ABSTRACT
Leptotrichia species are non-motile facultative anaerobic/anaerobic bacteria that are found mostly in the oral cavity and some other parts of the human body, in animals, and even in ocean sediments. Valid species include L. buccalis, L. goodfellowii, L. hofstadii, L. honkongensis, L. shahii, L. trevisanii, and L. wadei. Some species require serum or blood for growth. All species ferment carbohydrates and produce lactic acid that may be involved with tooth decay. Acting as opportunistic pathogens, they are involved in a variety of diseases, and have been isolated from immunocompromised but also immunocompetent individuals. Mucositis, oral lesions, wounds, and abscesses may predispose to Leptotrichia septicemia. Because identification of Leptotrichia species by phenotypic features occasionally lead to misidentification, genetic techniques such as 16S rRNA gene sequencing is recommended. Early diagnosis and treatment of leptotrichia infections is important for positive outcomes. Over the last years, Leptotrichia species have been associated with several changes in taxonomy and new associations with clinical diseases. Such changes are reported in this updated review.
Introduction
Leptotrichia is one of four genera within the family Leptotrichiaceae. Description of Leptotrichiaceae is based on phylogenetic analyses of the 16S rRNA gene sequences. Leptotrichia species are facultative anaerobic/anaerobic Gram-negative rods that inhabit the oral cavity, intestines, urogenital system, and female genital tract of humans [Citation1–Citation5]. They are non-motile and ferment carbohydrates to produce various organic acids, including lactic acid, and traces of acetic, formic, or succinic acid, depending on the substrates and species. Some species are fastidious, requiring serum or blood for growth [Citation1–Citation3]. L. buccalis was for centuries the only known Leptotrichia species, but new species have now been formally accepted, which include L. goodfellowii, L. hofstadii, L. shahii, L. trevisanii, and L. wadei () [Citation2,Citation4,Citation5] and L. hongkongensis [Citation6]. As with other members of the oral commensal microbiota, Leptotrichia species are also associated with periodontal diseases and oral cavity abscesses [Citation5,Citation7,Citation8], typically as opportunistic infections. However, isolation of Leptotrichia species from infective endocarditis patients with normally functioning immune systems has been also reported [Citation5,Citation9–Citation12]. Leptotrichia species can cause opportunistic infections that lead to bacteremia in neutropenic patients with oral mucosal injuries [Citation2,Citation5] and bacteremia due to L. trevisanii after an allogeneic bone-marrow transplant [Citation13]. Although systemic infections involving Leptotrichia species are infrequent, severe infections have been reported in immunocompromised patients [Citation2,Citation4,Citation7,Citation9,Citation10,Citation13–Citation19].
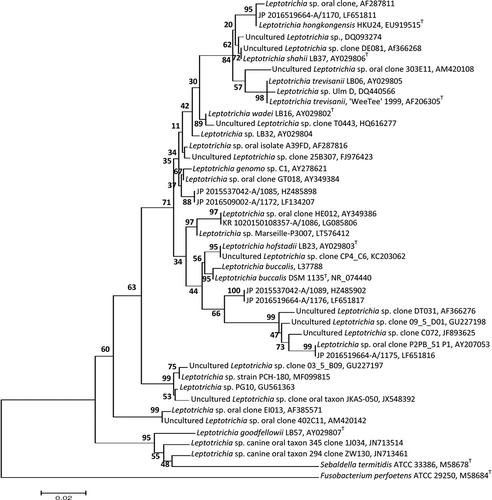
Figure 1. A phylogenetic tree obtained from the MEGA (www.megasoftware.net) program based on only sequences >800 bp by neighbor joining after ClustalW alignment. The analysis of the 16S rRNA gene sequences of the representative clones and reference strains of related Leptotrichia species and other members of Fusobacteriacea derived from GenBank is shown. Bootstrap values from 500 replicate trees are given at the nodes. Scale bar shows sequence divergence. T = type strain.
Some species have been recovered from the human oral cavity, while others such as L. buccalis and L. goodfellowii have been recovered from dog bites [Citation20] and guinea-pig wounds [Citation6,Citation21]. Based on 16S rDNAsequences comparisons Leptotrichia species were isolated from the hindgut of termites, fish, and even ocean sediments () [Citation3]. Most mammals may have their own versions of human oral species, which are typically host-species specific.
Table 1. Leptotrichia completed genome assembly sequences
Table 2. Update on reported Leptotrichia infections. Cases 1–54 were reported in a previous review by the authors [Citation2]
In most cases, the cause of Leptotrichia infections has been L. buccalis. Since previous reviews [Citation2,Citation3], Leptotrichia species have been reported in >124 cases [Citation4,Citation7,Citation16,Citation18–Citation69,Citation87–Citation93], whereby 30 cases involved L. buccalis [Citation4,Citation8,Citation15,Citation21,Citation34,Citation52,Citation56,Citation70–Citation79,Citation87], 24 cases L. wadei [Citation4,Citation20,Citation24,Citation34,Citation37,Citation42,Citation48,Citation56,Citation67,Citation80,Citation81,Citation90,Citation93], 16 cases L. trevisanii [Citation4,Citation5,Citation9,Citation10,Citation13,Citation14,Citation17,Citation37,Citation87], 14 cases L. hofstadii [Citation34,Citation40,Citation49,Citation56,Citation81–Citation84,Citation93], 10 cases L. goodfellowii [Citation4,Citation11,Citation12,Citation21,Citation56,Citation74,Citation85,Citation87], eight cases L. hongkongensis [Citation4,Citation6,Citation18,Citation45,Citation47,Citation56], and five L. shahii [Citation34,Citation56,Citation86]. L. trevisanii and L. wadei bacteremia are extremely rare; clinicians should consider these species in cases involving immunocompromised patients with oral lesions [Citation4,Citation5,Citation13,Citation17,Citation87]. The aim of the present review is to update the knowledge on the genus Leptotrichia as given in previous reports, adding information published after 2008 [Citation2,Citation3].
Taxonomy
Leptotrichia was recognized and described by van Leeuwenhoek in 1683, and the genus was established in 1879 by Trevisan [Citation2,Citation3]. Leptotrichia ferments carbohydrates, producing lactic acid as its major metabolic end product [Citation2,Citation3]. The primary habitat has been considered to be the oral cavity.
In Bergey’s Manual of 2005 [Citation95] and based on comparative analysis of 16S rDNA sequences [Citation31], the genus Leptotrichia is placed in the phylum Fusobacteria in the family II Leptotrichiaceae with Leptotrichia as the first genus. Other genera of this family include Sebaldella, Sneathia, and Streptobacillus [Citation3,Citation95].
The genus Leptotrichia comprises seven formally described species: L. buccalis is the type species of the genus, followed by L. goodfellowii, L. hofstadii, L. hongkongensis, L. shahii, L. trevisanii, and L. wadei () [Citation1–Citation3,Citation6,Citation96]. Their characteristics have been described in detail elsewhere [Citation1,Citation6,Citation95] and will not be repeated here. L. amnionii is not validly published [Citation2,Citation97]. However, based on 16S rRNA gene sequences, L. amnionii was suggested to be transferred to the genus Sneathia [Citation1,Citation2], and recently, a strain with similar resemblances and features was characterized, renamed, and transferred to the genus Sneathia as S. amnii [Citation98]. For this reason, L. amnionii will not be discussed in this review.
Genomics
The whole genomes of 12 Leptotrichia species have been completely sequenced [Citation99,Citation100]. A short description of these species and their genomic features are given in . In addition, a large variety of 16S rRNA gene Leptotrichia nucleotide sequences exists in various databases (e.g. in HOMD; www.homd.org), NCBI GenBank, RDP, DNA data Bank of Japan (DDBJ), and other private databases. For instance, a survey from the NCBI GenBank showed that >4,800 Leptotrichia nucleotide sequences were registered and deposited as of 7 August 2017. The sequences came from material collected from humans, animals, fish, and ocean sediment. A representative phylogenetic tree based on 4,800 Leptotrichia sequences showing the diversity of the species aligned by ClustalW is given in . The phylogenetic tree was generated by neighbor joining based on 500 bootstrap replicates and reconstructed with MEGA7 software (www.megasoftware.net).
Conserved proteins of the phylum Fusobacteria
Conserved signature inserts
Genome sequencing has provided insight into rich resources of molecular markers or signatures that are specific for different groups of bacteria. These novel molecular markers can be used to demarcate diverse bacterial taxa. An example is conserved signature inserts (CSIs) or deletions (i.e. indels) in protein sequences [Citation100].
Members of the family Leptotrichiaceae are easily distinguished based on concatenated sequences for conserved proteins. Comparative analysis of Fusobacteria identified CSIs in proteins involved in a broad range of functions specific for the phylum. Some of these CSIs important proteins are uniquely present in the protein homologs of all sequenced members of Fusobacteria and thereby provide potential molecular markers for this phylum, which includes the family Leptotrichiacaeae. Further, it has been suggested that these specific CSIs provide evidence that could be used as novel tools for identifying and distinguishing members of the families Fusobacteriaceae and Leptotrichiaceae and other bacteria [Citation100]. The gene sequences for many of the proteins containing these CSIs are highly conserved and based upon the conserved regions of the genes/proteins, for which PCR primers can be designed.
Clinical importance of Leptotrichia species
Eribe and Olsen [Citation2,Citation3] reported previously that the clinical importance of Leptotrichia species remains unclear due to difficulties in isolation and identification of the organisms [Citation2,Citation3,Citation70]. Recently, with modern molecular techniques and more awareness, more light has been shed on Leptotrichia species and their involvement in a variety of diseases. Leptotrichia species commonly colonize the mucous membrane of humans and animals, and are significant constituents of the microbiota of the human oral cavity, playing an important role in many diseases [Citation2,Citation3,Citation100]. , a continuation of previous [Citation2], depicts 176 cases of Leptotrichia species presented in the current review. It shows where Leptotrichia species were isolated, the various sources they came from, which Leptotrichia species were detected, the polymicrobial species they are associated with, as well as their frequencies. As can be seen, Leptotrichia species are commonly present in the human and animal gastrointestinal tract, in the periurethral region, and in the genitalia of women [Citation1–Citation3,Citation21,Citation54,Citation97].
In a previous review [Citation3], it was concluded that Leptotrichia species were isolated and recovered from various sources, including patients who had gingivitis, necrotizing ulcerative gingivitis, adult/juvenile periodontitis, ‘refractory periodontitis’, Vincent’s angina, noma, acute appendicitis, bacterial vaginosis, aortic aneurysms, cellulitis, phagedenic chancroid, saplpingitis, neutropenia, human immunodeficiency virus (HIV), leukemia, endocarditis, and human and animal infections [Citation2,Citation97]. It was suggested that Leptotrichia species are opportunistic pathogens. Current documentation and a review of the literature support this view.
Brief additional clinical information on Leptotrichia species
L. buccalis
Recently, L. buccalis has been isolated from irreversible pulpitis, pulp necrosis, apical periodontitis [Citation70], and dental plaques of both humans and guinea pigs with alveolar bone loss () [Citation21,Citation56,Citation71,Citation90]. It has also been recovered from root canals of patients with or without other oral diseases, tissue fluids and subgingival plaque samples, and exudate with cellulitis after a dog bite () [Citation8,Citation52,Citation72–Citation74,Citation77,Citation90]. Furthermore, it has been recovered from the blood and amniotic fluid of a female patient and from the amniotic fluid of an afebrile pregnant woman with acute chorioamnionitis [Citation4,Citation78] (). It has also been detected in saliva, on the mucosal surface of patients with removable partial dentures, in peri-implant crevicular fluids [Citation34,Citation76,Citation79], and in biofilms () [Citation75]. In addition, L. buccalis was isolated from the blood of an elderly woman who suffered from moderate normocytic anemia, acute myelogenous leukemia, and mucositis () [Citation15,Citation87].
L. goodfellowii
L. goodfellowii has been isolated from oral swabs of guinea pigs [Citation21] and the gastric fluid of patients who suffered spontaneous stillborn child expulsion [Citation85]. It has also been isolated from the blood of an amniotic fluid patient with a wound and respiratory difficulties [Citation4], from a wound exudate of a healthy person with cellulitis after a dog bite [Citation74], from saliva, plaque, and the mucosal surface of caries-active patients and diabetic smokers [Citation56,Citation90], and from the blood of patients with heart failure, diabetes, bladder cancer, pulmonary edema, and bronchopneumonia [Citation11]. L. goodfellowii has been recovered from an immunocompetent endocarditis patient with bioprosthetic pulmonic valve and an aortic valve homograft suffering from fever and chronic night sweats (diaphoretic) () [Citation12].
L. hofstadii
L. hofstadii has been isolated from subgingival samples and gingival crevicular fluid of periodontitis patients [Citation83], saliva, biofilm from caries [Citation49,Citation65], the mucosal surface of patients with removable partial dentures, and root canals of patients with or without disease [Citation34,Citation56,Citation84], tumor tissue [Citation40], and tongue coatings of halitosis patients () [Citation81,Citation82].
L. hongkongensis
L. hongkongensis has been isolated from the blood of metastatic breast carcinoma (MBC) patients [Citation6], the blood and amniotic fluid of a patient with a wound and respiratory difficulties [Citation4], plaque from dental caries [Citation45,Citation47,Citation56], saliva from pancreatic cancer patients and black pigmented stain caries patients () [Citation18,Citation63].
L. shahii
L. shahii has been recovered from the saliva and plaque of patients with active caries and the mucosal surface of patients with removable partial dentures () [Citation34,Citation56,Citation86].
L. trevisanii
L. trevisanii has been cultured from the blood of an immunocompetent patient, dental plaque and stool of patients with stomatitis, neutropenia, mucositis, peritonsillar abscess, blood progenitor-cell transplantation, catheter-related bloodstream infection, acute myelogenous leukemia, and redness and swelling in a tonsil incision wound [Citation5]. It has also been associated with mild liver dysfunction, normal renal function [Citation5], multiple myeloma, non-Hodgkin lymphoma (NHL), diffuse large B-cell lymphoma, post-transplant aplasia, neutropenic fever, myelodysplastic syndrome, mandibular tumor, esophageal carcinoma, and the wound and amniotic fluid of a patient with respiratory difficulties [Citation4,Citation5,Citation9,Citation10,Citation13,Citation14,Citation17,Citation37,Citation87].
L. wadei
L. wadei has been isolated from bronchoalveolar lavage fluid of a patient with leukocytosis, hypoxemia, and dyspnea [Citation24] and from the blood and amniotic fluid of a patient with a wound and respiratory difficulties () [Citation4]. Saliva, plaque, and the oral mucosal surface of caries patients [Citation34,Citation37,Citation56,Citation67] and the oral cavity and biofilms from oral epithelial cells of a patient with new-onset rheumatoid arthritis [Citation20,Citation48] all contained L. wadei. Patient material from tongue plaque, saliva, and the tongue coating of malodor and halitosis patients [Citation42,Citation81] was isolated with L. wadei present. This bacterium was even isolated from the antral gastric biopsy of a dyspeptic patient [Citation80], smokers’ plaque [Citation90], and oropharyngeal samples () [Citation93].
Unspecified Leptotrichia species
Leptotrichia species have been recovered from the blood of patients with liver abscesses, mucositis, neutropenic sepsis, diabetes, respiratory distress, community-acquired pneumonia (CAP), bilateral lung crackles, mild anemia, and vasculitis () [Citation7,Citation22,Citation33–Citation35]. They were also recovered from oral plaque of guinea pigs [Citation21] and feces of piglets [Citation54], dental plaque from healthy individuals, plaque and saliva from patients with various types of caries, gingivitis, chronic periodontitis, and peri-implantitis [Citation23,Citation25–Citation27,Citation34,Citation35,Citation37,Citation38,Citation44,Citation45,Citation47,Citation49,Citation52,Citation59,Citation60,Citation62,Citation66–Citation69,Citation91,Citation94], decayed tooth surfaces and discordant caries from intact enamel surfaces [Citation53]. Leptotrichia species were also isolated from bronchoalveolar lavage fluid, and patients with leukocytosis, hypoxemia, and dyspnea [Citation24]. Further, Leptotrichia species were recovered from healthy patients with oral cancer, premalignant oral lesion [Citation18,Citation28,Citation33,Citation56,Citation91], edentulous infants [Citation29], human vaginal fluid of sexually active and inactive individuals [Citation30,Citation32], HIV-seropositive and -seronegative patients [Citation46], pancreatic cancer patients [Citation18,Citation66], black pigmented stain caries patients [Citation63], and patients with halitosis () [Citation42,Citation65,Citation81,Citation82]. Besides, Leptotrichia species were isolated from the blood [Citation4,Citation5,Citation22,Citation74], the amniotic fluid of a patient with a wound and respiratory difficulties [Citation4], breast milk of obese women with gestational diabetes and normal weight [Citation36], oral samples of a patient with new-onset rheumatoid arthritis [Citation20], oral lichen planus patients [Citation88], and even from fermenting Lees liquor [Citation39]. Leptotrichia species were equally isolated from the blood and gastric fluid of patients with coronary artery disease (CAD), candidal esophagitis, chronic kidney disease, diabetic, duodenal ulcer, erythematous gastropathy, gastroesophageal reflux disease, gastric ulcer, hiatal hernia, reflux esophagitis, upper gastrointestinal bleeding, renal transplant, and sarcoidosis () [Citation16]. Also, Leptotrichia species were isolated from tumor tissues and sputum of patients with tuberculosis, acute exacerbation of chronic obstructive pulmonary disease, and feces of piglets with porcine epidemic diarrhea virus [Citation40,Citation41,Citation43,Citation50,Citation51]. They were also detected in patient material from tongue plaque with malodor [Citation42], biofilms of caries, oral epithelial cells [Citation48,Citation49], vaginal swabs with high-risk human papillomavirus, and from HIV-positive and -negative subjects [Citation55]. The guts of herbivorous, carnivorous, and omnivorous fish [Citation58], tumor tissues and saliva of patients with head and neck squamous-cell carcinoma human papillomavirus (HPV), oropharyngeal squamous-cell carcinoma HPV, and oral cavity squamous-cell carcinoma HPV [Citation19] all contained Leptotrichia species. They were also isolated from the bile aspirate of fish with cholelithiasis (gallstone diseases) and Opisthorchis felineus (fish-borne liver fluke infections), in pancreatitis and hepatitis C [Citation61], and in saliva from a Behçet’s disease patient [Citation64]. Wu et al. [Citation57] reported recovery of Leptotrichia species, together with Veillonella parvula and Peptostreptococcus species in low amounts in cigarette smokers’ mouthwash () [Citation57,Citation90,Citation91]. Also, human skin emanation samples and oropharyngeal samples of mite-food-sensitized children with rhinitis and asthma were found to contain Leptotrichia species [Citation31,Citation92].
Pathogenicity of Leptotrichia
The genus Leptotrichia consists of slow-growing, non-motile facultative anaerobic/anaerobic Gram-negative rods that reside in the oral cavity and the genitourinary and intestinal tract [Citation1]. Leptotrichia species were traditionally considered non-pathogenic but have recently been considered as opportunistic causes of human disease [Citation2,Citation3,Citation78]. Previously, Eribe and Olsen [Citation2] described a myriad of pathological conditions associated with Leptotrichia, including appendicitis, pneumonia, mucositis, and sepsis [Citation2,Citation78]. The concept that Leptotrichia infections are opportunistic is further supported in the current review. Leptotrichia species, primarily oral commensals, have been associated with infections, particularly in immunocompromised hosts () [Citation4,Citation9,Citation13–Citation17,Citation24,Citation30,Citation32,Citation46,Citation55,Citation74,Citation78,Citation97], but occasionally in immunocompetent persons [Citation5,Citation11,Citation12,Citation24,Citation33,Citation60,Citation74,Citation78,Citation85].
The cell surface of leptotrichia has protruding structures presumed fitted for adhesion [Citation2,Citation3]. Like any other Gram-negative rod that possesses lipopolysaccharide (LPS, endotoxin), Leptotrichia displays O-antigen linked to lipid-A. The latter may cause hemorrhage, fever, tumor necrosis, fatal shock, and septicemia [Citation4–Citation7,Citation9,Citation10,Citation12–Citation15,Citation17,Citation24,Citation33,Citation40,Citation85,Citation87] and may even lead to abortion, as observed in infection associated with L. goodfellowii [Citation85]. The virulence of L. buccalis was demonstrated experimentally in a rabbit model [Citation2,Citation3]. When Leptotrichia endotoxin was compared to Escherichia coli endotoxin in terms of a lethal dose for 50% survival, febrile response, and leukopenia, Leptotrichia endotoxin was 10–20% as active on a weight basis. In the same test, the endotoxin from L. buccalis proved more potent than Salmonella endotoxin.
Leptotrichia and proinflammatory mediators
It is known that the systemic release of endotoxin and proinflammatory mediators from infected host tissue can contribute directly or indirectly to the sepsis syndrome associated with Leptotrichia [Citation2,Citation3,Citation7]. Once activated, the immune system is hard to switch off, and sometimes it gets out of control, causing damage to other parts of the body. This ‘self-inflicted’ damage acts as trigger for various disease conditions [Citation101]. Many types of Gram-negative bacteria secrete LPS that stimulates the immune system. A study by Langfeldt et al. [Citation48] found that Leptotrichia was able to trigger the transcription level of proinflammatory interleukin (IL)-1β, IL-6, IL-8, and IL-10 in epithelial cells [Citation48]. This suggests that Leptotrichia may play a key role during the transition from health to disease [Citation54]. IL-1β modulates human cell differentiation, proliferation, and apoptosis, which regulate the release of other proinflammatory cytokines such as IL-6 and IL-8 [Citation48]. In addition, IL-6 and IL-8 have the capacity to attract granulocytes and lymphocytes, thereby inducing the host cellular immune response. In contrast, IL-10 is designated as an anti-inflammatory mediator that prohibits excessive immune response by suppressing pro-inflammatory cytokine production and the antigen-presenting capacity of monocytes, macrophages, and dendritic cells [Citation48]. Both pathogenic and commensal bacteria interfere with early host cell signalling for survival or promote bacterial infection by decreasing pro-inflammatory responses [Citation48]. In an in vitro multispecies biofilm model with or without major periodontal pathogens, it was documented that such biofilms can upregulate IL-8 expression in gingival epithelial cells. The presence of the ‘red-complex’ species (Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola) resulted in even greater upregulation [Citation48]. The data strongly argued that Leptotrichia may be crucially involved in the ‘fine-tune’ regulation of epithelial immune response to obtain homeostasis or propagate inflammatory response [Citation48]. Jang et al. [Citation102] reported that L. wadei, Fusobacterium nucleatum, and Campylobacter gracilis when co-cultured with human gingival fibroblasts highly upregulated the expression of antimicrobial chemokine peptides and the proinflammatory mediators IL-6 and IL-8, whereas the red-complex bacteria stimulated low levels or often suppressed expression of these factors [Citation102].
New-onset patients with chronic rheumatoid arthritis harbored high levels of several pathogens, including Gemella morbillorum, Propionibacterium acnes, Streptococcus gordonii, and L. buccalis. This indicated that L. buccalis can be more specifically associated with multiple disease activity than so far realized [Citation20,Citation52]. Irrespective of periodontal disease status, the Leptotrichia OTU 87 (L. wadei) clone and Prevotella OTU 60 (P. intermedia) clone were the only clones observed in increased amount in patients with new-onset rheumatoid arthritis but were absent in healthy controls [Citation20].
Leptotrichia species in bacteremia
Thirty-one cases of bacteremia and four cases of wound infections associated with Leptotrichia species have been reported (). Bacteremia caused by Leptotrichia species were found among neutropenic patients with various forms of predisposing diseases such as bone-marrow transplants, infective endocarditis, and sepsis associated with mucositis. The latter served as an oral or orodental portal of entry [Citation2,Citation3,Citation22]. In fact, neutropenic fever coupled with mucositis is an established predisposing factor for development of sepsis by Leptotrichia species [Citation4,Citation7,Citation87]. Peripheral blood stem-cell transplant patients (PBSCT) with fever due to mucosal disruptions and lesions have a portal of entry for bacteria that causes bacteremia [Citation5,Citation9,Citation22,Citation33]. Mucositis, esophageal ulcer, or diverticulitis are possible risk factors for infected patients [Citation7,Citation9,Citation13,Citation15–Citation17,Citation33,Citation85]. L. trevisanii was involved in 15 incidences of bacteremia. Eight cases each also involved Leptotrichia species and L. buccalis, six L. goodfellowii, three L. wadei, two L. hongkongensis, and one with L. shahii () [Citation4–Citation6,Citation9–Citation15,Citation17,Citation21,Citation22,Citation33,Citation42,Citation74,Citation78,Citation85–Citation88]. In cases involving L. trevisanii, seven were also associated with neutropenic fever [Citation5,Citation13,Citation14,Citation17,Citation87], while five were associated with PBSCT [Citation9,Citation10], four had acute myelogenous leukemia (AML) [5,7 9,10,15] and multiple myelomas (MM) [Citation4,Citation9,Citation13], two had stomatitis [Citation10,Citation14], three had NHL [Citation9,Citation87], and one had a catheter-related bloodstream infection [Citation17]. It is worth mentioning that L. goodfellowii has previously been associated with endocarditis. L. goodfellowii isolated from immunocompetent patients was found to be a pathogenic agent when associated with bacteremia [Citation11,Citation12,Citation33,Citation74,Citation85]. Lim et al. [Citation11] therefore hypothesized that L. goodfellowii could be secondary to pneumonia, as there was no other causative factor leading to bacteremia in their patient. In one of three cases, L. goodfellowii was even associated with a stillborn child, spontaneously expelled after 25 weeks of gestation [Citation4]. In three cases of L. hongkongensis bacteremia, one case was associated with amniotic fluid, fever, and MBC [Citation6]. L. wadei bacteremia was detected in wounds and amniotic fluid [Citation4].
Thus, recent reports have proven the pathogenicity of Leptotrichia species. Inappropriate clinical situations can affect the protective function of the indigenous bacterial flora, which can lead to disruption by broad-spectrum antibiotic therapy [Citation2–Citation4,Citation12,Citation69,Citation103], resulting in infection. Likewise, enhanced Leptotrichia proliferation and tissue invasion can culminate in bloodstream invasion and dissemination [Citation2,Citation3]. This occurs frequently when the patient’s immune system is comprised with Leptotrichia species such as with cases involving L. buccalis, L. trevisanii, L. wadei, and L. goodfellowii. These species have been reported to act as opportunistic pathogens responsible for bloodstream infections in immunocompromised patients [Citation2,Citation4,Citation5,Citation15,Citation17,Citation33,Citation74,Citation85,Citation87,Citation103].
L. buccalis has been associated with chorioamnionitis and child loss during pregnancy [Citation78]. The authors suggested that the development of chorioamnionitis was a result of hematogenous spread arising from the oral cavity [Citation78]. Unique to bacteremia from other Leptotrichia species, L. goodfellowii showed an association with bacteremia secondary to endocarditis [Citation11,Citation12]. In contrast to previously reported cases of Leptotrichia bacteremia, the patient in this report was immunocompetent and had no history of endocarditis. For the first time, a case of L. goodfellowii bacteremia was recently reported in a Korean patient [Citation11]. It is noteworthy that in a 62-month retrospective survey of 4,857 episodes of anaerobic bacteremia, Leptotrichia species were identified as the causative pathogens in 7.3% of cases [Citation12,Citation22].
Leptotrichia species in cancers
A few Leptotrichia species were related to 88 incidences of various cancers [Citation4–Citation7,Citation9–Citation11,Citation13–Citation15,Citation17–Citation19,Citation22,Citation28,Citation33,Citation40,Citation57,Citation60,Citation61,Citation66,Citation74,Citation80,Citation87,Citation91], of which 43 cases had neutropenia, sepsis, and fever [Citation4–Citation7,Citation9,Citation10,Citation12–Citation15,Citation17,Citation22,Citation24,Citation33,Citation87], 14 had transplant issues [Citation4,Citation9,Citation10,Citation13,Citation16,Citation17,Citation22,Citation33], 14 mucositis [Citation4,Citation7,Citation9,Citation13–Citation15,Citation17,Citation22,Citation87], 12 various lesions (6, 11, 27, 32, 37, 44, 56 64, 99), and five pneumonia [Citation5,Citation9,Citation11,Citation24,Citation33]. The suspected port of Leptotrichia entry included mucositis, abscesses, wound infections, gingivitis, diverticulitis, peritonitis, neutropenic sepsis, and ulcers ().
In an examination of the relationship of the oral microbiota with subsequent risk of pancreatic cancer in a large nested case-control study, the authors reported that the carriage of oral pathogens, P. gingivalis and Aggregatibacter actinomycetemcomitans, was associated with a higher risk of pancreatic cancer [Citation66]. They also found that a greater abundance of the phylum Fusobacteria was associated with decreased pancreatic cancer risk as well as its genus Leptotrichia [Citation66]. Their finding was inconsistent with a recent cross-sectional study of eight patients, which found higher abundances of Leptotrichia and Porphyromonas in the saliva of pancreatic cancer patients compared to controls and those with other diseases, including non-cancerous pancreatic disease [Citation18]. Torre et al. [Citation18] concluded that the Leptotrichia and Porphyromonas ratio may serve as a potential pancreatic cancer biomarker. Based on their findings, pancreatic cancer may be detected at early stages by sampling individuals’ saliva and looking at the ratios of Leptotrichia and Porphyromonas.
Leptotrichia in dental caries
Among the many microbial species residing in oral biofilms (plaque) at the tooth surface [Citation104], mutans streptococci have long been recognized as primary contributors in the etiology of dental caries [Citation104]. The pathogenicity of organisms such as Streptococcus mutans and S. sobrinus is attributable in part to (i) the capacity of these species to produce extracellular glucan(s) from dietary sucrose that facilitate microbial adherence to the tooth surface, and (ii) the fermentation of sucrose to lactic acid – the causative agent in the demineralization of tooth enamel [Citation104]. There is supporting evidence that the genus Leptotrichia is highly saccharolytic [Citation1–Citation3,Citation11,Citation104–Citation106], implying that it ferments a wide variety of mono- and disaccharides to lactic acid similar to S. mutans. This property may implicate the participation of Leptotrichia species in tooth decay [Citation1–Citation3,Citation11].
Association between Leptotrichia and halitosis
Leptotrichia has also been associated with halitosis (oral malodor) [Citation42,Citation65,Citation81,Citation82]. Most of the species within the core microbiome of the tongue-coating biofilm are Gram-negative anaerobic bacteria that are adaptable to the tongue-coating environment () [Citation81]. Malodor is foul-smelling breath from the oral cavity in humans [Citation42]. Most malodor originates from the host’s tongue plaque and is without any clear signs of disease, which is defined as physiologic oral malodor [Citation42]. Malodorants are produced by the tongue plaque resident on the large surface area of the tongue. Some bacteria inside tongue plaque can produce amino acids and peptide by-products as well as food debris to putrefy, thus producing malodorants [Citation42]. The unpleasant oral odor results from volatile sulfur compounds (VSCs), including hydrogen sulphide (H2S), methyl mercaptan (CH3SH), other thiols, and dimethyl sulphide ((CH3)2S) involved and associated with halitosis [Citation42]. Of the three major VSCs involved in oral malodor, (CH3)2S is the main contributor to halitosis [Citation81], whereas CH3SH is more pathogenic than H2S and is associated with periodontal disease [Citation81]. It has been inferred that the reason for halitosis might be cooperative polymicrobial activity, which includes Leptotrichia species interactions rather than the effect of a single pathogen [Citation81]. There is also evidence supporting that Leptotrichia species are present in increased abundances in people with oral malodor, despite a lack of H2S production [Citation81,Citation82]. Yang et al. reported that L. wadei was positively correlated with H2S concentrations [Citation42] and concluded that Leptotrichia spp. and Prevotella spp. were found to be strongly associated with oral malodour [Citation42], although direct proof of production was not provided. This bacterium was detected in relatively high abundance in all the halitosis tongue-coating samples and was inferred to be involved in halitosis [Citation81,Citation82], likewise L. hofstadii in some subjects [Citation81,Citation82]. Bacteria such as Peptostreptococcus stomatis and Prevotella shahii isolated from tongue coatings of diseased persons together with L. wadei were also suggested to be candidate halitosis pathogens [Citation81] ().
Leptotrichia in co-existence with other microbes
The human oral cavity has an indigenous microbiota known to include a robust community of microorganisms. Leptotrichia species are present in the salivary milieu and coexist with virus/bacteriophages in this environment, together with other microbes, for example Veillonella [Citation76]. Their interrelationships remain elusive. Leptotrichia, Clostridium, and Citrobacter were found as the most abundant bacteria in the herbivorous fish gut [Citation58]. Previous studies have reported that Clostridium, Citrobacter, Leptotrichia, Bacillus, and Enterobacter are important cellulose-degrading bacteria in herbivorous fish [Citation58]. It was suggested that these bacterial species might play significant roles in their host’s digestive system. Herbivorous fish harbored abundant cellulose-degrading bacteria, including Clostridium, Citrobacter, and Leptotrichia () [Citation58]. L. hofstadii was considered and reported as a potential biomarker for dental caries in association with Campylobacter showae and Parvimonas micra [Citation69,Citation84]. Leptotrichia species were found together with Fusobacterium and Campylobacter species in patients with colorectal carcinoma. This polymicrobial signature was associated with overrepresentation of numerous host genes, including the gene for encoding the proinflammatory chemokine IL-8 [Citation40].
Leptotrichia species were reported in close association with fungi, including species of Saccharomyces, Aspergillus, Zygosaccharomyces, Pichia, Saccharomycopsis, Talaromyces, Eurotium, Fomitopsis, Trichosporon, Candida albicans, C. parapsilosis, and C. tropicalis, and other species from liquor [Citation39], gastric fluid [Citation16], the saliva of HIV patients [Citation46], sputum [Citation50], blood, and saliva [Citation60] (). The importance of these associations remains unknown. Leptotrichia species, together with Delftia species and Actinobacteria species, were significantly correlated with individuals attacked by malaria mosquitoes [Citation31]. Leptotrichia species, L. wadei, and Streptococcus species were isolated together with C. albicans from dental plaque samples of patients with or without rampant caries [Citation67,Citation89]. The authors postulated that these pathogenic species and dysbiosis of the oral microbial community might have contributed to the pathogenesis of rampant caries in their patient. Leptotrichia spp. and Lautropia spp. were found to increase significantly in oral lichen planus (OLP) patients [Citation88]. The argument for this was that as OLP is an immune-related disease, the elevated colonization of these bacteria might be related to the local immune dysfunction of OLP, which again suggested that OLP is associated with dysbiosis of the oral microbiome [Citation88]. Kawanami et al. [Citation24] suggested that in a severe pneumonia patient, isolated L. wadei and other Leptotrichia species, together with mixed oral bacteria (Enterococcus faecalis, E. casseliflavus, Veillonella parvula, V. atypica, V. dispar, Prevotella nanceiensis, Streptococcus spp. clones, Delftia sp. clone, Lactobacillus sp. clone, Syntrophococcus sp. clone, Clostridium sp. clone, and Actinomyces sp. clone), played important roles () [Citation24].
Identification of Leptotrichia species
Identification of Leptotrichia species can be problematic in terms of culturing because some strains are strictly anaerobic or facultative anaerobic, while others prefer growth under the influence of CO2. Leptotrichia species usually stain Gram-negative, but fresh cells may be Gram-positive. Old cells may even stain both ways, leading to misclassification.
Due to the insufficiency of databases, identification of Leptotrichia species by conventional biochemical assays may be difficult and challenging, since most species are not recorded in databases. Most databases contain only one or two species known as L. buccalis or Leptotrichia species. Schrimsher et al. [Citation9] reported cases of misidentification of L. trevisanii sepsis where all the isolates were unidentified by biochemical tests. One of the isolates was misidentified as Sphingomonas paucimobilis [Citation9] and another as Clostridium acetobutylicum [Citation13]. A report from Lim et al. [Citation11] showed misidentification of L. trevisanii as Capnocytophaga spp. and L. buccalis by the Vitek 2 system [Citation11], or as unidentified using this system. In addition, the MALDI-TOF MS system may struggle in the identification of Leptotrichia species [Citation11]. The VITEK MS database has no known Leptotrichia species, making their identification impossible and underestimated. Lim et al. [Citation11], however, reported that the Bruker Biotyper System (Bruker Daltonics, Billerica, MA), which contains some Leptotrichia species in their database, gave successful identification [Citation11]. It is of general interest that more database development and strain accumulation are made to enable the precise identification of Leptotrichia species [Citation11]. To avoid misclassification of Leptotrichia species, application of 16S rRNA gene identification is recommended because of its reliability and feasability. HOMD with its large amount of genetic data from oral bacteria is probably the most reliable database to use.
Antimicrobial agents toward Leptotrichia
Leptotrichia species are susceptible to many antimicrobial agents such as penicillin, ampicillin, oxacillin, cephalothin, cefoxitin, cefotaxime, amoxicillin/sulbactam, ampicillin/sulbactam, amoxicillin/clavulanate, clindamycin, metronidazole, rifampicin, tetracycline, imipenem, and chloramphenicol. Strains have developed resistance to erythromycin, gentamycin, kanamycin, vancomycin, ciprofloxacin, tobramycin, amikacin, fluoroquinolones, and aminoglycosides [Citation2,Citation11,Citation70]. Some strains have been treated successfully while others have not with these antibiotics. L. goodfellowii bacteremia has been successfully treated with piperacillin/tazobactam, ceftriaxone/metronidazole, or amoxicillin/clavulanate, clindamycin, vancomycin, gentamycin, and imipenem [Citation11,Citation74]. L. goodfellowii was found resistant to tobramycin, amikacin, and ciprofloxacin [Citation74]. With antimicrobial susceptibility testing, prompt and adequate selection of antibiotics could be sufficient for treatment of L. goodfellowii bacteremia [Citation11]. Antibiotic treatment with piperacillin/tazobactam, moxifloxacin, piperacillin, erythromycin, levofloxacin, gentamycin, amikacin, and chloramphenicol was unsuccessful toward L. trevisanii [Citation13,Citation14,Citation17] and successful with meropenem [Citation14,Citation17], penicillin, amoxicillin, amoxicillin/clavulanate, cefoxitin, imipenem, clindamycin, tetracycline, metronidazole [Citation13,Citation14], cefotaxime, ceftazidime, piperacillin/tazobactam, and tigercycline [Citation14]. Severe pneumonia caused by L. wadei was successfully treated with imipenem/cilastin, minocycline, sulfametoxazole/trimethroprim, and clindamycin but not with cefcapene pivoxil or levofloxacin [Citation24].
Clustered regularly interspaced short palindromic repeats in Leptotrichia
There is evidence that almost all Archaea and about half of Bacteria possess clustered regularly interspaced short palindromic repeats (CRISPRs). These are segments containing short repetitions of base sequences. The unique sequences between the repeats match the DNA of the virus preying on the bacterium. CRISPRs are part of the bacterial immune system. CRISPR-associated proteins (Cas) are adaptive immune systems for Archaea and Bacteria defending microbes against foreign genetic elements (e.g. virus) via DNA or RNA-DNA interference [Citation107,Citation108]. Most Cas proteins are grouped into two functional modules: (i) the adaptation module, which delivers genetic materials into CRISPR arrays generating CRISPR RNAs (crRNAs); and (ii) the effector module, which is guided by crRNA that targets and cleaves invading nucleic acids [Citation107]. Up-to-date characterized CRISPR-Cas systems consist of Cas1 and Cas2, which are exclusively involved in spacer acquisition [Citation107]. C2c2 is the sole effector protein that uses a crRNA guide to achieve interference, targeting RNA [Citation107]. Targeting C2c2 to mRNA prevents gene expression [Citation107], suggesting that CRISPR-Cas systems and C2c2 can be used for development of a new molecular RNA-targeting tools [Citation107], including tools for Leptotrichiaceae. C2c2 from L. shahii was documented to provide interference against RNA phage [Citation108].
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Notes on contributors
Emenike R. K. Eribe
Emenike Ribs K. Eribe is a guest researcher at Department of Oral biology, Faculty of Dentistry, University of Oslo. BSc Biology from the University of Trondheim 1986. MSc Microbiology from the Faculty of Dentistry, University of Oslo in 2001 and PhD Microbiology in 2004. Co-supervisor of many MSc and PhD students.
Ingar Olsen
Ingar Olsen is professor emeritus and guest researcher at Department of Oral Biology, Faculty of Dentistry, University of Oslo. Senior Research Investigator, Department of Molecular Genetics, Forsyth Institute, Cambridge, MA. DDS from the Faculty of Dentistry, University of Oslo in 1966. Dr. odont. in 1976. Professor in oral microbiology 1988. Dean for Research 2002-2008. Previously, main supervisor of more than 20 PhD students.
References
- Eribe ERK, Paster BJ, Caugant DA, et al. Genetic diversity of Leptotrichia and description of Leptotrichia goodfellowii sp. nov., Leptotrichia hofstadii sp. nov., Leptotrichia shahii sp. nov. and Leptotrichia wadei sp. nov. Int J Syst Evol Microbiol. 2004;54:1–31.
- Eribe ERK, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14:131–137.
- Eribe ERK, Olsen I. Leptotrichia and Leptotrichia-like organisms. In: Liu D, editor. Molecular detection of human bacterial pathogens. Section III. Baceroidetes, Chlamydiae, and Fusobacteria, Chapter 49. Boca Raton, London, New York: CRC Press: Taylor & Francis Group; 2011. p. 555–566.
- Couturier MR, Slechta ES, Goulston C, et al. Leptotrichia bacteremia in patients receiving high-dose chemotherapy. J Clin Microbiol. 2012;50:1228–1232.
- Kumagai J, Takiguchi Y, Shono K, et al. Acute myelogenous leukemia with Leptotrichia trevisanii bacteremia. Intern Med. 2013;52:2573–2576.
- Woo PCY, Wong SSY, Teng JLL, et al. Leptotrichia hongkongensis sp. nov., a novel Leptotrichia species with the oral cavity as its natural reservoir. J Zhejiang Univ Sci B. 2010;11:391–401.
- Muttaiyah S, Paviour S, Buckwell L, et al. Anaerobic bacteraemia in patients admitted to Auckland City Hospital: its clinical significance. N Z Med J. 2007;120:U2809.
- Sassone L, Fidel R, Figueiredo L, et al. Evaluation of the microbiota of primary endodontic infections using checkerboard DNA-DNA hybridization. Oral Microbiol Immunol. 2007;22:390–397.
- Schrimsher JM, McGuirk JP, Hinthorn DR. Leptotrichia trevisanii sepsis after bone marrow transplantation. Emerg Infect Dis. 2013;19:1690–1691.
- Higurashi Y, Tatsuno K, Fujimoto F, et al. Two cases of bacteremia caused by Leptotrichia trevisanii in patients with febrile neutropenia. J Infect Chemother. 2013;19:1181–1184.
- Lim YK, Kweon OJ, Kim HR, et al. Leptotrichia goodfellowii infection: case report and literature review. Ann Clin Lab Sci. 2016;46:83–86.
- Matias WR, Bourque DL, Niwano T, et al. Subacute bacterial endocarditis with Leptotrichia goodfellowii in a patient with a valvular allograft: a case report and review of the literature. Case Rep Infect Dis. 2016;2016:3051212.
- Sabater Cabrera C, Fernández Blázquez A, García Carús E. Bacteremia due to Leptotrichia trevisanii after an allogeneic bone marrow transplant. Enferm Infect Microbiol Clin. 2016. DOI:10.1016/j.eimc.2016.09.010. pii: S0213-005X(16)30315-9. Spanish.
- Cooreman S, Schuermans C, Van Schaeren J, et al. Bacteraemia caused by Leptotrichia trevisanii in a neutropenic patient. Anaerobe. 2011;17:1–3.
- Baracaldo R, Bourbeau P. Photo quiz: an 80-year-old female with acute myeloid leukemia and induction-associated neutropenic fever. J Clin Microbiol. 2013;51:389, 737.
- Von Rosenvinge EC, Song Y, White JR, et al. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7:1354–1366.
- Martın-Gutierrez G, Rodrıguez N, Lepe JA, et al. Rapid identification of a Leptotrichia trevisanii catheter-related bloodstream infection using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. JMM Case Reports. 2015;1–4. DOI:10.1099/jmmcr.0.000036
- Torres PJ, Fletcher EM, Gibbons SM, et al. Characterization of the salivary microbiome in patients with pancreatic cancer. Peer J. 2015;3:e1373.
- Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7:51320–51334.
- Scher JU, Ubeda C, Equinda M, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64:3083–3094.
- Boot R, Van De Berg L, Reubsaet FAG, et al. Positive Streptobacillus moniliformis PCR in guinea pigs likely due to Leptotrichia spp. Vet Microbiol. 2008;128:395–399.
- Blairon L, De Gheldre Y, Delaere B, et al. A 62-month retrospective epidemiological survey of anaerobic bacteraemia in a university hospital. Clin Microbiol Infect. 2006;12:527–532.
- Preza D, Olsen I, Aas JA, et al. Bacterial profiles of root caries in elderly patients. J Clin Microbiol. 2008;46:2015–2021.
- Kawanami T, Fukuda K, Yatera K, et al. Severe pneumonia with Leptotrichia sp. detected predominantly in bronchoalveolar lavage fluid by use of 16S rRNA gene sequencing analysis. J Clin Microbiol. 2009;47:496–498.
- Ling Z, Kong J, Jia P, et al. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol. 2010;60:677–690.
- Jiang W, Jiang Y, Li C, et al. Investigation of supragingival plaque microbiota in different caries status of Chinese preschool children by denaturing gradient gel electrophoresis. Microb Ecol. 2011;61:342–352.
- Huang S, Yang F, Zeng X, et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health. 2011;11:33.
- Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788.
- Cephas KD, Kim J, Mathai RA, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503.
- Pépin J, Deslandes S, Giroux G, et al. The complex vaginal flora of West African women with bacterial vaginosis. PLoS One. 2011;6:e25082.
- Verhulst NO, Qiu YT, Beijleveld H, et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS One. 2011;6:e28991.
- Fethers K, Twin J, Fairley CK, et al. Bacterial vaginosis (BV) candidate bacteria: associations with BV and behavioural practices in sexually-experienced and inexperienced women. PLoS One. 2012;7:e30633.
- Lo TS. A cavitary pneumonia caused by Leptotrichia species in an immunocompetent patient. Infect Dis Rep. 2012;4:e24.
- Zhu X, Wang S, Gu Y, et al. Possible variation of the human oral bacterial community after wearing removable partial dentures by DGGE. World J Microbiol Biotechnol. 2012;28:2229–2236.
- Kumar PS, Mason MR, Brooker MR, et al. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol. 2012;39:425–433.
- Cabrera-Rubio R, Collado MC, Laitinen K, et al. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–551.
- Wolff D, Frese C, Maier-Kraus T, et al. Bacterial biofilm composition in caries and caries-free subjects. Caries Res. 2013;47:69–77.
- Ling Z, Liu X, Wang Y, et al. Pyrosequencing analysis of the salivary microbiota of healthy Chinese children and adults. Microb Ecol. 2013;65:487–495.
- Xiang W, Li K, Liu S, et al. Microbial succession in the traditional Chinese Luzhou-flavor liquor fermentation process as evaluated by SSU rRNA profiles. World J Microbiol Biotechnol. 2013;29:559–567.
- Warren RL, Freeman DJ, Pleasance S, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16.
- Cheung MK, Lam WY, Fung WYW. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One. 2013;8:e54574.
- Yang F, Huang S, He T, et al. Microbial basis of oral malodor development in humans. J Dent Res. 2013;92:1106–1112.
- Wu J, Liu W, He L, et al. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS One. 2013;8:e83445.
- Belstrøm D, Fiehn N-E, Nielsen CH, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–375.
- Xu H, Hao W, Zhou Q, et al. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. PLoS One. 2014;9:e89269.
- Li Y, Saxena D, Chen Z, et al. HIV infection and microbial diversity in saliva. J Clin Microbiol. 2014;52:1400–1411.
- Fernandez Y Mostajo M, Van Der Reijden WA, Buijs MJ, et al. Effect of an oxygenating agent on oral bacteria in vitro and on dental plaque composition in healthy young adults. Front Cell Infect Microbiol. 2014;4:95.
- Langfeldt D, Neulinger SC, Stiesch M, et al. Health- and disease-associated species clusters in complex natural biofilms determine the innate immune response in oral epithelial cells during biofilm maturation. FEMS Microbiol Lett. 2014;360:137–143.
- Lif Holgerson P, Öhman C, Rönnlund A, et al. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10:e0128534.
- Su J, Liu H-Y, Tan X-L. Sputum bacterial and fungal dynamics during exacerbations of severe COPD. PLoS One. 2015;10:e0130736.
- Card RM, Mafura M, Hunt T, et al. Impact of ciprofloxacin and clindamycin administration on Gram-negative bacteria isolated from healthy volunteers and characterization of the resistance genes they harbor. Antimicrob Agents Chemother. 2015;59:4410–4416.
- Arvikar S, Hasturk H, Nguyen D, et al. Elevated subgingival levels of periodontal pathogens in rheumatoid arthritis patients, particularly Leptotrichia species in new-onset disease. Abstract Number: 2721 2015 ACR/ARHP Annual Meeting; 2015 Sep 29. Available from: http://acrabstracts.org/abstract/elevated-subgingival-levels-of-periodontal-pathogens-in-rheumatoid-arthritis-patients-particularly-leptotrichia-species-in-new-onset-disease/
- Zhang M, Chen Y, Xie L, et al. Pyrosequencing of plaque microflora in twin children with discordant caries phenotypes. PLoS One. 2015;10:e0141310.
- Liu S, Zhao L, Zhai Z, et al. Porcine epidemic diarrhea virus infection induced the unbalance of gut microbiota in piglets. Curr Microbiol. 2015;71:643–649.
- Dareng EO, Ma B, Famooto AO, et al. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol Infect. 2016;144:123–137.
- Johansson I, Witkowska E, Kaveh B, et al. The microbiome in populations with a low and high prevalence of caries. J Dent Res. 2016;95:80–86.
- Wu J, Peters BA, Dominianni C, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10:2435–2446.
- Liu H, Guo X, Gooneratne R, et al. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep. 2016;6:24340.
- Huang S, Li Z, He T, et al. Microbiota-based signature of gingivitis treatments: a randomized study. Sci Rep. 2016;6:24705.
- Rashid M-U, Rosenborg S, Panagiotidis G, et al. Ecological effect of solithromycin on normal human oropharyngeal and intestinal microbiota. Antimicrob Agents Chemother. 2016;60:4244–4251. pii: AAC.00461-16.
- Saltykova IV, Petrov VA, Logacheva MD, et al. Gallstone disease and infection with Opisthorchis felineus. PLoS Negl Trop Dis. 2016;10:e0004809.
- Xiao C, Ran S, Huang Z, et al. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front Microbiol. 2016;7:1145.
- Li Y, Zou CG, Fu Y, et al. Oral microbial community typing of caries and pigment in primary dentition. BMC Genomics. 2016;17:558.
- Coit P, Mumcu G, Ture-Ozdemir F, et al. Sequencing of 16S rRNA reveals a distinct salivary microbiome signature in Behçet’s disease. Clin Immunol. 2016;169:28–35.
- Ren W, Zhang Q, Liu X, et al. Supragingival plaque microbial community analysis of children with halitosis. J Microbiol Biotechnol. 2016;26:2141–2147.
- Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2016. pii: gutjnl-2016-312580. DOI:10.1136/gutjnl-2016-312580.
- Hu X-Y, Yao Y-F, Cui B-M, et al. [Analysis of causes and whole microbial structure in a case of rampant caries]. Nan Fang Yi Ke Da Xue Xue Bao. 2016;36:1328–1333. Chinese.
- Han CS, Martin MA, Dichosa AEK, et al. Salivary microbiomes of indigenous Tsimane mothers and infants are distinct despite frequent premastication. Peer J. 2016;4:e2660.
- Jiang S, Gao X, Jin L, et al. Salivary microbiome diversity in caries-free and caries-affected children. Int J Mol Sci. 2016;17:1978.
- Ruviére DB, Leonardo MR, Da Silva LAB, et al. Assessment of the microbiota in root canals of human primary teeth by checkerboard DNA-DNA hybridization. J Dent Child (Chic). 2007;74:118–123.
- Persson GR, Yeates J, Persson RE, et al. The impact of a low frequency chlorhexidine rinsing schedule on the subgingival microbiota (the TEETH clinical trial). J Periodontol. 2007;78:1751–1758.
- Sassone LM, Fidel R, Faveri M, et al. Microbiological evaluation of primary endodontic infections in teeth with and without sinus tract. Int Endod J. 2008;41:508–515.
- Adriaens LM, Alessandri R, Spörri S, et al. Does pregnancy have an impact on the subgingival microbiota? J Periodontol. 2009;80:72–81.
- Guiu A, Domingo D, Correa A, et al. [Leptotrichia goodfellowii wound infection after a dog bite]. Rev Esp Quimioter. 2012;25: 220–221. Spanish.
- Kapferer I, Beier US, Jank S, et al. Randomized controlled trial: lip piercing: the impact of material on microbiological findings. Pediatr Dent. 2013;35:e23–e28.
- Robles-Sikisaka R, Ly M, Boehm T, et al. Association between living environment and human oral viral ecology. ISME J. 2013;7:1710–1724.
- Murad CF, Sassone LM, Faveri M, et al. Microbial diversity in persistent root canal infections investigated by checkerboard DNA-DNA hybridization. J Endod. 2014;40:899–906.
- Smid MC, Dotters-Katz SK, Plongla R, et al. Leptotrichia buccalis: a novel cause of chorioamnionitis. Infect Dis Rep. 2015;7:5801.
- Renvert S, Widén C, Persson RG. Cytokine and microbial profiles in relation to the clinical outcome following treatment of peri-implantitis. Clin Oral Implants Res. 2016. DOI:10.1111/clr.12927
- Yang I, Woltemate S, Piazuelo MB, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep. 2016;6:18594.
- Ren W, Xun Z, Wang Z, et al. Tongue coating and the salivary microbial communities vary in children with halitosis. Sci Rep. 2016;6:24481.
- Takeshita T, Suzuki N, Nakano Y, et al. Relationship between oral malodor and the global composition of indigenous bacterial populations in saliva. Appl Environ Microbiol. 2010;76:2806–2814.
- Asikainen S, Doğan B, Turgut Z, et al. Specified species in gingival crevicular fluid predict bacterial diversity. PLoS One. 2010;5:e13589.
- Luo AH, Yang DQ, Xin BC, et al. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 2012;18:595–601.
- Bouvet P, Grégory A, Bellon L, et al. [Fetal Leptotrichia goodfellowii bacteremia]. Med Mal Infect. 2012;42:174–175. French.
- Morou-Bermudez E, Rodriguez S, Bello AS, et al. Urease and dental plaque microbial profiles in children. PLoS One. 2015;10:e0139315.
- Cho EH, Park KS, Yang M, et al. Laboratory identification of Leptotrichia species isolated from bacteremia patients at a single institution. Ann Lab Med. 2017;37:272–276.
- He Y, Gong D, Shi C, et al. Dysbiosis of oral buccal mucosa microbiota in patients with oral lichen planus. Oral Dis. 2017;23:674–682.
- Janus MM, Crielaard W, Volgenant CM, et al. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J Oral Microbiol. 2017;9:1270613.
- Ganesan SM, Joshi V, Fellows M, et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11:2075–2089.
- Mok SF, Karuthan C, Cheah YK, et al. The oral microbiome community variations associated with normal, potentially malignant disorders and malignant lesions of the oral cavity. Malays J Pathol. 2017;39:1–15.
- Chiu C-Y, Chan Y-L, Tsai Y-S, et al. Airway microbial diversity is inversely associated with mite-sensitized rhinitis and asthma in early childhood. Sci Rep. 2017;7:1820.
- Lopes Dos Santos Santiago G, Brusselle G, Dauwe K, et al. Influence of chronic azithromycin treatment on the composition of the oropharyngeal microbial community in patients with severe asthma. BMC Microbiol. 2017;17:109.
- Duan X-B, Wu T-X, Guo Y-C, et al. Marginal bone loss around non-submerged implants is associated with salivary microbiome during bone healing. Int J Oral Sci. 2017;9:95–103.
- Krieg NR, Staley JT, Brown DR, et al. Bergey’s Manual of Systematic Bacteriology. In: Staley JT, Whitman WB, editors. The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatiminadetes, Lentisphaeraae, Verrucomicrobia, Chlamydiae, and Planctomycetes. 2nd ed. Vol. 4. New York: Springer; 2005. p. 766–769.
- Tee W, Midolo P, Janssen PH, et al. Bacteremia due to Leptotrichia trevisanii sp. nov. Eur J Clin Microbiol Infect Dis. 2001;20:765–769.
- Shah HN, Olsen I, Bernard K, et al. Approaches to the study of the systematics of anaerobic, gram-negative, non-sporeforming rods: current status and perspectives. Anaerobe. 2009;15:179–194.
- Harwich MD Jr, Serrano MG, Fettweis JM, et al. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics. 2012;13:S4.
- Ivanova N, Gronow S, Lapidus A, et al. Complete genome sequence of Leptotrichia buccalis type strain (C-1013-b). Stand Genomic Sci. 2009;1:126–132.
- Gupta RS, Sethi M. Phylogeny and molecular signatures for the phylum Fusobacteria and its distinct subclades. Anaerobe. 2014;28:182–198.
- Sandle T. Bacteria in the blood could trigger dozens of diseases. In: Science. 2016. Available from: http://www.digitaljournal.com/tech-and-science/science/bacteria-in-the-blood-could-trigger-dozens-of-diseases/article/474337?
- Jang JY, Song IS, Baek KJ, et al. Immunologic characteristics of human gingival fibroblasts in response to oral bacteria. J Periodontal Res. 2016. DOI:10.1111/jre.12410
- Decroix V, Goudjil S, Kongolo G, et al. ‘Leptotrichia amnionii’, a newly reported cause of early onset neonatal meningitis. J Med Microbiol. 2013;62:785–788.
- Thompson J, Pikis A. Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol Oral Microbiol. 2012;27:34–44.
- Birkeland NK, Hofstad T. Oligosaccharides obtained by partial hydrolysis of lipopolysaccharides from Leptotrichia buccalis. Scand J Dent Res. 1985;93:432–435.
- Hofstad T, Jantzen E. Fatty acids of Leptotrichia buccalis: taxonomic implications. J Gen Microbiol. 1982;128:151–153.
- Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397.
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573.
