ABSTRACT
Objective: To characterize the salivary microbiota of patients with aggressive periodontitis, patients with chronica periodontitis and orally healthy individuals.
Methods: A total of 81 unstimulated saliva samples from aggressive periodontitis patients (n = 31), chronic periodontitis patients (n = 25), and orally healthy controls (n = 25) were examined. The V1-V3 region of the 16S rDNA gene was sequenced with Illumina® MiSeqTM, and sequences were annotated to the expanded Human Oral Microbiome Database (eHOMD).
Results: A mean percentage of 97.6 (range: 89.8–99.7) of sequences could be identified at species level. Seven bacterial species, including Porphyromonas gingivalis, were identified with significantly higher relative abundance in saliva from aggressive periodontitis patients than in saliva from orally healthy controls. Salivary abundance of P. gingivalis could discriminate aggressive (AUC: 0.80, p = 0.0001) and chronic periodontitis (AUC: 0.72, p = 0.006) from healthy controls. Likewise, salivary presence of P. gingivalis was significantly associated with aggressive (p < 0.0001, RR: 8.1 (95% CI 2.1–31.2)) and chronic periodontitis (p = 0.002, RR: 6.5 (95% CI: 1.6–25.9)).
Conclusion: Salivary presence and relative abundance of P. gingivalis associate with aggressive and chronic periodontitis, but do not discriminate between aggressive and chronic periodontitis.
Introduction
When entering the oral cavity saliva is sterile [Citation1]. However, when sampled from the oral cavity, one milliliter of saliva contains hundred millions of bacteria [Citation2]. Thus, the salivary microbiota is believed to be a compilation of bacteria shed from oral surfaces [Citation3,Citation4], which is why salivary microbiota may potentially be used as a biomarker of oral health status. In oral health, the salivary microbiota is closely related to microbiotas found on the tongue and the oral mucosa [Citation5]. However, salivary carriage of specific putative periodontal bacterial species, including Porphyromonas gingivalis, has previously been reported to associate with periodontitis [Citation6,Citation7].
Periodontitis is a prevalent, multifactorial inflammatory disease, induced by a biofilm on tooth surfaces, and the associated breakdown of tooth-supporting tissues is driven by a chronic inflammation within the periodontal tissues [Citation8]. Periodontitis is diagnosed clinically and may present in either an aggressive form with early onset and rapid progression, affecting only 5%, or a chronic form characterized by slower breakdown of tooth-supporting tissues and onset later in life, affecting approximately 40% of the adult population [Citation9,Citation10]. A 40-year follow-up study in male plantation workers from Sri Lanka, without access to dental care, reported different trajectories of periodontal breakdown [Citation11]. Consequently, the immune responses towards the periodontal microbiota are believed to distinguish aggressive from chronic forms of periodontitis [Citation8].
By means of various molecular techniques, cross-sectional analyses have reported that the salivary microbiota of patients with periodontitis differs from that of orally healthy controls [Citation12–Citation14]. Furthermore, several studies have demonstrated correlation of subgingival and salivary levels of specific bacteria proposed to be present in the periodontium of patients with periodontitis [Citation15–Citation18]. Finally, two longitudinal studies have shown that non-surgical periodontal treatment impacts salivary levels of proposed periodontal bacteria [Citation19,Citation20]. Thus, salivary carriage of bacteria associated with periodontitis seems to reflect the periodontal status. However, to the best of our knowledge, the characterization of the salivary microbiota in aggressive periodontitis patients remains to be performed.
The purpose of the present study was to characterize the salivary microbiota of patients with aggressive periodontitis and compare it with that of chronic periodontitis and of orally healthy controls. The hypothesis to be tested was that composition of the salivary microbiota in patients with aggressive periodontitis is distinct from those of patients with chronic periodontitis and orally healthy controls. Such salivary characteristics might hold potential for use as biomarkers for detection of periodontitis patients with rapidly progressing periodontitis.
Methods
Sample size
Sample sizes were calculated using one-way ANOVA with estimated levels of relevant biomarkers (controls µ1 = 10,000, and periodontitis patients µ2 = 15,000) with a predicted standard deviation of 6,250, α = 0.05 and a power of 0.80. Based on the sample size calculation each group should consist of a minimum of 25 participants. The sample size was adjusted to allow as much as 20% of the participants to withdraw their consent, leaving 30 participants in each group. Because of the novelty of the methods applied for sequencing, we accounted for unforeseen technical difficulties during analyses of the samples, by inclusion of an additional aggressive periodontitis patient, as these were the primary population of interest.
Study population
In the period from 8 February 2017 to 12 October 2018, a total of 90 individuals were recruited at the Department of Odontology, University of Copenhagen. All participants were medically healthy and assigned in accordance with the periodontal disease classification system from 1999 American Academy of Periodontology [Citation9]. Participants were divided into three groups: Aggressive periodontitis, chronic periodontitis and periodontally healthy controls on this basis. The inclusion criteria were:
Aggressive periodontitis patients:
19–40 years of age.
Interproximal attachment loss at minimum 3 teeth that were neither first molars nor incisors.
Clinical attachment loss at minimum 10 sites, also characterized by bleeding and appearance of pus upon probing.
Radiographic bone loss.
Chronic periodontitis patients:
50–60 years of age.
Interproximal attachment loss at minimum three teeth besides molars and incisors.
Clinical attachment loss at minimum 10 sites, also characterized by bleeding on probing.
Radiographic bone loss.
Orally healthy controls:
19–61 years of age.
No interproximal attachment loss.
No clinical attachment loss at sites with bleeding on probing.
No radiographic bone loss.
No sign of other inflammatory lesions in the oral mucosa.
General exclusion criteria included pregnant and breastfeeding women, systemic antibiotic treatment within 6 months, systemic diseases, and hematologic anomalies or syndromes. All participants signed an informed consent, and the study was approved by the regional ethical committee (The Capital Region of Denmark, protocol: H-1602473) and registered by the Danish Data Authorization (approval number: P-2019–18), and registered at clinicaltrials.gov: NCT03225950.
Clinical registrations
Background information of the study population including gender, age, ethnicity, and smoking status were recorded (). Clinical registrations were recorded at six sites per tooth (3rd molars excluded), including pocket depth (PD), clinical attachment loss (CAL), bleeding on probing (BOP), plaque index (PI), number of teeth excluding 3rd molars, and number of decayed, missing, filled teeth (DMFT). For details see .
Table 1. Study population characteristics. All parameters tested were checked for normality. Metadata of the study population, which followed a Gaussian distribution were compared using t-test, chi-square test and ANOVA, whereas non-parametric data were compared by Mann-Whitney U test, Friedman test and Kruskall-Wallis H test. All statistics were computed with GraphPad Prism (Graphpad Software, San Diego, CA).
Table 2. Clinical data of the study population. Pocket depth (PD), clinical attachment loss (CAL), bleeding on probing (BOP), plaque index (PI), number of teeth excluding 3rd molars, number of decayed, missing, filled teeth (DMFT). All parameters tested were checked for normality. Metadata of the clinical data of the study population, which followed a Gaussian distribution were compared using t-test, chi-square test and ANOVA, whereas non-parametric data were compared by Mann-Whitney U test, Friedman test and Kruskall-Wallis H test. All statistics were computed with GraphPad Prism (Graphpad Software, San Diego, CA).
Saliva sample collection
Two ml of unstimulated saliva samples were collected in Oragene DNA tubes (Cat.#OG-500, DNA Genotek Inc., Ontario, Canada) as previously described [Citation21] and stored immediately at −80°C until further analysis. Clinical registrations on healthy controls and sample collection were performed by the same examiner (AKD). All clinical registrations on patients with periodontitis were confirmed by a clinical instructor at the Section of Periodontology, Department of Odontology, University of Copenhagen.
DNA extraction
One hundred µl of a 25 mg/ml lysozyme (Cat.#90082, ThermoFisher, Roskilde, Denmark) were mixed with 400 µl of Oragene saliva sample, incubated for 2 h at 37°C, and the entire sample was subsequently purified using the Maxwell 16 Cell DNA Purification Kit (Cat.# AS1020, Promega, Wisconsin). Purified DNA was measured using the Qubit dsDNA high-sensitivity kit (Cat.#Q32854, ThermoFisher, Roskilde, Denmark) and diluted to 20 ng/ul.
Sequencing and data analysis
Bacterial 16S rRNA gene targeted amplicon sequencing was performed using a custom dual-index protocol [Citation22]. Custom 16S primers amplified the V1-V3 region of the 16S rRNA gene and were designed to provide the best coverage of the 16S rRNA gene while maintaining high sensitivity. The sample libraries were prepared using a 22 cycle PCR reaction to reduce chimera formation (unless otherwise noted). The final PCR products were purified using Ampure XP beads, pooled in equal amounts, and gel purified using the QIAGEN MinElute Gel Extraction Kit (Qiagen, Hilden, Germany). Purified, pooled libraries were quantified using the NEBNext Library Quant Kit for Illumina. Final libraries were sequenced on Illumina® MiSeq™ with a v2 reagent kit (500 cycles). The sequencing was performed at a 10 pM loading concentration with >20% PhiX spike-in. The DADA2 R package [Citation23] was used to identify and quantify amplicon sequencing reads on the FastQC files obtained after demultiplexing with the Illumina MiSeq software. Briefly, reads were trimmed and filtered to remove sequences with low quality. Quality of the trimmed and filtered reads was assessed using FastQC. Results of FastQC were compiled using MultiQC [Citation24]. The trimmed and filtered reads were then processed through the denoising, concatenating read1 and read2 with a 10 N spacer, and chimera removal steps of DADA2 to identify and quantify true amplicon sequence variants (ASV) present in the sample. Taxonomy of the identified ASVs was assigned using the RDP classifier algorithm [Citation25] implemented in the DADA2 package with a training dataset developed at The Forsyth Institute, Cambridge, MA and based on the Extended Human Oral Microbe Database (eHOMD).
Statistics
All parameters tested were checked for normality. Metadata of the study population, clinical data and sequence metadata, which followed a Gaussian distribution were compared using t-test, chi-square test and ANOVA, whereas non-parametric data were compared by Mann-Whitney U test, Friedman test and Kruskall-Wallis H test. The core salivary microbiota was defined as bacterial genera and species present with a mean relative abundance >1% across all samples. The salivary microbiota were characterized and compared by means of predominant genera and species, relative abundance, Shannon index, principal component analysis and correspondence analysis. Data on relative abundance were corrected for multiple dependent associations using Benjamini-Hochbergs correction [Citation26]. All statistics were computed with MeV [Citation27] and GraphPad Prism (GraphPad Software, San Diego, CA).
Results
Study population
One participant has been excluded after the recruitment, due to missing clinical registrations. A total of six samples failed DNA extraction, and two samples failed sequencing. Thus, at a total of 81 saliva samples from aggressive periodontitis patients (n = 31), chronic periodontitis patients (n = 25) and orally healthy controls (n = 25) were included in the analysis. Metadata of the study population (n = 81) are detailed in .
A significant different mean age across groups was observed (p < 0.05), whereas a comparable gender distribution was noted. A significantly higher number of current smokers was observed in the aggressive periodontitis group and the chronic periodontitis group, as compared to the orally healthy controls (p < 0.05). A higher proportion of subjects with both aggressive periodontitis and chronic periodontitis had African and Asian descent than observed in the healthy control group (p < 0.05).
Clinical data
Clinical data are presented in . A significant different mean BOP across groups was observed (p < 0.05), whereas comparable numbers of teeth were noted. As expected, mean CAL and mean PD were significantly higher for both aggressive and chronic periodontitis patients than for orally healthy controls (p < 0.05). Aggressive and chronic periodontitis patients had a significantly higher PI than orally healthy controls (p < 0.05) Aggressive periodontitis patients had a lower DMFT than both orally healthy and chronic periodontitis patients (p < 0.05).
Sequencing metadata
Sequencing generated >2.9 million sequences, which passed quality control (mean: 35,516, range: 10,810–72,698). Based on a BLAST with eHOMD a total of different 574 OTUs were identified, with a mean number of 170 (87–253) OTUs per sample. A total of 99.5% (93.9–100) and 97.6% (89.8–99.7) of all sequences could be referenced at genus and species level, respectively. No differences in mean number of sequences, and percentages of sequences identified at genus and species level were observed in the three groups investigated.
Core salivary microbiota in periodontitis and oral health
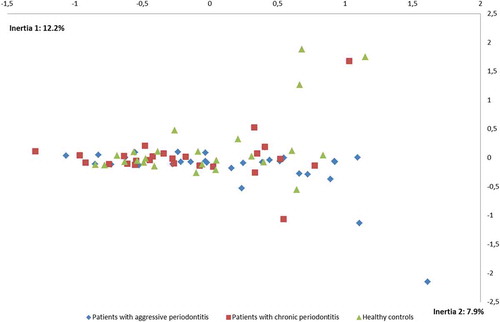
No differences were observed in the composition of the core salivary microbiota in aggressive periodontitis, chronic periodontitis and oral health. Relative abundance of predominant genera is visualized in , from which it can be seen that Prevotella, Neisseria, Streptococcus and Veillonella species constituted approximately 60% of the salivary microbiota in all groups. Furthermore, comparable α-diversities, as determined by the Shannon-index, were recorded in patients with aggressive periodontitis (3.4), chronic periodontitis (3.4) and oral health (3.5). Finally, data reduction using principal component analysis () and correspondence analysis () showed completely random distribution with no tendency of sample clustering based on periodontal disease status.
Species-specific associations with periodontitis and oral health
Seven bacterial species (six periodontitis-associated and one health-associated) were identified with a statistically significant difference in relative abundance in patients with aggressive periodontitis, patients with chronic periodontitis and orally healthy individuals (adjusted p-value <0.05, ). Furthermore, nine bacterial species (seven aggressive periodontitis-associated and two health-associated) were identified with significant differences in relative abundance in aggressive periodontitis patients and orally healthy individuals (adjusted p-value <0.05, ). No bacterial species were identified with a statistically significant relative abundance when comparing aggressive periodontitis patients with chronic periodontitis patients.
Table 3. Bacterial species with significantly different relative abundance in salivary samples from patients with periodontitis and oral health. The salivary microbiotas were characterized and compared by means of predominant genera and species, relative abundance, Shannon index, principal component analysis and correspondence analysis. Data on relative abundance were corrected for multiple dependent associations using Benjamini-Hochbergs correction [Citation26]. All statistics were computed with MeV [Citation27].
Table 4. Bacterial species with significant different relative abundance in salivary samples from patients with aggressive periodontitis. The salivary microbiotas were characterized and compared by means of predominant genera and species, relative abundance, Shannon index, principal component analysis and correspondence analysis. Data on relative abundance were corrected for multiple dependent associations using Benjamini-Hochbergs correction [Citation26]. All statistics were computed with MeV [Citation27].
Salivary abundance of P. gingivalis could discriminate aggressive (AUC: 0.80, p = 0.0001) and chronic periodontitis patients (AUC: 0.72, p = 0.006) from healthy controls. Likewise, salivary presence of P. gingivalis was significantly associated with aggressive periodontitis (p < 0.0001, RR: 8.1 (95% CI 2.1–31.2)) and chronic periodontitis (p = 0.002, RR: 6.5 (95% CI: 1.6–25.9)). No statistically significant differences in presence (p = 0.42, RR: 1.2 (95% CI: 0.8–2.0)) or relative abundance (AUC: 0.61, p = 0.15) of P. gingivalis were observed between patients with aggressive and chronic periodontitis.
Discussion
To the best of our knowledge, this is the first study to characterize the salivary microbiota in patients with aggressive periodontitis.
The most prominent finding was that salivary presence of P. gingivalis was significantly associated with aggressive periodontitis and chronic periodontitis, as compared to orally healthy controls (). Specifically, salivary abundance of P. gingivalis could discriminate aggressive (AUC: 0.80, p = 0.0001) and chronic periodontitis (AUC: 0.72, p = 0.006) from healthy controls. Likewise, salivary presence of P. gingivalis was significantly associated with aggressive (p < 0.0001, RR: 8.1 (95% CI 2.1–31.2)) and chronic periodontitis (p = 0.002, RR: 6.5 (95% CI: 1.6–25.9)). Subgingival colonization by P. gingivalis is considered a risk factor periodontitis [Citation28], which is why subgingival presence of P. gingivalis has been used as a screening biomarker of periodontitis [Citation6,Citation29]. Interestingly, several cross-sectional studies based on different molecular methods have reported a strong correlation in subgingival and salivary levels of P. gingivalis [15–18]. Thus, data from the present study supports the use of P. gingivalis in saliva as a screening biomarker of periodontitis. However, further studies are needed to determine if salivary presence of P. gingivalis precedes clinical signs of periodontal tissue breakdown.
The main clinical difference distinguishing aggressive from chronic periodontitis is the early onset and rapid progression of aggressive periodontitis. Furthermore, loss of attachment is often observed in aggressive periodontitis despite sufficient plaque control [Citation30].
In the present study, patients with aggressive periodontitis had a notably high PI. All participants were included prior to periodontal treatment; thus, the rapidity of disease progression was estimated by relating the attachment loss to the age of the participant. The patients with aggressive periodontitis were all aged under 40 years and presented with absence of medical diseases, but with severe attachment loss (mean 3.5 mm) at the time of inclusion.
Subgingival colonization of virulent periodontal bacteria, such as P. gingivalis may drive the destructive immune responses towards the periodontal microbiota. In line herewith, abundance of P. gingivalis have been suggested as a possible explanation to the different clinical phenotypes observed in aggressive and chronic periodontitis [Citation31,Citation32]. The anaerobic environment found in inflamed periodontal pockets offers nutritional requirements, which favors growth of especially P. gingivalis. Interestingly, we found no differences between the salivary microbiota of patients with aggressive periodontitis patients and that of patients with chronic periodontitis (–). Accordingly, this may be explained by the similar clinical characteristics observed in the aggressive periodontitis and chronic periodontitis group in the present study (). However, virulence mechanisms may vary within a species, and P. gingivalis can be divided according to fimA genotype. While P. gingivalis fimA type II is the most prevalent in periodontitis and in particular in patients with aggressive periodontitis [Citation33,Citation34], P. gingivalis fimA type I is exclusively found in periodontally healthy subjects [Citation35]. HOMINGS cannot discriminate P. gingivalis according to fimA genotypes, which may explain the present comparable findings of P. gingivalis in aggressive and chronic periodontitis. Nevertheless, the finding of comparable salivary microbiotas in patients with aggressive and chronic periodontitis may also support the assumption of the immune response towards the periodontal microbiota as the decisive factor behind the clinical presentations of periodontitis [Citation8].
Another notable finding was that salivary presence and relative abundance of Prevotella aurantiaca was associated with aggressive periodontitis, see . P. aurantiaca, which is a member of the Prevotella intermedia-group [Citation36], was isolated from the oral cavity in 2010 [Citation37]. To the best of our knowledge, this is the first study that associates P. aurantiaca with aggressive periodontitis, which is, why further studies are needed to evaluate this particular finding.
In the present study comparable core salivary microbiotas were observed in patients with periodontitis and orally healthy controls (–). Accordingly, the salivary microbiota is believed to be a conglomerate of bacteria shed from oral surfaces, with the tongue, throat and tonsils as the primary donor sites [Citation5]. Therefore, salivary presence and abundance of P. gingivalis recorded in the present study could potentially originate from either the tongue or the subgingival pocket. Previous studies have reported that periodontal bacteria associated with periodontitis may be found on the tongue of orally healthy individuals [Citation38]. It is therefore interesting that P. gingivalis was observed in saliva from 64% and 52% of patients with aggressive and chronic periodontitis, respectively, as compared to only 8% of orally healthy controls (). Therefore, P. gingivalis identified in saliva from patients with aggressive and chronic periodontitis in the present study were probably spill-over from concomitant periodontal lesions in these patients rather than remnants from the tongue.
Some limitations apply to the present study, including that six samples failed DNA extraction and two samples failed sequencing, which is why only data from 81 participants were included in downstream analyses. Furthermore, saliva samples were collected throughout the day, which means that the data presented could potentially be influenced by diurnal variation. However, previous studies have reported a minimal impact of collection method and sampling time on the composition of the salivary microbiota [Citation21,Citation39]. Finally, as the new classification of periodontal diseases from 2018 [Citation40], were introduced during the study period it was not possible to use this classification in the present study. However, it appears that the aggressive periodontitis patients included comprising periodontitis, grade C, generalized, leaving only the staging undescribed. The main strength of the present study was the capability of the new HOMINGS protocol, which targets the V1-V3 region of the 16S rRNA gene to identify 97.6% of all sequences at species level. To the best of our knowledge, this is the first study to use next-generation sequencing to achieve >97% coverage at species level.
In conclusion, salivary presence and relative abundance of P. gingivalis associate with aggressive and chronic periodontitis compared to oral health, but do not discriminate between aggressive and chronic periodontitis. Based on the findings of the present study, P. gingivalis is relevant as a proxy for deep periodontal pockets and thus periodontitis in clinical trials. However, future investigations would benefit from data discriminating P. gingivalis according to fimA genotypes, to allow differentiation according to the virulence of the strains.
Figure 1. Predominant bacterial genera. Mean relative abundance of predominant bacterial genera in patients with aggressive periodontitis, chronic periodontitis and orally healthy controls. The core salivary microbiota was defined as species/genus with a mean relative abundance >1% across sample. The predominant species/genera were compared using Mann-Whitney U test and Kruskall-Wallis test with Benjamini-Hochberg correction.
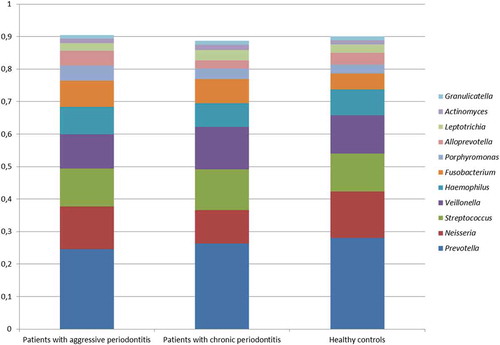
Figure 2. Principal component analysis. Principal component analysis visualized two-dimensionally with axes expressed as two principal components values accounting for a cumulative value of (38.0%). Sample denotation: aggressive periodontitis patients: blue, chronic periodontitis patients: red, healthy controls: green.
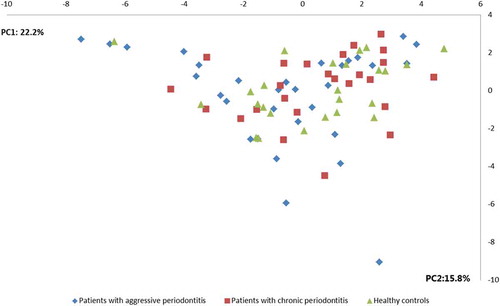
Figure 3. Correspondence analysis. Correspondence analysis visualized two-dimensionally with axes expressed as the two foremost inertia values accounting for a cumulative inertia of (20.1%). Sample denotation: aggressive periodontitis patients: blue, chronic periodontitis patients: red, healthy controls: green.
Data availability
Access to all data, including raw sequence data, will be granted upon request ([email protected]).
Acknowledgments
Sequencing was performed at the Forsyth Institute, Cambridge, MA by the Sequencing and Bioinformatics Core.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Schroder SA, Bardow A, Eickhardt-Dalboge S, et al. Is parotid saliva sterile on entry to the oral cavity? Acta Otolaryngol. 2017;137(7):762–9.
- Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10(4):302–306.
- Kilian M, Chapple IL, Hannig M, et al. The oral microbiome – an update for oral healthcare professionals. Br Dent J. 2016;221(10):657–666.
- Krishnan K, Chen T, Paster BJ. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017;23(3):276–286.
- Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42.
- Liljestrand JM, Gursoy UK, Hyvarinen K, et al. Combining salivary pathogen and serum antibody levels improves their diagnostic ability in detection of periodontitis. J Periodontol. 2014;85(1):123–131.
- Paju S, Pussinen PJ, Suominen-Taipale L, et al. Detection of multiple pathogenic species in saliva is associated with periodontal infection in adults. J Clin Microbiol. 2009;47(1):235–238.
- Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 2013;62(1):203–217.
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4(1):1–6.
- Eke PI, Dye BA, Wei L, et al. Prevalence of periodontitis in adults in the USA: 2009 and 2010. J Dent Res. 2012;91(10):914–920.
- Ramseier CA, Anerud A, Dulac M, et al. Natural history of periodontitis: disease progression and tooth loss over 40 years. J Clin Periodontol. 2017;44(12):1182–1191.
- Belstrom D, Constancias F, Liu Y, et al. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes. 2017;3:23.
- Belstrom D, Jersie-Christensen RR, Lyon D, et al. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. PeerJ. 2016;4:e2433.
- Belstrom D, Paster BJ, Fiehn NE, et al. Salivary bacterial fingerprints of established oral disease revealed by the Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) technique. J Oral Microbiol. 2016;8:30170.
- Boutaga K, Savelkoul PH, Winkel EG, et al. Comparison of subgingival bacterial sampling with oral lavage for detection and quantification of periodontal pathogens by real-time polymerase chain reaction. J Periodontol. 2007;78(1):79–86.
- He J, Huang W, Pan Z, et al. Quantitative analysis of microbiota in saliva, supragingival, and subgingival plaque of Chinese adults with chronic periodontitis. Clin Oral Investig. 2012;16(6):1579–1588.
- Nickles K, Scharf S, Rollke L, et al. Comparison of two different sampling methods for subgingival plaque: subgingival paper points or mouthrinse sample? J Periodontol. 2017;88(4):399–406.
- Haririan H, Andrukhov O, Bertl K, et al. Microbial analysis of subgingival plaque samples compared to that of whole saliva in patients with periodontitis. J Periodontol. 2014;85(6):819–828.
- Kageyama S, Takeshita T, Asakawa M, et al. Relative abundance of total subgingival plaque-specific bacteria in salivary microbiota reflects the overall periodontal condition in patients with periodontitis. PLoS One. 2017;12(4):e0174782.
- Belstrom D, Grande MA, Sembler-Moller ML, et al. Influence of periodontal treatment on subgingival and salivary microbiotas. J Periodontol. 2018;89(5):531–539.
- Belstrom D, Holmstrup P, Bardow A, et al. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J Oral Microbiol. 2016;8:30112.
- Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120.
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583.
- Ewels P, Magnusson M, Lundin S, et al. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048.
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267.
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–818.
- Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193.
- Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–144.
- Kinney JS, Morelli T, Braun T, et al. Saliva/pathogen biomarker signatures and periodontal disease progression. J Dent Res. 2011;90(6):752–758.
- Albandar JM. Aggressive and acute periodontal diseases. Periodontol 2000. 2014;65(1):7–12.
- Kononen E, Muller HP. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65(1):46–78.
- Kulkarni C, Kinane DF. Host response in aggressive periodontitis. Periodontol 2000. 2014;65(1):79–91.
- Amano A, Kuboniwa M, Nakagawa I, et al. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79(9):1664–1668.
- Zhao L, Wu YF, Meng S, et al. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodontal Res. 2007;42(6):511–517.
- Miura M, Hamachi T, Fujise O, et al. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40(2):147–152.
- Fteita D, Könönen E, Gürsoy M, et al. Quorum sensing molecules regulate epithelial cytokine response and biofilm-related virulence of three Prevotella species. Anaerobe. 2018;54:128–135.
- Sakamoto M, Suzuki N, Okamoto M. Prevotella aurantiaca sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2010;60:500–503.
- Tanner AC, Paster BJ, Lu SC, et al. Subgingival and tongue microbiota during early periodontitis. J Dent Res. 2006;85(4):318–323.
- Belstrom D, Holmstrup P, Bardow A, et al. Temporal stability of the salivary microbiota in oral health. PLoS One. 2016;11(1):e0147472.
- Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Clin Periodontol. 2018;45(Suppl 20):S162–S170.
