ABSTRACT
The yeast Candida albicans is an oral commensal microorganism, occurring in the oral cavity of 50–70% of healthy individuals. Its effect on oral ecology has mostly been studied using dual-species models, which disregards the complex nature of oral biofilms. The aim of this study was to culture C. albicans in a complex model to study its effect on oral biofilms. Biofilms, inoculated using pooled stimulated saliva with or without addition of C. albicans, were grown under anaerobic, aerobic, or aerobic +5% CO2 conditions. Red autofluorescence was quantified using a spectrophotometer and visualized in fluorescence photographs. The microbiome of 5 h biofilms was determined using 16S rDNA sequencing. C. albicans was only able to proliferate in biofilms grown under aerobic conditions. After 48 h, C. albicans did not induce differences in total biofilm formation, lactic acid accumulation (cariogenic phenotype) or protease activity (periodontitis phenotype). In vitro, anaerobically grown biofilms developed red autofluorescence, irrespective of inoculum. However, under aerobic conditions, only C. albicans–containing biofilms showed red autofluorescence. Facultative or strict anaerobic Veillonella, Prevotella, Leptotrichia, and Fusobacterium genera were significantly more abundant in biofilms with C. albicans. Biofilms without C. albicans contained more of the aerobic and facultative anaerobic genera Neisseria, Rothia, and Streptococcus. The presence of C. albicans alters the bacterial microbiome in early in vitro oral biofilms, resulting in the presence of strictly anaerobic bacteria under oxygen-rich conditions. This in vitro study illustrates that C. albicans should not be disregarded in healthy oral ecosystems, as it has the potential to influence bacteria significantly.
Introduction
The healthy oral cavity represents a very diverse niche [Citation1] that is colonized by >500 bacterial species [Citation2]. In addition, >100 fungal species were identified in the oral cavity of healthy individuals [Citation3,Citation4]. With oral diseases being among the most prevalent in Western society [Citation5], it is not surprising that the relation between microbial colonization and the development of oral diseases is well-established both for bacteria and for fungi. Oral diseases develop due to an imbalance between microbial colonization and the host. According to the ecological plaque hypothesis [Citation6,Citation7], changes in environmental conditions can lead to long-term changes in microbial ecology. Such an imbalance can lead to tooth decay (i.e. caries) or inflammation of the soft tissues of the mouth (e.g. gingivitis). Ecological balance, the net result of all inter-microbe and host–microbe interactions, is essential in maintaining health.
Candida albicans is a polymorphic yeast and an opportunistic pathogen. In the oral cavity, it is associated with early childhood caries [Citation8], and it can cause infections of the oral soft tissues, ranging from superficial overgrowth to deep-seeded invasion, resulting in disseminated disease [Citation9]. Nevertheless, C. albicans colonizes the oral cavity as a commensal in 50–70% of individuals [Citation3,Citation4]. It has the ability to interact with many bacterial species on different levels [Citation10]. For instance, the physical interaction between C. albicans and Streptococcus gordonii enhances hyphal formation of C. albicans and increases the biomass of the dual-species biofilms [Citation11]. C. albicans was also found to have a complex interaction with the cariogenic organism S. mutans. The latter produces glucan that binds to the cell wall of C. albicans. The yeast thus provides adhesion sites for the bacterium, resulting in increased biofilm formation of the dual-species biofilms [Citation12]. In a rodent model, these dual-species biofilms resulted in higher numbers of severe caries lesions compared with infection with either of the species alone [Citation12].
In addition to the physical interaction, C. albicans also interacts with bacteria via chemical signals in a process called quorum sensing. For instance, Aggregatibacter actinomycetemcomitans produces autoinducer-2 (AI-2), which is a small signaling molecule. AI-2 is sensed by C. albicans, which then responds to this signal, resulting in lower biofilm formation [Citation13]. Similarly, S. mutans produces a small peptide called competence-stimulating peptide (CSP), which inhibits C. albicans hyphal formation in the early stages of biofilm formation [Citation14].
Finally, metabolic interactions are found between C. albicans and S. mutans. Willems et al. showed that despite higher abundance of S. mutans in 72 h dual-species biofilms, the pH of the spent medium remained higher compared with S. mutans biofilms alone [Citation15]. The lactic acid produced by S. mutans can be metabolized by C. albicans [Citation16], and C. albicans can increase the external pH by producing ammonia [Citation17], leading to less cariogenic conditions.
Interactions between C. albicans and bacteria have been studied in dual-species biofilm models. It is clear that these interactions affect the behavior of both microbes involved. While studies on bacterial biofilms have evolved to multi-species or even microcosm model systems, mixed kingdom biofilm studies have not. The aim of this study was to introduce C. albicans in an in vitro model for complex oral biofilm in order to study its effect on the microbiome and pathology-related phenotypes of these biofilms.
Materials and methods
Inoculum collection and strains
Saliva used in this study was obtained in accordance with the ethical principles of the 64th WMA Declaration of Helsinki. The Medical Ethical Committee of the VU Medical Center, The Netherlands (2011/236) determined that Dutch law concerning Medical Research Involving Human Subjects Act (WMO) was not applicable. Stimulated saliva from 10 self-reported systemically and orally healthy donors was collected on ice and pooled [Citation18]. C. albicans SC5314 [Citation19] was grown overnight, centrifuged, and resuspended in fresh buffered semi-defined McBain medium [Citation20] to OD600 = 5.0. An inoculum was created by diluting pooled saliva 50-fold in fresh McBain medium supplemented with 0.2% sucrose, or by diluting both pooled saliva and C. albicans 50-fold in fresh McBain medium supplemented with 0.2% sucrose (mixed inoculum).
Biofilm formation
Biofilms were grown in vitro in buffered semi-defined McBain medium [Citation20] supplemented with 0.2% sucrose using the Amsterdam Active Attachment Model (AAA model) [Citation20], with 12 mm glass coverslips (Menzel, Braunsweig, Germany). Glass cover slips were used as model substrates because glass resembles the physical properties (smoothness and surface characteristics) of teeth without demineralization related to microbial acid production during growth. Biofilms were generated with medium refreshment twice a day [Citation20]. The initial microbial seeding used pooled saliva, a mixed inoculum, or C. albicans alone. Biofilms were incubated at 37°C anaerobically, aerobically, or aerobically with 5% CO2.
For the dynamic analysis of red autofluoresence, biofilms were grown up to 72 h with medium refreshment twice a day.
Confocal scanning laser microscopy
Prior to confocal scanning laser microscopy (CLSM), 48 h biofilms were fixed for 30–60 min with 4% formaldehyde, washed with phosphate-buffered saline (PBS), and stained with SYTO9 (Life Technologies Europe BV, Bleiswijk, The Netherlands) for 15 min in the dark. CLSM images of two biofilms per condition were captured with a LEICA TCS SP2 microscope using a 20× objective and the FITC filter settings (excitation 488 nm, emission 510–540 nm).
DNA isolation and quantitative polymerase chain reaction of Candida
DNA isolation and purification were performed as previously described [Citation18]. To determine the amount of Candida in the biofilms, quantitative polymerase chain reaction (qPCR) was performed using four biofilms per condition. Primers and a probe for the 28S large-subunit ribosomal RNA gene were used (F: GCA TAT CAA TAA GCG GAG GAA AAG, R: TTA GCT TTA GAT GAT TTAC CAC C, Probe: 6FAM-CGG CGA GTG AAG CGG SAA RAG CTC-BHQ1) [Citation21]. Reactions contained LightCycler® 480 Probes Master (Roche Diagnostics, Basel, Switzerland), 0.5 pmol primers, 0.3 pmol probe, and 3 µL DNA in a total reaction volume of 20 µL. qPCR was performed using the Light cycler 480-II (Roche Diagnostics) with the following protocol: 10 min pre-incubation at 95°C, followed by 50 cycles of denaturation (95°C, 10 s), annealing (60°C, 30 s), extension (72°C, 30 s), and cooling at 37°C for 10 s. The concentration of Candida DNA (ng/µL) was determined from standard curves derived from C. albicans genomic DNA.
Quantifying biofilm formation
Biofilms (48 h) were dispersed in PBS by sonication [Citation18]. To estimate the amount of biofilm formation, total anaerobic colony forming units (CFUs) were determined [Citation20] for four biofilms per condition.
Cariogenic potential
Lactic acid production is a key virulence factor of oral biofilms with regard to caries [Citation6,Citation22]. To assess the cariogenic potential of the 48 h biofilms, lactic acid accumulation of four biofilms per condition was determined prior to harvesting [Citation20]. Biofilms were incubated anaerobically for 3 h at 37°C in sterile Buffered Peptone Water (BPW) supplemented with 0.2% sucrose. Lactic acid accumulation (in mM per biofilm) was determined using a colorimetric assay [Citation23].
Protease activity
Increased protease activity, and especially gingipain activity, is related to periodontitis [Citation24]. Total and gingipain-specific protease activity was measured to determine the periodontitis activity of the 48 h biofilms. The protease activity was measured using a fluorescence resonance energy transfer (FRET) assay, as described previously [Citation18,Citation24]. Briefly, spent medium of four biofilms per condition was filter sterilized and mixed 1:1 with TRIS-buffered saline (TBS). A FRET-probe PEK-054 ([FITC]-NleKKKKVLPIQLNAATDK-[KDbc]) [Citation25] was used to measure total protease activity and gingipain-specific protease activity [Citation24]. The probe was added at a final concentration of 16 µM and fluorescence (excitation at 485 nm and emission at 530 nm) was measured every 2 min for 2 h using a fluorescence microplate reader (Fluostar Galaxy; BMG Laboratories, Offenburg, Germany). Protease activity was defined in relative fluorescence per min (RF/min).
Red autofluorescence
Red autofluorescence is a fast and direct, non-invasive, quantifiable biofilm phenotype. This autofluorescence is related to older oral biofilms [Citation26,Citation27]. It has been suggested that red autofluorescence is also related to oral disease, especially gingival inflammation [Citation28] and caries [Citation26,Citation27,Citation29].
Red autofluorescence of four biofilms per condition was assessed after 8, 24, 32, 48, 56, and 72 h by measuring the emission spectra (excitation 405 ± 20 nm) with a fiber-optic spectrophotometer (USB4000–FL, reflection probe, Ø 600 µm; Ocean Optics, Inc., Dunedin, FL), which consists of six illumination fibers around one read fiber. Measurements were performed in a dark room 3 mm from the surface of the glass coverslips (three measuring points per biofilm) [Citation27]. Thereafter, biofilms were photographed for visual fluorescence assessment using a quantitative light-induced fluorescence digital (QLF-D) SLR camera system (Inspektor Research Systems BV, Amsterdam, The Netherlands). The photographs were taken with fixed camera settings (white light photographs: shutter speed 1/30th; aperture value 8.0; ISO 1600; QLF-D photographs: shutter speed 1/30th; aperture value 5.6; ISO 1600).
Analysis of initial biofilm formation
To increase total biomass of the initial biofilms compared with initial studies, a roughly 5× larger surface area was used for the 5 h biofilms. Sterilized microscope slides (Menzel-Gläser, 7.6 × 2.6 cm) were placed in a 50 mL Greiner tube, filled with 50 ml: pooled saliva inoculum or mixed inoculum, and incubated under aerobic +5% CO2 conditions at 37°C.
After 5 h, the slides were aseptically transferred to a Petri dish filled with 25 mL BPW, and attached microbes were dislodged using a cell scraper. Subsequently, 5 mL BPW was serial diluted for CFU determination, and microbes in the remaining volume were harvested by centrifugation and frozen at −20°C for DNA isolation, qPCR, and sequencing.
Microbiome analysis
Seven 5 h-old initial biofilms for both saliva and mixed inoculum were used for microbiome analysis. Bacterial DNA concentration after purification was determined by qPCR using a universal primer-probe set to target the 16S rRNA gene [Citation30].
The V4 hypervariable region of the 16S rRNA gene was amplified using 100 pg DNA, as described previously [Citation30], except that 33 amplification cycles were performed. The generated amplicons were pooled in equimolar quantities and then purified from agarose gels (Qiagen, Roermond, The Netherlands) to remove non-specific PCR products prior to sequencing. The Illumina MiSeq platform and Illumina MiSeq reagent kit V2 (Illumina, Inc., San Diego, CA) were used for paired-end sequencing of the amplicons to generate 200-bp paired-end reads.
Sequence processing and analysis
The sequence data were processed as described previously [Citation31].
Data normalization and statistical analysis
Fluorescence spectra were normalized to the amplitude and angle of inclination of the excitation peak (420–430 nm) using dedicated software (SP1 v1.0.0.10; Inspektor Research Systems BV; zero compensation 345–380 nm). Gaussian curve fitting was performed to calculate the separate fluorescence peaks for peak wavelength, amplitude, and full width at half maximum [Citation27].
A two-sided t-test was used for statistical comparison of lactate production, protease activity, or biofilm formation in different groups. qPCR data were log10-transformed prior to performing a two-sided unpaired t-test. Groups were considered statistically different at p < 0.05.
The OTU table was randomly subsampled at 10,500 reads/sample. The Shannon diversity index was calculated using PAST software v3.01 [Citation32]. The OTU data set was log2 transformed and ordinated by principal component analysis (PCA) into two dimensions using PAST. One-way permutational multivariate analysis of variance was performed on the Bray–Curtis Similarity Index to calculate the significance of the compositional differences between the saliva biofilms and mixed biofilms. Groups were considered statistically different at p < 0.05.
Linear discriminant analysis effect size (LEfSe) [Citation33] was used to identify OTUs that differed in relative abundance between the saliva and mixed biofilms. The alpha values were kept at the default of 0.05, and the LDA (spell in full) threshold was kept at 2.0.
Results
C. albicans integrates in complex in vitro oral biofilms in the AAA model
The AAA biofilm model was inoculated using saliva or saliva + C. albicans and cultured at 37°C under anaerobic, aerobic, and aerobic +5% CO2 conditions. Under both aerobic conditions, the presence of C. albicans was readily observed using CLSM ((a and b)). Under elevated CO2 conditions, C. albicans formed hyphae, while under aerobic conditions, hyphal formation was less extensive. In contrast, anaerobic growth conditions resulted in biofilms without visible C. albicans ((c)).
Figure 1. Confocal images of (a) aerobic biofilm, (b) aerobic +5% CO2 biofilm, and (c) anaerobic biofilm. White arrows indicate visible Candida albicans cells; the size bar indicates 25 µm. (d) Quantitative polymerase chain reaction (qPCR) data in ng/µL, where white bars are saliva biofilms and black bars are mixed biofilms. The differences in C. albicans presence between the saliva only and mixed inoculum were all significant (p < 0.05) within the same condition. C.albicans was integrated in the microcosm biofilm when O2 was available.
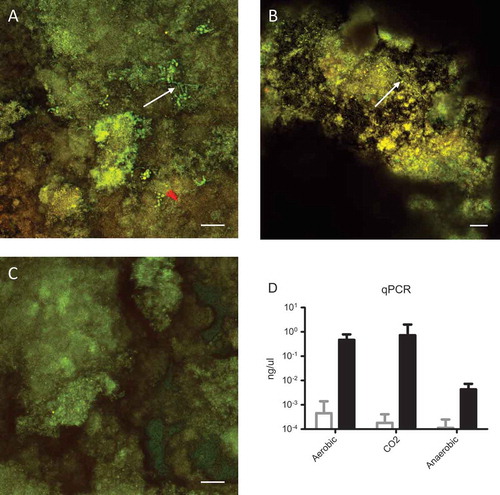
Since biofilms are three-dimensional structures, it is possible that C. albicans was present yet undetectable using CLSM. Therefore, the presence of C. albicans was analyzed using qPCR. As expected, the saliva-inoculated biofilms showed no detectable presence of C. albicans in all conditions ((d)). In line with the CLSM data, significant presence of C. albicans was observed in biofilms cultured in the presence of oxygen. Under anaerobic conditions the presence of C. albicans was 100-fold lower than under either aerobic condition. In conclusion, C. albicans can be introduced in the AAA model for oral microcosm biofilms when cultured under aerobic conditions.
Effect of C. albicans on phenotypes of oral biofilms
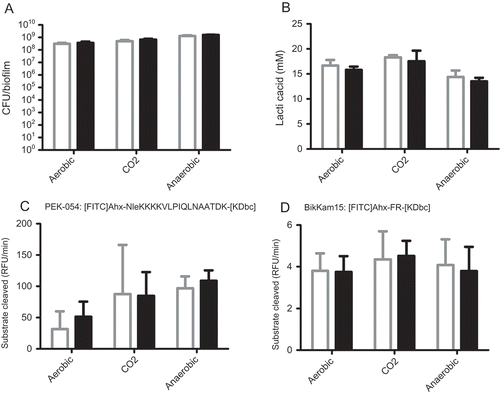
Oral biofilms consist mainly of oral bacteria with fungi probably representing <0.1% of viable cells. Therefore, the effect of C. albicans on total biofilm formation was evaluated by counting bacterial CFUs. For both aerobic conditions, the mixed biofilms resulted in similar total CFUs compared to biofilms inoculated with saliva alone ((a)), indicating a similar amount of biofilm formation. For the anaerobic biofilms, the total CFUs were statistically different (1.3 × 109 vs. 1.6 × 109, p = 0.36). Since the difference is much less than one log, this difference is most likely not biologically relevant. Pathology-related phenotypes [Citation18] were assessed by measuring lactic acid production (cariogenic phenotype) and protease activity (periodontitis phenotype). The presence of C. albicans did not affect lactic acid accumulation ((b)), and neither did it affect total or specific protease activity of the 48 h biofilms ((c and d)).
Figure 2. Phenotypes of 48 h biofilms. White bars represent saliva biofilms; black bars represent mixed biofilms. (a) Colony forming units (CFU) counts in CFU per biofilm. (b) Lactic acid accumulation in mM per biofilm. (c) Total protease activity in RFU/min. (d) Specific protease activity in RFU/min. No statistical differences were found between mixed and saliva biofilms. All conditions resulted in statistically different CFUs.
Dynamic analysis of red autofluorescence
In vivo oral biofilms produce red autofluorescence under certain conditions [Citation28]. In vitro saliva biofilms, grown for 48 h in the presence of oxygen, showed reduced red autofluorescence in some experiments (end-point measurement) compared with mixed biofilms (data not shown). To obtain more insight into the development of this red autofluorescence, the dynamics of its development with time were studied.
Red autofluorescence was analyzed after 8, 24, 32, 48, 56, and 72 h for all biofilms. Anaerobically grown biofilms always showed red autofluorescence from 32 h onwards, irrespective of the presence of C. albicans in the inoculum ((c and f)). This was in contrast to both aerobic conditions, where clear differences appeared in red autofluorescence between the saliva and mixed biofilms. In saliva biofilms, red autofluorescence was absent or considerably reduced for all time points, while mixed biofilms were red autofluorescent from 32 h onwards ((a, b, d, and e)). C. albicans thus induces red autofluorescence in mixed biofilms under aerobic growth conditions, mimicking the anaerobically grown biofilms.
Figure 3. Red autofluorescence of biofilms grown under aerobic, anaerobic, and CO2 conditions in time. The biofilms were inoculated with pooled stimulated saliva + C. albicans (mixed) or with saliva alone. Representative quantitative light-inducted fluorescence pictures of (a) biofilms grown aerobically, (b) biofilms grown aerobically +5% CO2, and (c) biofilms grown anaerobically. Red autofluorescence in RFU after curve fitting for (d) biofilms grown aerobically, (e) biofilms grown aerobically +5% CO2, and (f) biofilms grown anaerobically.
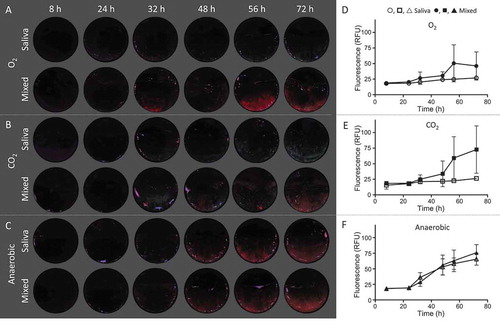
The pattern of the red autofluorescence in the biofilms was also different between the three conditions. In anaerobic biofilms, red autofluorescence was always evenly distributed throughout the biofilms, while in both aerobic conditions, spots of red autofluorescence were visible, explaining the high standard deviations in the red fluorescence spectra ((d, e, and f)).
Effect of C. albicans on microbiome of early biofilms
Hyphae provide sites of adhesion for many bacteria that interact with C. albicans. Since hyphae were present predominantly in the CO2 grown conditions (as observed by CLSM, vide supra), the effect of C. albicans presence during initial (5 h) biofilm formation was analyzed for CO2 conditions. Based on qPCR, the saliva biofilms resulted in hardly detectable C. albicans, very close to the detection limit of the qPCR. In the mixed biofilms, C. albicans was a detectable part of the biofilm ((a)) and more than 4 log10 higher than in the saliva control. The same amount of C. albicans was found in the 5 h biofilms as in the 48 h biofilms (0.73 ng/µL vs. 9.8 ng/µL). Since the surface covered by the 5 h biofilms was roughly 25 times larger than the surface covered by the 48 h biofilms, this indicates that C. albicans not only blends in the biofilm, but also is able to proliferate.
Figure 4. Microbiome analysis of initial (5 h) biofilms. (a) qPCR of C. albicans in ng/µL. White bar represents saliva biofilms; black bar represents mixed biofilms. (b) Principal component analysis plot of initial (5 h) biofilms where ┚ are saliva biofilms and ● are mixed biofilms. The data were randomly subsampled and log2 transformed. (c) Visualization of most significant OTUs that differentiate between saliva and mixed biofilms, ranked by the effect size in LEfSe. White bars represent OTUs more abundant in saliva biofilms; black bars represent OTUs more abundant in mixed biofilms.
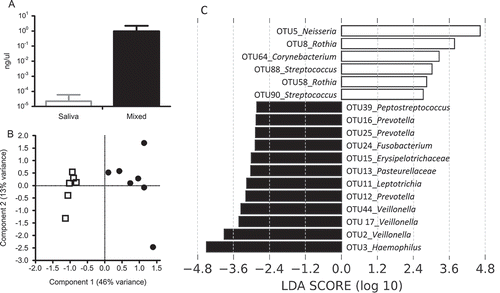
Since not all bacterial species interact with C. albicans, the introduction of this yeast during initial biofilm formation potentially leads to differences in the bacterial microbiome of the biofilm. This microbiome was analyzed using 16S rDNA sequencing of the 5 h biofilms. The complete OTU list is available in Supplementary Table S1. The Shannon diversity index was statistically significantly higher (p = 0.02) for mixed biofilms (3.6) than it was for saliva biofilms (3.4). A clear difference in species composition was observed between the saliva biofilms and mixed biofilms (p = 0.0004, F = 8.4), which is also visible in the PCA plot ((b)). LEfSe [Citation33] was used to detect the OTUs responsible for this separation, and the relative abundance of the OTUs uncovered by LEfSe is plotted in Supplementary Figure S1. A total of 18 OTUs were significantly different in relative abundance between the 5 h saliva and mixed biofilms ((c)). The most prominent differences were Neisseria (OTU 5), Rothia (OTU 8, OTU 58), and Streptococcus (OTU 90) that were more abundant in saliva biofilms. Biofilms inoculated with C. albicans and saliva contained relatively more Haemophilus (OTU 3), Veillonella (OTU 2, OTU 17, OTU 44), Prevotella (OTU 12, OTU 16, OTU 25), Leptotrichia (OTU 11), and Fusobacterium (OTU 24). These OTUs are in agreement with the first component of the PCA, which explains 46% of the variance and separates the two groups. Concluding, biofilms inoculated with a mixture of saliva and C. albicans showed a significantly different bacterial species composition and a higher diversity compared with biofilms inoculated with saliva alone.
Discussion
Oral biofilms are very complex, diverse communities with >500 species of bacteria, most of which are unculturable [Citation2]. Candida is present in low abundance in the oral cavity of 50–70% of healthy individuals [Citation3,Citation4], and thus can be considered a commensal microorganism. This study aimed to introduce C. albicans in a microcosm in vitro model to study the effect of C. albicans on the phenotype and microbiome of complex oral biofilms.
In order to study the fundamentals of oral biofilm development, one should aim to use in vitro models that mimic the oral situation as well as possible. The AAA model, inoculated with saliva, grown in artificial saliva medium, is such a model [Citation20]. Usually, this model, like many other oral biofilm models, is incubated anaerobically to preserve the strictly anaerobic species typical for oral biofilms. Since the oral cavity is not strictly anaerobic [Citation7], this approach may result in a biased microbial population with aerobic species being underrepresented. For example, while generally present in the initial inoculum, C. albicans was not present in biofilms grown in the AAA model under standard anaerobic conditions. Although C. albicans is able to grow under anaerobic conditions, growth rates are very low [Citation34]. C. albicans is probably outcompeted by (facultative) anaerobic bacteria that grow more efficient in the absence of oxygen. In the present study, this problem was overcome by culturing the AAA model in the presence of oxygen. C. albicans successfully integrated in the microcosm biofilms in the aerobically incubated AAA model. When cultured under aerobic conditions, C. albicans was detectable both by microscopy and qPCR. Under aerobic conditions with 5% CO2, hyphae were formed, which is consistent with reports showing that CO2 is a potent inducer of hyphae formation [Citation35]. It may be expected that aerobic incubation will prevent anaerobic bacteria from colonizing the model, which indeed seemed to occur in the saliva biofilms. However, this did not happen in the presence of C. albicans, where strict anaerobic bacteria colonized the AAA model.
The presence of C. albicans did not affect the total number of culturable microorganisms (CFU counts) in the biofilm or the pathology-related phenotypes of oral biofilms that were defined previously [Citation18]. Lactic acid accumulation in the biofilms was not different between saliva and mixed biofilms. This was unexpected, as C. albicans has been suggested to be a cariogenic microbe, especially in early childhood caries [Citation36]. It was also previously reported that C. albicans enhanced the virulence of S. mutans [Citation15]. The discrepancy between the study by Willems et al. [Citation15] and this study is probably related to the complex nature of the biofilms compared with previously used dual-species models (give at least a couple of references). In complex biofilms, metabolic interaction of its members is not uncommon [Citation37]. One example is the production of lactic acid by saccharolytic bacteria, which is consumed by Veillonella [Citation38]. End products of a dual-species biofilm are therefore not necessarily waste products to a complex biofilm, making the produced metabolites of complex biofilms very different from dual-species biofilms. The findings support the importance of using more complex, realistic models to study the role of microbes in a biofilm.
The occurrence of red autofluorescence of the in vitro biofilms was affected by the presence of C. albicans. C. albicans is known to autofluoresce orange/red when exited with 405 nm (blue-violet) light [Citation29]. However, no studies have reported the effect of C. albicans presence on red autofluorescence of oral biofilms. This is, to the authors’ knowledge, the first study describing red autofluorescence of biofilms grown under different oxygen conditions. In vivo, red autofluorescent plaque is indicative of older plaque [Citation26,Citation27], which contains more anaerobic species than young plaque does [Citation37]. In line with this, red autofluorescence was observed in vitro when saliva biofilms were cultured under anaerobic conditions, and it was rarely detected when cultured under aerobic conditions. However, the presence of C. albicans induced red autofluorescence under aerobic conditions. The induced red autofluorescence might therefore be an indication that C. albicans supports the growth of anaerobic bacteria in the early stage of biofilm formation.
Interactions between C. albicans and bacteria can occur on physical, chemical, and metabolic levels [Citation10]. In many cases, physical interactions (e.g. adhesion) occur with the hyphae [Citation39,Citation40]. Therefore, 5 h biofilms were grown with 5% CO2 to stimulate hyphae formation. Community analysis of 5 h-old biofilms confirmed the presence of more anaerobic bacteria in mixed biofilms. In the presence of C. albicans, more strict anaerobic genera were found, namely Veillonella, Leptotrichia, Prevotella, and Fusobacterium, compared with more aerobic genera in saliva biofilms, namely Neisseria and Rothia. This is in agreement with a recent study reporting that C. albicans biofilms allow anaerobic bacteria to grow under aerobic culture conditions [Citation41]. It should, however, be noted that in contrast to the Fox study [Citation41], in the present study, C. albicans biofilms were not preformed. The depletion of oxygen seems to be rapid and not necessarily biofilm dependent. Oxygen consumption by C. albicans is high and rapid [Citation42], and the presence of C. albicans could quickly create anaerobic micro-niches. In these micro-niches, strict anaerobic bacteria could survive and proliferate. As a consequence of the presence of C. albicans, young oral biofilms that would normally consist of aerobic or facultative anaerobic bacteria can now also contain strict anaerobes. This is supported by the induced red autofluorescence observed in the present study. The amount of oxygen removed, and the extent of anaerobic niches formed, would be dependent on the number of C. albicans cells present and their individual respiratory rate.
All OTUs that determine the difference between saliva and mixed biofilms were identified as part of the healthy oral core microbiome [Citation2]. However, many of these organisms are to some extent associated with oral infectious diseases. For example, Leptotrichia and Prevotella were reported to be related to the presence of active caries lesions [Citation43,Citation44]. Prevotella and Fusobacterium species are part of the so-called orange complex, which is a consortium of bacteria related to periodontitis [Citation45]. Veillonella species are also associated with caries lesions, presumably as a result of high concentrations of lactic acid [Citation6]. Yet, they are mostly considered beneficial to the oral cavity due to their lactic acid consumption [Citation46], which reduces the caries activity.
Concluding, a model was established to incorporate C. albicans in microcosm oral biofilms. Using this model, it was shown that C. albicans does not elevate disease-associated phenotypes (i.e. lactic acid accumulation and proteolytic activity) of the biofilms, but it did induce survival and proliferation of strictly anaerobic bacteria under aerobic culture conditions. As such, the presence of C. albicans in initial in vitro oral biofilms changes the community composition significantly.
Supplementary Table
Download MS Word (48.6 KB)Supplementary Figure
Download PDF (166.9 KB)Acknowledgments
The authors thank Canan Unal, Djawid Hossainian, Mark Buijs, Wendy de Wit, and Caroline Bosch-Tijhof for their technical support.
Disclosure statement
MHvdV is a co-inventor of several patents related to quantitative light-induced fluorescence. The authors declare that there is otherwise no conflict of interest pertaining to the data presented in this article.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
Notes on contributors
M. M. Janus
M. M. Janus is a PhD fellow at the department of Preventive Dentistry, at the Academic Center for Dentistry Amsterdam. She received her MSc Engineers degree at the Delft University of Technology in 2010. Her research focuses on studying the ecology and phenotypes of in vitro oral biofilms.
W. Crielaard
W. Crielaard is Professor in Molecular Biology and head of the Department of Preventive Dentistry at the Academic Centre of Dentistry Amsterdam (ACTA). He received his PhD in Molecular Microbiology from the University of Groningen (NL) in 1989, performed a post-doctoral EMBO fellowship at the University of Sheffield (UK) and held assistant and associate professorships at the Science Faculty of the University of Amsterdam. Since he has joined ACTA 8 years ago he has used his expertise in studying the molecular biology of multi-species oral microbial communities in relation to the role these communities play in maintaining oral health and causing oral infections.
C. M. C. Volgenant
C. M. C. Volgenant is a dentist and post-doc researcher at the Academic Center for Dentistry Amsterdam (ACTA). She received her PhD at the department of Preventive Dentistry at ACTA on the validation of red autofluorescence from biofilms in the oral cavity.
M. H. van der Veen
M. H. van der Veen is Associate Professor at the Academic Center of Dentistry Amsterdam (ACTA). She received her PhD in physics from the University of Groningen in 1995 on the development of a method for root caries assessment by use of tissue optics. She held a post-doctoral fellowship at Indiana University-Purdue University Indianapolis where she was principal investigator on a research program concerning the development of early caries assessment methods and a position as senior lecturer in plaque related diseases at the dental school of the University of Liverpool. Her current research focusses on the link between plaque ecology and supra and sub-gingival plaque fluorescence in health and disease.
B. W. Brandt
B. W. Brandt received his PhD in mathematical modelling of biodegradation from the VU University Amsterdam in 2002. Since then, he has been working as a bioinformatician in different areas of biology, where he applied and developed tools in the field of biological data analysis. He currently works on sequence analysis, metagenomics, and the relation between (oral) health and the (oral) microbiome.
B. P. Krom
B. P. Krom is Associate Professor at the Academic Center of Dentistry Amsterdam (ACTA). He received his PhD in Molecular Microbiology from the University of Groningen in 2002. He held a post-doctoral fellowship at Georgetown University Medical Centre before being appointed assistant professor at the Department of Biomedical engineering of the University Medical Center Groningen. His main focus is on the role of interspecies interactions in development of (oral) biofilms. Since 2011 he is working at the Academic Center of Dentistry Amsterdam (ACTA).
References
- Simón-Soro A, Tomás I, Cabrera-Rubio R, et al. Microbial geography of the oral cavity. J Dent Res. 2013;92(7):1–10. DOI:10.1177/0022034513488119
- Zaura E, Keijser BJ, Huse SM, et al. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. DOI:10.1186/1471-2180-9-259
- Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. Plos Pathog. 2010;6(1):e1000713. DOI:10.1371/journal.ppat.1000975
- Dupuy AK, David MS, Li L, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. Plos One. 2014;9(3):e90899. DOI:10.1371/journal.pone.0090899
- WHO. World Health Organization: oral health fact sheet N°318. WHO Media Centre; 2012. www.who.int/mediacentre/factsheets/fs318/en/
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8(2):263–271.
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149(Pt 2):279–294. DOI:10.1099/mic.0.26082-0
- Raja M, Hannan A, Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010;44(3):272–276. DOI:10.1159/000314675
- Odds FC. Candida and candidosis. London: Bailliere Tindall; 1988.
- Krom BP, Kidwai S, Ten Cate JM. Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res. 2014;93(5):445–451. DOI:10.1177/0022034514521814
- Silverman RJ, Nobbs AH, Vickerman MM, et al. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78(11):4644–4652. DOI:10.1128/IAI.00685-10
- Falsetta ML, Klein MI, Colonne PM, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82(5):1968–1981. DOI:10.1128/IAI.00087-14
- Bachtiar EW, Bachtiar BM, Jarosz LM, et al. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infectmicrobiol. 2014;4:94.
- Jarosz LM, Deng DM, van der Mei HC, et al. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8(11):1658–1664. DOI:10.1128/EC.00070-09
- Willems M, Kos K, Jabra-Rizk MA, et al. Candida albicans in oral biofilms could prevent caries. Pathog Dis. 2016;74:ftw039. DOI:10.1093/femspd/ftw039
- Metwalli KH, Khan SA, Krom BP, et al. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. Plos Pathog. 2013;9(10):e1003616. DOI:10.1371/journal.ppat.1003616
- Vylkova S, Carman AJ, Danhof HA, et al. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2(3):e00055–11. DOI:10.1128/mBio.00055-11
- Janus MM, Keijser BJ, Bikker FJ, et al. In vitro phenotypic differentiation towards commensal and pathogenic oral biofilms. Biofouling. 2015;31(6):503–510. DOI:10.1080/08927014.2015.1067887
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134(3):717–728.
- Exterkate RA, Crielaard W, Ten Cate JM. Different response to amine fluoride by Streptococcus mutans and polymicrobial biofilms in a novel high-throughput active attachment model. Caries Res. 2010;44(4):372–379. DOI:10.1159/000316541
- Vollmer T, Störmer M, Kleesiek K, et al. Evaluation of novel broad-range real-time PCR assay for rapid detection of human pathogenic fungi in various clinical specimens. J Clin Microbiol. 2008;46(6):1919–1926. DOI:10.1128/JCM.02178-07
- Geddes DA. Acids produced by human dental plaque metabolism in situ. Caries Res. 1975;9(2):98–109.
- van Loveren C, Buijs JF, Ten Cate JM. The effect of triclosan toothpaste on enamel demineralization in a bacterial demineralization model. J Antimicrob Chemother. 2000;45(2):153–158.
- Kaman WE, Galassi F, de Soet JJ, et al. Highly specific protease-based approach for detection of porphyromonas gingivalis in diagnosis of periodontitis. J Clin Microbiol. 2012;50(1):104–112. DOI:10.1128/JCM.05313-11
- Cummings RT, Salowe SP, Cunningham BR, et al. A peptide-based fluorescence resonance energy transfer assay for Bacillus anthracis lethal factor protease. Proc Natl Acad Sci U S A. 2002;99(10):6603–6606. DOI:10.1073/pnas.062171599
- Kim Y-S, Lee E-S, Kwon H-K, et al. Monitoring the maturation process of a dental microcosm biofilm using the Quantitative Light-induced Fluorescence-Digital (QLF-D). J Dent. 2014;42(6):691–696. DOI:10.1016/j.jdent.2014.03.006
- Volgenant CM, Hoogenkamp MA, Buijs MJ, et al. Red fluorescent biofilm: the thick, the old, and the cariogenic. J Oral Microbiol. 2016;8:30346. DOI:10.3402/jom.v8.30346
- van der Veen MH, Volgenant CM, Keijser B, et al. Dynamics of red fluorescent dental plaque during experimental gingivitis-A cohort study. J Dent. 2016;48:71–76. DOI:10.1016/j.jdent.2016.02.010
- Koenig K, Hibst R, Meyer H, et al., editors. Laser-induced autofluorescence of carious regions of human teeth and caries-involved bacteria. Europto biomedical optics’ 93. International Society for Optics and Photonics; 1993. DOI:10.1117/12.166180
- O’Donnell LE, Robertson D, Nile CJ, et al. The oral microbiome of denture wearers is influenced by levels of natural dentition. Plos One. 2015;10(9):e0137717. DOI:10.1371/journal.pone.0137717
- Koopman JE, Buijs MJ, Brandt BW, et al. Nitrate and the origin of saliva influence composition and short chain fatty acid production of oral microcosms. Microb Ecol. 2016;72:479–492. DOI:10.1007/s00248-016-0775-z
- Hammer Ø, Harper D, Ryan P. PAST-PAlaeontological STatistics, ver. 1.89. Palaeontol Electron. 2001;4(1):1–9.
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. DOI:10.1186/gb-2011-12-10-r102
- Dumitru R, Hornby JM, Nickerson KW. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob Agents Chemother. 2004;48(7):2350–2354. DOI:10.1128/AAC.48.7.2350-2354.2004
- Bahn Y-S, Mühlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9(6):572–578. DOI:10.1016/j.mib.2006.09.003
- Klinke T, Guggenheim B, Klimm W, et al. Dental caries in rats associated with Candida albicans. Caries Res. 2011;45(2):100–106. DOI:10.1159/000324809
- Kolenbrander PE, Palmer RJ Jr., Rickard AH, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. DOI:10.1111/j.1600-0757.2006.00187.x
- Distler W, Kröncke A. Acid formation by mixed cultures of cariogenic strains of Streptococcus mutans and Veillonella alcalescens. Arch Oral Biol. 1980;25(10):655–658.
- Brand A, Barnes JD, Mackenzie KS, et al. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol Lett. 2008;287(1):48–55. DOI:10.1111/j.1574-6968.2008.01301.x
- Peters BM, Ovchinnikova ES, Krom BP, et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology. 2012;158(Pt 12):2975–2986. DOI:10.1099/mic.0.062109-0
- Fox EP, Cowley ES, Nobile CJ, et al. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24(20):2411–2416. DOI:10.1016/j.cub.2014.08.057
- Wesolowski J, Hassan RY, Hodde S, et al. Sensing of oxygen in microtiter plates: a novel tool for screening drugs against pathogenic yeasts. Anal Bioanal Chem. 2008;391(5):1731–1737. DOI:10.1007/s00216-008-1947-6
- Johansson I, Witkowska E, Kaveh B, et al. The microbiome in populations with a low and high prevalence of caries. J Dent Res. 2016;95(1):80–86. DOI:10.1177/0022034515609554
- Nadkarni MA, Caldon CE, Chhour K-L, et al. Carious dentine provides a habitat for a complex array of novel Prevotella-like bacteria. J Clin Microbiol. 2004;42(11):5238–5244. DOI:10.1128/JCM.42.11.5238-5244.2004
- Gomar-Vercher S, Cabrera-Rubio R, Mira A, et al. Relationship of children’s salivary microbiota with their caries status: a pyrosequencing study. Clin Oral Investig. 2014;18(9):2087–2094. DOI:10.1007/s00784-014-1200-y
- Kara D, Luppens SB, Cate JM. Differences between single- and dual-species biofilms of Streptococcus mutans and Veillonella parvula in growth, acidogenicity and susceptibility to chlorhexidine. Eur J Oral Sci. 2006;114(1):58–63. DOI:10.1111/j.1600-0722.2006.00262.x
