods
2.1. Patients and criteria
Between 2018 and 2020, from a cohort of 176 BC patients followed up at the National Cancer Institute (NCI) in Sabratha, Libya, 36 BC patients belonging to 33 unrelated families were selected. Following the guidelines of v2.2022 of the NCCN [Citation18], patients were selected based on the following criteria:
Woman diagnosed with BC when 45 years old or younger;
Woman with BC diagnosed between the ages of 46 and 50 with any of the following:
multiple primary BC
one or more close blood relative with breast, ovarian, pancreatic, or prostate cancer at any age
Woman diagnosed with BC at the age of 51 years and one or more close blood relative with BC at the age of 50 years or younger;
Woman of any age with triple-negative BC;
BC patient of any age with one or more male close blood relative with BC.
In these 33 families, 20 women (18 with BC and two unaffected women) underwent genetic testing for variants in BRCA1 exons 5, 11, and 20. At the outset of sample collection, all the participants provided informed consent for participation in the research study and genetic analysis, and provided relevant information about their personal and familial cancer histories and geographic origins. Medical records and pathology reports were used to collect clinical and pathological characteristics. The study was approved by the local Research Ethics Committee of the National Cancer Institute (NCI), Sabratha, Libya.
Based on specific characteristics, tumors were classified into four molecular subtypes as described by Goldhirsch et al. (2011). These subtypes are luminal A (ER+ and/or PR+, Ki67 low, and HER2-), luminal B (ER+ and/or PR+, Ki67 high, and/or HER2+), HER2-positive (ER-, PR-, and HER2+), and triple-negative (ER-, PR-, and HER2-) [Citation19]. To determine the expression levels of ER, PR, HER2, and Ki67, medical records were reviewed. ER and PR were considered positive or negative when less than 1% of tumor cells showed positive nuclear staining. High Ki67 expression was identified in patients with more than 20% positive nuclei, while those with 20% or less positive nuclei were considered to have low Ki67 expression. A HER2 positivity was indicated by a score of + 3. Histological tumors were graded using the modified Scarf-Bloom-Richardson grading system (SBR), and the tumor stage was determined according to the 7th edition of the AJCC cancer staging manual 2010 [Citation20].
2.2. Extraction of genomic DNA
The samples were examined at the Libyan Biotechnology Research Center’s Department of Genetic Engineering in Tripoli, Libya. The QIAamp® Blood Mini Kit (Qiagen) was used to extract genomic DNA from peripheral blood collected on EDTA, according to the manufacturer’s instructions. The quantity and the quality of genomic DNA were determined with a NanoDrop spectrophotometer and visualized by 1.5% agarose electrophoresis.
2.3. Polymerase chain reaction (PCR) of the targeted BRCA1 exons
Exons 5, 11 and 20 of the BRCA1 gene were PCR amplified in a total reaction volume of 25 µL containing 12.5 µL of 1×QIAamp® Green Master Mix (Promega), 0.3 µL (8.3 µM) upstream and downstream primers, 2.5 µL of nuclease-free water, and 2 µL of 30 ng/µL DNA template. PCR was performed in a thermal cycler (Applied Biosystems Fisher thermocycler) with initial denaturation at 95°C for 2 min followed by 35 cycles of 95°C for 40 sec, annealing at temperatures specific for each primer pair, and extension at 72°C 30 sec. The primers were designed by Centre Jean Perrin [Citation21]. To be able to analyze the large exon 11 completely, the exon was divided into nine overlapping parts (Supplementary Table S1). PCR products were verified by electrophoresis in 1.2% agarose gel containing GoTaq® (Biotium) and visualized with UV transillumination. GelRed® SV Gel and the PCR Clean-Up System (Promega) were used to purify PCR products. Samples from 18 BC patients and 2 unaffected women produced bands of sufficient quality for Sanger sequencing.
2.4. DNA sequencing
The purified amplicons were sequenced using the Applied 152 Biosystems BigDye V3.1 cycle sequencing kit. Cycle sequencing started with 1 minute at 96°C followed by 25 cycles of 10 seconds at 96°C, 5 seconds at 50°C, and 4 minutes at 60°C. Following ethanol/EDTA precipitation according to the manufacturer’s instructions, Sanger sequencing was performed on a 3730×l DNA Analyzer (Applied Biosystems) using trace2ps, V 2.72 software.
2.5. Nomenclature and variants analysis and in-silico prediction
Unipro UGENE software was used to analyze ABI files generated from sequencing [Citation22]. Then, samples were aligned using the National Center for Biotechnology Information (NCBI) BLASTN tool (https://blast.ncbi.nlm.nih.gov/blast.cgi).
When variants were discovered, reference SNPs (rsID) were reported. Furthermore, genomic position, allele frequency, transcript (NM_007294.4), and protein (NP_009225.1) locations, as well as ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) clinical significance, were presented. ClinVar classifies the variants as recommended by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines into six categories: pathogenic, likely pathogenic, uncertain significance, variant of unknown significance, likely benign, and benign [Citation23]. Additionally, to facilitate the analysis of variants of unclear clinical significance detected in our patients, we utilized the VarSome (https://varsome.com/) and Franklin Genoox (https://franklin.genoox.com) variant analysis/interpretation tools. The VarSome tool integrates multiple prediction algorithms with clinical knowledge to provide more informed assessments of variant pathogenicity. Furthermore, we used the protein data base InterPro (https://www.ebi.ac.uk/interpro), which provides functional analysis of proteins by classifying them into families and predicting domains and important sites were used.
We also generated a Variant Call Format (VCF) to facilitate further analysis of the variants. We employed the Ensemble Variant Effect Predictor (VEP; https://www.ensembl.org/info/docs/tools/vep/index.html) to analyze the effects of the new variants we identified. We utilized three primary scoring systems for our analysis: Sorting Intolerant from Tolerant (SIFT), which assesses the conservation of amino acid residues across species to predict the impact of amino acid substitutions on protein structure and function; Polymorphism Phenotyping v2 (PolyPhen-2), which evaluates the impact of variations on protein function based on various features, such as evolutionary conservation, physicochemical properties, and structural information; and combined annotation‐dependent depletion (CADD), which integrates multiple types of annotations and scores the relative deleterious effects of variants. Deleterious thresholds used in the above-mentioned tools were as follows: SIFT < 0.05 [Citation23], PolyPhen-2 > 0.5 [Citation24]. For CADD, we used the highest Phred-like score cutoff of 20, as recommended by Rentzsch et al.(2019) [Citation25,Citation26].
3. Results
3.1. Characteristics of the families and patients’ cohort
Out of 36 patients, 33 had unilateral BC (6 of whom had multifocal BC), one had bilateral BC, one had both breast and ovarian cancer, and the right-side breast was affected in 17 women (54.8%). The age at diagnosis ranged from 20 to 67 years, with a mean age of 45 years. Luminal B was the most common subtype (52.8%) followed by luminal A and TNBC (19.4%), and HER2+ (3.8%). Based on the data, invasive ductal carcinoma (IDC) is the most frequent type of BC (83.3%), and the majority of our patients have tumors with histological grades 2 and 3, as well as TNM stages II and III (). These patients belonged to 33 different families from various Western regions in Libya. Supplementary table S2 shows that 21 out of the 33 proband cases were diagnosed at the age of 45 years or younger, and that 84.9% (28/33) had at least one first- or second-degree relative with breast or ovarian cancer, while 18.2% (6/33) had a first- or second-degree relative with male BC. Another 12.1% (4/33) had no family history of BC or others. Five of the 33 (15.2%) probands were over the age of 50, and four of them reported a family history of BC in at least one close relative under the age of 50.
Table 1. Demographic and pathological features of the patients’ cohort.
3.2. Identification of BRCA1 variants
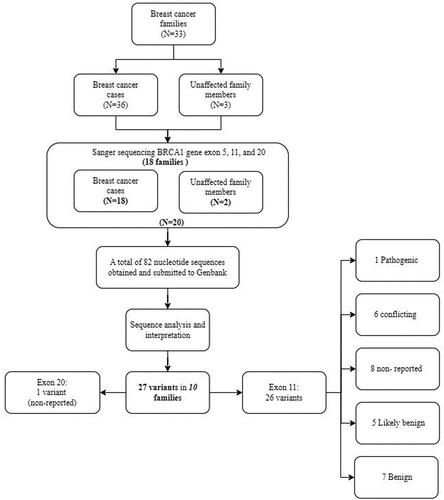
Out of the 33 selected families, 20 cases belonging to 18 families were analyzed for BRCA1 variants (exons 5, 11, and 20). Twenty-seven variants (1 in exon 20 and 26 in exon 11), were found in 10 of the 18 families (55.6%). present the distribution of these variants. According to the ClinVar classification, one (3.7%) was pathogenic, one novel allele (3.7%), 7 (25.9%) were of unknown clinical significance, 6 (22.2%) had conflicting interpretations of pathogenicity (classified as VUS or likely benign), 5 (18.5%) were likely benign, and 7 (25.9%) were benign. Furthermore, 19 variants were missense, 7 were synonymous and 1 was deleterious (). Out of all detected variations, 26 (96%) were heterozygous alleles. Additionally, based on the data from the 1000 Genome Project, all of the identified variants are considered rare with a population minor allele frequency of ≤ 1%. A flowchart of our study is shown in .
Table 2. Pathogenicity status of BRCA1 germline variants identified in breast cancer families in this study and reporting status in ClinVar.
Table 3. Benign status of BRCA1 variants in this study.
4. Pathogenic, likely benign variants, and variants with conflicting interpretations or unknown clinical significance
The BRCA1 c.2643del (p.Glu881fs), a frameshift pathogenic variant located on exon 11 (, and ), was discovered in a proband with breast cancer at 37 years, and with local recurrence. This proband woman had variants in exon 11 [Citation1]: synonymous likely benign variant c.1353A>G (p.Ser451=) [Citation2], novel allele c.1366A>G (p.Ile456Val) [Citation3], missense variant c.1158T>A (p.Phe386Leu) with unknown clinical significance, and [Citation4] missense variant c.1054 G>A (p.Glu352Lys) with conflicting interpretations of pathogenicity (). The last variant was frequent finding in four other unrelated probands, with an allele frequency of 71.7% (, and ).
Figure 2. Pedigrees of families with pathogenic, conflicting pathogenicity, likely benign, and non-reported clinically significant variants. A blackened circle represents an affected individual with breast cancer, while a divided circle represents an unaffected individual carrying variations. An affected individual with leukemia is represented by a green square, and one with colon cancer by an orange circle. The proband is indicated by an arrow.
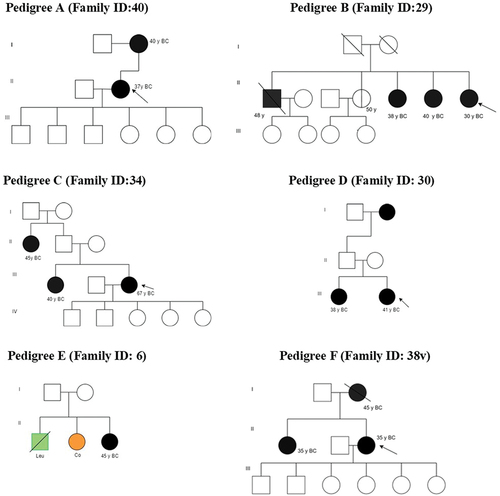
Table 4. Clinicopathological and molecular characteristics of BRCA1 variants.
In a proband diagnosed with triple-negative breast cancer (TNBC) at age 50 years and with 10 years of follow-up, we found three BRCA1 mutations on exon 11 with conflicting interpretations of pathogenicity: c.3700 G>C (p.Val1234Leu), c.3572 G>A (p.Ser1191Asn), and c.3460T>A (p.Leu1154Ile). Additionally, three other missense variants of unknown clinical significance were detected: c.3630 G>T (p.Glu1210Asp), c.3599A>T (p.Gln1200Leu), and c.3400 G>C (p.Glu1134Gln). Two likely benign BRCA1 variants were also identified: c.3563 G>T (p.Arg1188Met) and 1629T>C (p.Gly543=). These variants are listed in .
On exon 11, we also identified two other synonymous, likely benign BRCA1 variants, c.1173A>G (p.Glu391=>G) and c.2733A>G (p.Gly911=), in a proband who had a family history of male and female breast cancer. Moreover, the unaffected sister of the proband also tested positive for the likely benign c.2733A>G ( and ).
Interestingly, two more missense conflicting pathogenicity c.1246C>G (p.Leu416Val) andc.1344C>G(p.His448Gln) variants and two missense unknown clinical significance c.1346C>G (p.Ser449Cys) and c.1174C>G (p.Leu392Val) variants (exon 11) were observed in a 41-year-old proband at diagnosis. The patient had mucinous carcinoma, locally recurrent, and metastasis in bone and liver. This proband had a family history of female and male breast cancer (, and ).
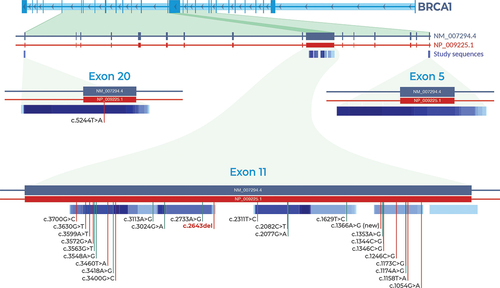
When screening exon 20 of BRCA1, c.5244T>A variant (p.Gly1748=) detected in a bilateral breast cancer proband diagnosed at the age of 45 years who had bone metastasis, but the clinical significance of this variant was not clear (, and ). The distribution and locations of all variants identified in this study are illustrated in .
Figure 3. Distribution and locations of identified BRCA1 variants. Only one variant was detected in exon 20. Exon 11 had 26 variants. One is pathogenic (c.2643del) and one is a novel allele (c.1366A>G). No variants were detected in exon 5. Vertical red indicate pathogenicity or conflicting interpretations of pathogenicity. Vertical green vertical lines indicate benign or likely benign variants.
4.1. Analysis and in-silico prediction of variants of unknown clinical significance or having conflicting interpretations
Our analysis identified seven genetic variants with no reported clinical significance in the literature and major reference databases. They were either absent from ClinVar or not accompanied by any interpretation of their clinical significance. To further investigate these variants, we used in-silico prediction tools and interpretation databases, such as VarSome and Franklin Genoox.
Among these, the pathogenicity of three variants (c. 1366A>G, c. 3630 G>T, and c. 3400 G>C) were defined as ‘not-reported’ on ClinVar, classified as likely benign on the VarSome database, and as VUS on the Franklin Genoox database. The Phred scores for these variants are 20.5, 12.9, and 22.8, respectively. However, the variants were predicted to be likely deleterious by SIFT and possibly damaging by polyphen2 (). c.3630 G>T and c.3400 G>C variants are also located within the region of unclassified membrane-bound proteins, according to the InterPro protein database. Another variant in exon 11 (c.1158T>A) has not provided clinical significance on ClinVar, but the InterPro protein database placed it within the serine-rich domains of amino acids 345–508. However, VarSome and Franklin Genoox declared it as VUS, while PolyPhen-2 predicted it as possibly damaging with a Phred score of 23.4 ().
Table 5. Unclassified BRCA1 variants: frequency, predicted impact, and classification discrepancies.
Furthermore, the c.3599A>T variant in exon 11 has not been reported on ClinVar, and according to the InterPro protein database, it is located within the consensus disorder predicted region. The variant was classified as likely pathogenic by VarSome and VUS by the Franklin Genoox. Nonetheless, the SIFT tool predicted that it is deleterious ().
Two variants in exon 11 of BRCA1, c.3460T>A (Phred score 20.6) and c.3572 G>A (Phred score 11.3), have conflicting interpretations of pathogenicity. Both variants were classified as VUS by VarSome and the Franklin Genoox databases. However, SIFT predicted them to be deleterious, while PolyPhen2 classified them as possibly damaging. Another exon 11 variant, c.3700 G>C, was predicted with a Phred score of 17.8 and classified as VUS by the Franklin Genoox database (). Interestingly, variant c.3563 G>T (exon 11), which is classified as likely benign in the ClinVar database, was predicted to be deleterious by SIFT and possibly damaging by Polyphen2, with a Phred score of 23.
5. Benign BRCA1 variants
Seven unrelated probands were diagnosed with BC and one unaffected sister of one proband carried both the c.2082C>T and c.3548A>G variants. These variants were observed in 42.9% of the cases screened for heterozygosity and 14.3% of those screened for homozygosity (). Variant c.2311T>C was detected in five unrelated probands and one unaffected woman with an allele frequency of 28.6% for heterozygosity and 14.3% for homozygosity, while variant c.2077 G>A was frequent in unrelated probands and one unaffected relative with an allele frequency of 28.6%. One proband and her unaffected sister also had variant 3024 G>A with an allele frequency of 20% ().
6. Discussion
No surveys have investigated BRCA1/2 gene variants in the Libyan population [Citation4]. Therefore, our understanding of the significance of BRCA1/2 variants in Libyan women with familial BC remains largely unexplored. In this study, we screened three exons [Citation5,Citation11], and [Citation20] of the BRCA1 gene in a cohort of 18 BC families from West Libya. These exons have been previously examined in North African countries neighboring Libya in the West (Tunisia and Algeria), where 22 pathogenic variants have been reported [Citation4]. Specifically, three pathogenic variants were identified in exon 5 [Citation21,Citation23,Citation24], seventeen in exon 11 [Citation6,Citation7,Citation11,Citation21,Citation27–29], and two in exon 20 [Citation7,Citation30].
Exon 11 of the BRCA1 gene is especially significant. It accounts for roughly 60% of the entire BRCA1 coding sequence. This exon contains more than half (57%) of the BRCA1 variants identified in our study, which emphasizes its significance in the function of the BRCA1 gene and the development of diseases such as hereditary breast and ovarian cancer [Citation31]. Our analysis revealed a higher frequency of BRCA1 mutations within exon 11 compared to exons 5 and 20. This finding aligns with a 2014 Tunisian study for exons 5 and 20, but differs regarding the prevalence of mutations in exon 11 [Citation10].
Our study in the Libyan population further underscores the remarkable diversity of germline BRCA1/2 variants across ethnicities, a point echoed by recent research [Citation32,Citation33]. These studies highlight how genetic predispositions to cancer can vary significantly based on factors such as race, ethnicity, and even geographic location. This is particularly evident in Africans, where BRCA1/2 pathogenic variants tend to be allelic heterogeneous and occur at low frequencies within the population [Citation32].
Our study identified the c.2643del pathogenic variant. This variant is considered rare based on population cohorts in the Genome Aggregation Database (gnomAD), but it might be more common in certain understudied groups, such as Libyans. This variant was previously found in a patient who was diagnosed with BC at the age of 40 and had a history of familial BC [Citation23]. It was also identified in another patient diagnosed with ovarian cancer [Citation24]. In our study, we recorded the third patient with this deletion: a woman diagnosed with IDC at a young age with a high histological grade and with local recurrence one year after diagnosis. She reported having a mother with BC at age 40. The c.2643delA gene, located at position 43,092,888‒43092889, involves a point mutation disrupting the reading frame and introducing a premature stop codon (E881fs), resulting in loss of a substantial portion of the BRCA protein. This protein truncation, as highlighted by ClinVar, likely leads to loss of function due to premature protein termination or nonsense-mediated mRNA decay. Consequently, this variant is classified as disease-causing.
The variant c.1366A>G was not found in the gnomAD and it met Rule PM2 (pathogenic moderate) according to the proposed criteria for interpretation of sequence variants by ACMG/AMP [Citation22]. This variant could be considered a novel alteration, as it involves the change of isoleucine to valine (p.Ile456Val) in the non-cytoplasmic domain of the protein (region of a membrane-bound protein). Furthermore, it is located within the region of amino acid sequences (341–748) required for interaction with Rad50 and recruits Rad50/MreII/Nbs1 complex to facilitate DNA repair [Citation23]. Both isoleucine and valine are hydrophobic, with valine being more acidic, but the significance of this alteration is unknown. Its location is crucial for DNA repair and warrants further functional investigation.
Unclassified BRCA1 variants pose a significant challenge. Our understanding of their impact on protein function and their true prevalence in the population remains incomplete, making classification difficult [Citation32,Citation33]. This study identified several such variants, including c.3630 G>T, c.3400 G>C, and c.1158T>A, adding further complexity. While some databases classify them as VUS or likely benign, prediction tools like SIFT and PolyPhen2 suggest potential harm due to variable effects on protein function. However, their rarity and limited association with BRCA1 individuals make their true role unclear. The c.3630 G>T and c.3400 G>C variants are exceptionally rare in gnomAD. Similarly, the c.1158T>A variant was not found in gnomAD. These variants, c.3630 G>T and c.3400 G>C, result in amino acid changes (p.Glu1210Asp and p.Glu1134Gln) within a region of unclassified membrane-bound proteins. Moreover, the c.1158T>A variant substitutes phenylalanine with leucine at position 386, located within the serine-rich domains of amino acids 345–508.
The c.3599A>T variant presented is particularly intriguing. Its allele frequency was not found on the gnomAD entry, and ClinVar offers no verdict, but SIFT and VarSome lean towards deleterious or pathogenic interpretations. Furthermore, our study identified this variant in women with TNBC. The resulting p.Gln1200Leu alteration is potentially significant because it replaces nonacidic, polar, and hydrophilic glutamine with hydrophobic leucine. This variant may affect the protein structure and function as it takes place within the consensus disorder predicted region. Therefore, it may be considered disease-causing.
Despite thorough analysis using various resources, including prediction tools, databases, information on different alleles at the same location, allele frequencies, population frequencies, location within functional domains, and relevant literature [Citation32], definitively concluding the pathogenicity of these unclassified variants remains elusive. These findings highlight the limitations of current knowledge and tools, particularly for under-represented populations like Africans. Further research, including functional assays, is crucial to bridge the gap in our understanding and determine the clinical implications of these variants.
Two BRCA1 variants, c.3460T>A and c.3572 G>A, were interpreted as likely benign/VUS on ClinVar, with varying interpretations across databases. Neither were found in population databases (gnomAD) or linked to BRCA1-related conditions in the literature. c.3460T>A replaces leucine with isoleucine at codon 1154. While the leucine residue is moderately conserved, leucine and isoleucine have a small physicochemical difference [Citation32]. c.3572 G>A resides in a ‘coldspot’ region where pathogenic missense variants are rare [Citation22]. It replaces serine with asparagine, both highly similar and located in a well-conserved region. While some missense variants in BRCA1/2 coldspots can be pathogenic, their rarity allows us to initially classify most uncharacterized variants in these regions as likely benign. However, careful classification still requires consideration of three key factors: conservation of the variant position, functional data, and clinical context [Citation22]. Ultimately, both variants lack sufficient evidence for definitive classification. Functional studies are crucial to confirming clinical significance.
Although the prevalence of HER2 positivity in BC patients with BRCA1 gene variants is poorly documented and highly variable, ranging from 0% to 10% [Citation33], our study suggests a potentially significant association. We identified the c.1054 G>A variant in one of three HER2-positive BC patients. This variant affects codon 352 of the BRCA1 protein, substituting glutamic acid with lysine (p.Glu352Lys). Notably, it was absent from population databases like gnomAD and the existing literature on BRCA1-related conditions. However, 16.7% of our BC cancer cohort, encompassing primary, recurrent, and metastatic cases, harbored this homozygous missense variant. Despite its presence in our study, ClinVar submissions classified c.1054 G>A as conflicting, with interpretations ranging from ‘VUS’ to ‘likely benign.’ While the glutamic acid residue is moderately conserved, the substitution with lysine introduces a slight physicochemical difference.
Furthermore, we note that the BRCA1 variants detected in the Libyan population differed from those seen in other parts of North Africa in the same exons. Such findings about the geographic differences in the distribution of the mutational spectrum of BRCA1/2 genes have already been reported in various populations in various countries [Citation33,Citation34].
While our study represents a significant first step in exploring BRCA1 variants in the Libyan population, it faced resource constraints. Focusing on just three BRCA1 exons and a smaller patient cohort likely underestimates the true prevalence of variants. PCR-Sanger sequencing offered a preliminary picture compared to the comprehensive view next-generation sequencing could provide. These limitations highlight the need for future research with larger sample sizes and advanced sequencing techniques to fully uncover the spectrum of BRCA1/2 variants in Libyan BC patients.
However, including data from all 36 patients strengthens our analysis by allowing us to examine valuable demographic, clinical, and familial characteristics. This data is particularly important due to the scarcity of information on familial breast cancer epidemiology in Libya. This comprehensive dataset provides a more informative picture of this understudied population.
Despite these limitations, a key strength of our study was the discovery of rare BRCA1 variants not previously described in North African populations, including a novel allelic variant (c.1366A>G). This significantly expands our understanding of the mutational spectrum in this region. Furthermore, all identified variants were observed in patients with confirmed family and clinical history, providing valuable context for their potential clinical significance. These findings are particularly valuable, providing preliminary insights into the BRCA1 variants potentially involved in hereditary BC within the Libyan population. However, further research is necessary to understand the functional consequences of these unclassified variants, elucidate their risk implications, and ultimately refine personalized management strategies. This could involve co-segregation analysis, functional studies, and population-based studies.
7. Conclusion
In conclusion, this study has shed light on unique BRCA1 variants in the Libyan population, marking a significant step forward in understanding hereditary breast cancer in this demographic. These variants, previously unreported in North African patients, offer preliminary insights into the potential role of BRCA1 in hereditary BC in Libya. However, the clinical significance of these variants remains unclear, necessitating further functional, computational, and population-based analyses to fully comprehend their impact on BC risk. This research also underscores the importance of comprehensive screening of BRCA1 and BRCA2 genes in a substantial patient cohort. Such efforts will help determine the frequency, spectrum, contribution, and prevalence of BRCA1/2 gene variations and facilitate the implementation of genetic testing. Ultimately, these findings could enhance the clinical management and risk assessment of hereditary breast cancer in the Libyan population, paving the way for more personalized treatment strategies.
Author contributions
EE: Conceptualization, data collection, experiments, formal analysis, in silico analysis, Writing – Original Draft, and data interpretation, and drafted the manuscript. IA: data interpretation and Writing – Review & Editing, AMR: analysis, in silico analysis, data interpretation and Writing – Review & Editing; ME: Writing – Review & Editing, experiments; FE: Writing – Review & Editing, Resources, clinical investigation of patients; AE: Resources, Writing – Review & Editing; and EH: Supervision, Writing – Review & Editing. All authors read and approved the final manuscript.
Ethics statement
The study was approved by the Research Ethics Committee of the National Cancer Institute (NCI), Sabratha, Libya. Written informed consent was obtained from each patient before recruitment in the study. This study was conducted in accordance with the ethical principles of the Helsinki Declaration.
supplemntary 3.xlsx
Download MS Excel (78.3 KB)supplementary1 .docx
Download MS Word (19.9 KB)Supplementary 2.docx
Download MS Word (16.5 KB)Acknowledgments
Authors are grateful to the Libyan Biotechnology Research Center in Tripoli, Libya, and to the Oncology Department of the National Cancer Institute at Sabratha, Libya, for their support. We are also grateful to all the participants and their family members for taking part in the study. We would like to express our profound gratitude to Madam Sallouha Gabbouj at the Laboratory of Molecular Immuno-Oncology, Faculty of Medicine, Monastir, Tunisia, for her invaluable assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All sequence data have been submitted to GenBank and provided with accession numbers (OR610584- OR610663) https://www.ncbi.nlm.nih.gov/nuccore/OR610584.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19932820.2024.2356906
Additional information
Funding
References
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, & Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca A Cancer J Clinicians. 2024;10(3): Advance online publication. 229–14. doi: 10.3322/caac.21834
- Arnold M, Morgan E, Rumgay H, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast [Internet]. 2022;66(August):15–23. Available from 10.1016/j.breast.2022.08.010
- Feizi H, Alizadeh M, Nejadghaderi SA, et al. The burden of chronic obstructive pulmonary disease and its attributable risk factors in the Middle East and North Africa region, 1990–2019. Respir Res [Internet]. 2022;23(1):1–15. Available from 10.1186/s12931-022-02242-z
- Elbiad O, Laraqui A, El BF, Mounjid C, Lamsisi M, Bajjou T, et al. Prevalence of specific and recurrent/founder pathogenic variants in BRCA genes in breast and ovarian cancer in North Africa. BMC Cancer [Internet]. 2022;22(1):1–19. Available from doi: 10.1186/s12885-022-09181-4
- Wang SM. A global perspective on the ethnic-specific BRCA variation and its implication in clinical application. J Natl Cancer Cent. 2023;3(1):14–20. doi: 10.1016/j.jncc.2022.12.001
- Mighri N, Hamdi Y, Boujemaa M, et al. Identification of Novel BRCA1 and RAD50 mutations associated with breast cancer predisposition in Tunisian patients. Front Genet. 2020;11(November). doi: 10.3389/fgene.2020.552971
- Fourati A, Louchez M, Fournier J, et al. Screening for common mutations in BRCA1 and BRCA2 genes: interest in genetic testing of Tunisian families with breast and/or. Bull Cancer. 2014;101(11):E36–E40. doi: 10.1684/bdc.2014.2049
- Abdulrashid K, Alhussaini N, Ahmed W, et al. Prevalence of BRCA mutations among hereditary breast and/or ovarian cancer patients in Arab countries: systematic review and meta-analysis. BMC Cancer. 2019;19(1):1–12. doi: 10.1186/s12885-019-5463-1
- Rotimi SO, Rotimi OA, Salhia B. A review of cancer genetics and genomics studies in Africa. Front Oncol. 2021;10(February):1–24. doi: 10.3389/fonc.2020.606400
- Hamdi Y, Mighri N, Boujemaa M, et al. Identification of eleven novel BRCA mutations in Tunisia: Impact on the clinical management of BRCA related cancers. Front Oncol. 2021;11(August). doi: 10.3389/fonc.2021.674965
- Ayed-Guerfali D B, Kridis-Rejab W B, Ammous-Boukhris N, Ayadi W, Charfi S, Khanfir A, et al. Novel and recurrent BRCA1/BRCA2 germline mutations in patients with breast/ovarian cancer: a series from the south of Tunisia. J Transl Med [Internet]. 2021;19(1):1–10. Available from doi: 10.1186/s12967-021-02772-y
- Abulkhair O, Al BM, Makram O, et al. Prevalence of BRCA1 and BRCA2 mutations among high-risk Saudi patients with breast cancer. J Glob Oncol. 2018;2018(4):1–9. doi: 10.1200/JGO.18.00066
- Wang SM. Journal of the National cancer center a global perspective on the ethnic-specific BRCA variation and its implication in clinical application. J Nat Cancer Cent. 2023;3(1):14–20. doi: 10.1016/j.jncc.2022.12.001
- Triki-Fendri S, Sánchez-Diz P, Rey-González D, et al. Paternal lineages in Libya inferred from Y-chromosome haplogroups. Am J Phys Anthropol. 2015;157(2):242–251. doi: 10.1002/ajpa.22705
- Najem F. N15: pHD Tribe, Islam and State in Libya ; analytical study of the roots. London, UK: University of Westminster; 2004.
- Clark SL, Rodriguez AM, Snyder RR, Hankins GDV, Boehning D. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J [Internet]. 2012;1(1):e201204005. Available from doi: 10.5936/csbj.201204005
- Ismail T, Alzneika S, Riguene E, et al. BRCA1 and its Vulnerable C-Terminal BRCT domain: Structure, function, genetic mutations and links to Diagnosis and treatment of breast and ovarian cancer. Pharmaceuticals. 2024;17(3):333. doi: 10.3390/ph17030333
- NCCN clinical practice guidelines in oncology (NCCN Guidelines). Genetic/familial high-risk assessment: breast, ovarian and Pancreatic. National Comprehensive Cancer Network. Version 2.2022. [citied 2022 Mar 9] Available from: https://www.melbournebreastcancersurgery.com.au/wp-content/themes/ypo-theme/pdf/nccn-clinical-practice-genetic.pdf
- Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St gallen international expert consensus on the primary therapy of early breast cancer 2011. Annals Oncology. 2011;2011(8):1736–1747. doi: 10.1093/annonc/mdr304
- AJCC Cancer Staging Manual. AJCC cancer staging manual. 2010.
- Mahfoudh W, Bouaouina N, Ben AS, et al. Hereditary breast cancer in Middle Eastern and North African (MENA) populations: Identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Mol Biol Rep. 2012;39(2):1037–1046. doi: 10.1007/s11033-011-0829-8
- Dines JN, Shirts BH, Slavin TP, et al. Systematic misclassification of missense variants in BRCA1 and BRCA2 “coldspots. Genet Med. 2020;22(5):825–830. doi: 10.1038/s41436-019-0740-6
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med [Internet]. 2015;17(5):405–424. Available from doi: 10.1038/gim.2015.30
- Hamdi Y, Mighri N, Boujemaa M, et al. Identification of eleven novel BRCA mutations in Tunisia: Impact on the clinical management of BRCA related cancers. Front Oncol. 2021 Aug;11:3181. doi: 10.3389/fonc.2021.674965
- Rentzsch P, Witten D, Cooper GM, et al. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94. doi: 10.1093/nar/gky1016
- Sokolenko AP, Sultanova LV, Stepanov IA, et al. Strong founder effect for BRCA1 c.3629_3630delAG pathogenic variant in Chechen patients with breast or ovarian cancer. Cancer Med. 2023;12(3):3167–3171. doi: 10.1002/cam4.5159
- Laraqui A, Uhrhammer N, Lahlou-Amine I, et al. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int J Med Sci. 2012;10(1):60–67. doi: 10.7150/ijms.5014
- Henouda S, Bensalem A, Reggad R, et al. Contribution of BRCA1 and BRCA2 germline mutations to early Algerian breast cancer. Dis Markers. 2016;2016:1–7. doi: 10.1155/2016/7869095
- Troudi W, Uhrhammer N, Ben RK, et al. Complete mutation screening and haplotype characterization of BRCA1 gene in Tunisian patients with familial breast cancer. Cancer Biomarkers. 2008;4(1):11–18. doi: 10.3233/CBM-2008-4102
- Cherbal F, Bakour R, Adane S, et al. BRCA1 and BRCA2 germline mutations screening in Algerian breast/ovarian cancer families. Dis Markers. 2010 Jan;28(6):377–384. doi: 10.1155/2010/585278
- El KM, Diakite B, Hamzi K, et al. Screening of exon 11 of BRCA1 gene using the high resolution melting approach for diagnosis in Moroccan breast cancer patients. BMC Cancer. 2015;15(1):1–5. doi: 10.1186/s12885-015-1040-4
- Purnomosari D, Pals G, Wahyono A, et al. BRCA1 and BRCA2 germline mutation analysis in the Indonesian population. Breast Cancer Res Treat. 2007;106(2):297–304. doi: 10.1007/s10549-006-9493-4
- Blay P, Santamaría I, Pitiot AS, et al. Mutational analysis of BRCA1 and BRCA2 in hereditary breast and ovarian cancer families from Asturias (Northern Spain). BMC Cancer. 2013;13(1):1–10. doi: 10.1186/1471-2407-13-243
- Mehemmai C, Cherbal F, Hamdi Y. BRCA1 and BRCA2 germline mutation analysis in hereditary breast/ovarian cancer families from the aures region (Eastern Algeria. Pathol Oncol Res. 2020;26(2):715–726. doi: 10.1007/s12253-019-00586-4