ABSTRACT
Introduction: Periodontitis is an inflammatory dysbiotic disease. Among putative dysbiosis causes, transmission of Porphyromonas gingivalis between individuals of the same family remains unclear. The aim of this systematic review and meta-analysis is to assess the likelihood of shared detection of Porphyromonas gingivalis among cohabiting family members.
Methods: A literature search was conducted on different databases up to September 2018. Articles assessing the presence of P.gingivalis between members of the same family were screened. Only English literature was retrieved, whereas no limits were applied for bacterial sampling and detection methods.
Results: Overall, 26 articles published between 1993 and 2017 met the inclusion criteria. Of these, 18 articles were used for meta-analyses. Based on bacterial culture, the likelihood of an intra-familial transmission of P.gingivalis once a member of the family harbors the bacterium is estimated at 63.5% (n = 132 pairs of family members); this drops to 45% when pooling together culture and Polymerase-Chain-Reaction (n = 481 pairs), whereas it is estimated at 35.7% when genotyping is applied (n = 137 pairs).
Conclusion: Pooled results suggest that the likelihood of detecting P.gingivalis within within family members is moderately frequent. Personalized periodontal screening and prevention may consider intra-familial co-occurrence of P.gingivalis as feasible.
Introduction
Periodontitis is a multifactorial inflammatory disease associated with dysbiotic subgingival microbiota and characterized by progressive destruction of the tooth-supporting apparatus [Citation1]. The transition from periodontal health to periodontal disease is associated with the shift from a symbiotic microbial community, mostly composed of facultative anaerobic bacterial genera like Actinomyces and Streptococci, to a dysbiotic microbial community composed of anaerobic genera from the phyla Firmicutes, Proteobacteria, Spirochaetes, Bacteroidetes and Synergistetes [Citation2,Citation3]. This shift is likely induced by pathogenic bacteria able to trigger quantitative and qualitative alterations in the commensal communities and consequently initiate the destructive inflammatory processes at the level of the periodontium. The most studied ‘keystone pathogen’ for periodontitis is Porphyromonas gingivalis (P. gingivalis), a highly virulent Gram-negative asaccharolytic bacterium [Citation4,Citation5].
Thus, colonization of the mouth by putative pathogens may lead to periodontal disease in susceptible recipients. The infectious etiology of periodontitis is consistent with the familial clustering of the disease, although the classical meaning of infection transmission cannot be applied in periodontitis given the complexity of the multifactorial aspect of oral microbiome and host interactions [Citation6,Citation7]. However, the possibility of an inter-individual transfer of oral microbiota remains relevant and investigating how and how often the main periodontal pathogens can be transmitted may provide further information to assess the patient’s risk profile.
The present systematic review and meta-analysis aims to answer the following question: what is the likelihood of shared detection of P. gingivalis among cohabiting family members?
Materials and methods
Study design
This is a systematic review of studies focusing on the intra-familial co-occurring detection of P. gingivalis between spouses, parents-infants, and among siblings. The PRISMA statement checklist was followed in the reporting of this systematic review.
Eligibility criteria for study inclusion
Clinical trial, longitudinal studies (retrospective or prospective), cross-sectional studies, case–control studies, case-series (reporting about at least four families cases) were eligible for inclusion. Review articles, case report, and studies treating of a possible intrauterine transmission of periodontal pathogens or transmission between humans and animals were not considered. Studies focusing on periodontal bacteria other than Porphyromonas gingivalis were also excluded.
The eligibility criteria by applying the PICO framework were the following:
Population: Family members living together with at least one member carrying P. gingivalis (so-called proband), with or without periodontitis.
Intervention: All microbiological detection methods were considered (e.g. culture, polymerase chain reaction (PCR), serotyping, genotyping, ribotyping).
Comparator: Not applicable
Outcomes: Likelihood of intra-familial detection of P. gingivalis.
In the majority of the studies, the co-occurring detection of P. gingivalis between two family members (concordance rate) was used to support the hypothesis of a direct intra-familial transfer, namely horizontal transmission when occurring between spouses and vertical transmission when occurring between parent and infant. In the present study, we avoid to talk about bacterial transmission, we rather estimate the likelihood of a simultaneous detection of the bacterium in the proband (carrier of Porphyromonas gingivalis) and his/her family relative.
Information sources
The literature search for the present systematic review was conducted on the following online available databases: MEDLINE (through PubMed), EMBASE, Cochrane Oral Health Group Specialized Register, ProQuest Dissertations and Thesis Database. A grey literature search was also performed by searching the OpenGrey database. Studies meeting the selection criteria were reviewed if written in English. The study protocol was begun in November 2017; literature search was performed on September 2018. The systematic review protocol was registered in Prospero on 17 April 2018 (registration number: ID = CRD42018092737).
Search strategy
A specific research equation was formulated for each different database, using the following keywords and/or MeSH terms: transmission, aggregation, Porphyromonas gingivalis, family/families, spouse, periodontitis, oral bacterial colonization, oral bacteria, microbiome. In addition, the reference lists of eligible studies and relevant review articles (not included in the systematic review) were crosschecked to identify other relevant studies.
Study selection and quality assessment
Studies were selected by two independent reviewers (M.B. and M.C.C.). At first, the titles and abstracts of the retrieved studies were independently and blindly screened for relevance. To enhance sensitivity, records were removed only if both reviewers excluded them at the title/abstract level. Subsequently, both reviewers performed a full-text analysis of the selected articles. Disagreements about inclusion or exclusion of a study were resolved by consensus. The two reviewers independently assessed the risk of bias, using appropriate tools according to the study design. Most of the study were observational studies, thus the quality assessment was carried out by using the star template of the Newcastle-Ottawa Scale (NOS) tool. The NOS scores of 1 to 3, 4 to 6, and 7 to 9 were judged for low, moderate, and high quality of studies, respectively.
Data extraction and analysis
Data from the selected studies were processed for quality synthesis. Relevant findings and outcomes were extracted from the original studies and summarized in tables. Extracted data included first author, year of publication, patient numbers, study design, periodontal status of the probands, bacteria sampling, detection methods, transmission rate. Whenever the transmission rate was not provided or not estimable from the article data, corresponding authors were contacted by email to obtain data on the proportion of family members sharing P. gingivalis. Consequently, whenever possible, studies were grouped by detection method and type of familial relationship, and a proportion meta-analysis was run. The meta-analysis and forest plots were derived by using MedCalc software (version 17.9 for Windows). The pooled proportion of likelihood of co-occurrence of P. gingivalis was analyzed with the estimation of 95% confidence interval (CI). Random effects model (DerSimonian and Laird method) [Citation8] was used. Heterogeneity of the studies was tested by Cochran’s Q statistic, and I2. Sensitivity analysis was also performed by sequentially excluding studies that may be responsible for heterogeneity. Funnel plots were used to examining the presence of publication bias.
Results
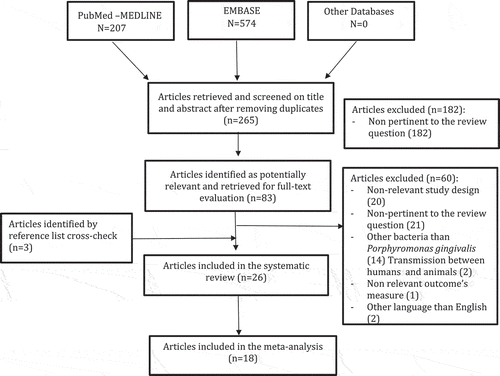
Article search and selection
Overall, 265 articles were initially identified; of these, 182 were rejected upon titles and abstract because not relevant for the review topic. The remaining 83 articles were screened at the full-text level; 26 were selected for the systematic review. Of these, 18 articles were used for pooled data analyses. Overall, the selected articles were published between 1993 and 2017. The flow chart of the study selection process is shown in .
Study characteristics
All the selected studies were observational studies, including 7 cross-sectional, 10 case series, 5 case-control, and 4 cohort studies. Co-occurrence of P. gingivalis between spouses was explored in 18 studies, including 334 couples; co-occurrence between siblings was explored in 3 studies. Detection concordance was explored between parent and infant(s) in 14 studies, including 625 pairs of parents-children. Details of study characteristics according to the type of familial relationship are displayed in –.
Table 1. Summary of the included studies assessing the simultaneous detection of Porphyromonas gingivalis (Pg) between adult couples of the same family
Table 2. Summary of the included studies assessing the simultaneous detection of Porphyromonas gingivalis (Pg) between parents and children of the same family
Table 3. Summary of the included studies assessing the simultaneous detection of Porphyromonas gingivalis (Pg) between siblings
Regarding the clinical periodontal status of the proband, the classical periodontal parameters, i.e. periodontal pocket depth, clinical attachment level, gingival inflammation, and bleeding were recorded in some studies. In few of them, radiographic parameters were also considered. Classification of periodontitis according to the AAP Classification 1999 defining chronic or aggressive periodontitis was used in three studies [Citation9–Citation12]; adult periodontitis was described in four studies, whereas advanced periodontitis in three. Several studies did not provide a clear definition of the periodontal disease used.
Bacterial sampling was carried out on supra-gingival and/or subgingival plaque, stimulated saliva, or sampled from dorsum of the tongue, buccal mucosa, or tonsillar area. Detection methods included culture in 10 studies (38.4%), and Polymerase Chain Reaction (PCR) in 11 studies (42.3%). DNA restriction enzyme analysis (REA-DNA) was used in 4 studies (15.3%), pulsed field gel electrophoresis (PFGE) in 1 study, arbitrarily primed polymerase chain reaction (AP-PCR) in 2 (7.6%) studies, amplified fragment length polymorphism (AFLP-PCR) in 3, strain-specific identification of P. gingivalis I Isi 1126 PCR in 1, and Fim A genotyping in 3 studies. Serotyping characterization and ribotyping were reported in 2 studies, respectively.
Quantitative analyses
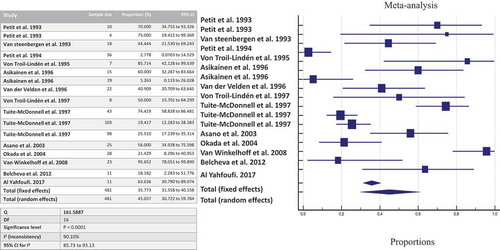
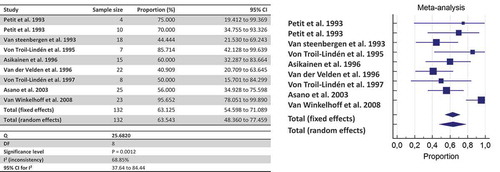
Forest plots were built pooling together intra-familial data according to the detection method used. Concerning the co-occurring detection between adult partners or spouses living together, the meta-analysis showed that the likelihood of detecting P. gingivalis in the partner/spouse when a proband was harboring P. gingivalis was 53.58% (95%CI: 44.98%-62.95%; I2: 0%; n = 99 pairs of partner/spouse) by culture, and 58.88% (95%CI: 48.02%-69.32%; I2: 39.6%; n = 142 pairs) when considering culture + PCR.
By applying genotyping, the likelihood of detecting the same strain of P. gingivalis between the members of the couple dropped at 29.02% (95%CI: 17.68%-41.87%; I2: 43.2%; n = 91 pairs). Using Fim A genotyping method, the same Fim A was retrieved in 57.15% (95%CI: 40.87%-72.68%; I2: 0.2%; n = 33 pairs) of the cases. Overall, the same ribotype of P. gingivalis was found in 64.7% (95%IC: 38.83%-86.63%; I2: 0%; n = 12 pairs) of couples when both adults were positive for P. gingivalis.
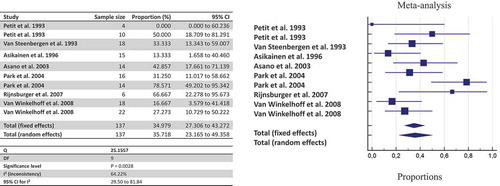
Concerning the parent-infant co-occurrence, when at least one parent was colonized by P. gingivalis, the likelihood that also one child harbored P. gingivalis was estimated at 20.15% when considering three studies using culture (95%CI: 0.31%-58.89%; I2: 90.4%, n = 65 pairs of parent-infant) [Citation13–Citation15], and 5% when considering two studies (95%CI: 0.91%-12.11%; I2: 0%; sensitivity analysis on 55 pairs) [Citation13,Citation15]. The detection of P. gingivalis using PCR amplification showed a likelihood of 22.46% (95%CI: 17.46%-27.90%; I2: 0%, n = 240 pairs), whereas when combining both detection methods (culture + PCR), the estimated likelihood was 24.05% (95%CI: 13.41%-36.64%; I2: 80.5%, n = 316 pairs) [Citation13–Citation19]. Based on two studies only, the same P. gingivalis genotype of the parents was shared in 64.95% of children harboring P. gingivalis (95%CI: 37.73–87.74%; I2: 50.5%; n = 24 pairs) [Citation12,Citation14].
By pooling together all data about shared detection both between-spouses and parent-infant, the likelihood of detecting P. gingivalis once a family member harbors the bacterium is estimated at 63.54% by culture (), 45.03% by culture + PCR (), and 35.71% by genotyping ().
Figure 2. The likelihood of co-occurrence of P. gingivalis once a family member harbors P. gingivalis assessed by culture
Study quality assessment
Two reviewers (M.B. and M.C.C.) scored the methodological qualities of the included studies. The NOS score varies from 3 to 5, and over 9; thus, the studies were qualified as low (n = 17) and moderate (n = 9) quality studies. Detailed information regarding the quality assessment of the included studies is reported in Supplement Table 1.
Discussion
The present systematic review and meta-analysis focuses on the likelihood of detecting one of the major periodontal pathogens, namely P. gingivalis, among cohabiting family members, and demonstrates that the co-occurrence of P. gingivalis among couples, children, or siblings ranges between 42% and 62% when a family member of is carrying P. gingivalis (the so-called proband). However, only in 35% of cases, these members are sharing the same genotype of the bacterium.
These data quantify the likelihood of co-occurring detection and support the hypothesis that an intra-familial bacterial transfer may occur. However, this appears to be highly variable both between spouses or couples and between parents and children despite their intimate cohabitation for long periods of time. The observed variability could be explained in several ways. First of all, bacterial transfer and detection does not necessarily mean persistent colonization up to the detectable levels of the pathogens but could be caused by repeated inoculation [Citation6]. Indeed, bacterial transmission results from a combination of a sufficiently large and concentrated inoculum enabling bacterial survival during colonization, with a favorable oral environment of the recipient, which is dependent on the resident microbiota and the host defenses and characteristics [Citation6,Citation20]. Moreover, several behavioral and environmental patterns may influence the likelihood of bacterial colonization, such as hygiene habits, proximity, and intimacy differences among family members. However, as observed for other body sites, like skin or fecal microbiota, the family unit have a strong effect on human microbial community composition: indeed, family membership may explain a large proportion of the variability in bacterial diversity, with family members tending to harbor similar microbiota [Citation7,Citation21,Citation22]. Indeed, the chance of sharing the same bacterial genotype appears to be greater among related individuals than unrelated ones or if P. gingivalis was randomly distributed in the population [Citation13,Citation19,Citation23].
Sampling and detection of porphyromonas gingivalis
Although the possible ways for transferring of P. gingivalis remains unclear, the role of saliva as a vehicle of bacterial spread is probable and it is supported by the fact that P. gingivalis can be cultured from salivary samples, indicating that this bacterium survives in the saliva during transportation to a new host [Citation24]. Indeed, the probability of inoculation appears to be directly related to the salivary bacterial load, with a greater risk of colonization in the recipient for greater bacterial loads [Citation20,Citation24]. Then, P. gingivalis is able to spread intra-orally and colonize supra-gingival and sub-gingival plaque at sites with and without periodontal attachment loss, although, it is more likely to find P. gingivalis in deep pockets rather than shallow ones [Citation25]. Consequently, the eventual eradication (i.e., a bacterial load under the detection level) of this pathogen by efficacious periodontal treatments may prevent its spread among individuals [Citation6,Citation24].
It must be noted that the detection of periodontal pathogens is drastically influenced by the methods applied to sample and recover them (e.g., culture, PCR, DNA probe checkerboard) [Citation26]. As deemed from and , the sampling and detection methods applied in the studies included in the present systematic review are highly heterogeneous. This implies to analyze data by sub-groups of comparable detection methods while avoiding global comparisons. However, no distinction could be made according to the bacterial sampling site (e.g. saliva, subgingival plaque) because almost all studies provided pooled results of all samples examined (although multiple sites were sampled in most cases). Moreover, we must highlight that the included studies were conducted in a time span of 24 years during which microbiological techniques have drastically evolved as well as our knowledge about the role of specific periodontal pathogens and the complexity of the oral microbiota [Citation27].
Several studies used culture to detect and quantify P. gingivalis, but no distinction between clones was made. The sensitivity of bacterial culturing is rather low, with detection limits averaging at 103–104 bacterial cells [Citation26,Citation28]. On the contrary, methods based on immune diagnosis (serotyping) and molecular analysis, such as PCR and ribotyping are highly sensitive and specific [Citation26]. No study to date investigated the concordance of bacterial colonization and the likelihood of sharing the same periodontal microbiota between family members by applying modern methods, such as high throughput sequencing methods.
Clinical implications of familial porphyromonas gingivalis sharing
If an intra-familial transmission of P. gingivalis is possible and probable, the available studies present a design and an overall level of quality that do not allow to conclude about ta specific intra-familial transmission pattern. Nevertheless, they provide evidence about the likelihood of a shared detection of P. gingivalis among family members.
A key question is whether the sharing of P. gingivalis can affect the periodontal health of the individuals becoming colonized. Some studies found that spouses of patients with advanced periodontitis have a worse periodontal status than spouses of periodontal healthy individuals [Citation29]. Others did not demonstrate that the periodontal condition of the spouse was influenced by that of the partner [Citation15,Citation19]. When the relationship between the duration of marriage and the chance of transmission was explored, the study of Tuite-Mcdonnel et al. found no relationship between the frequency of co-occurring detection and the length of marriage, suggesting that cross-colonization likely occurred in the early years of marriage and remains stable over time [Citation19].
When the frequency of presence of P. gingivalis in spouses of colonized proband was compared to unrelated patients, the spouses were significantly more frequently colonized by P. gingivalis than what would be expected if P. gingivalis was randomly distributed in the population [Citation19,Citation23]; but this was not observed in all studies [Citation13].
Indeed, transfer of P. gingivalis from an individual to another does not necessarily translate into stable colonization and periodontal breakdown. It may persist an equilibrium between the host and the resident microbiota despite the repeated inoculation of P. gingivalis, especially in periodontally healthy individuals. However, we may consider P. gingivalis colonization as a potential risk factor, as it is known that this bacterium is causal in periodontitis initiation, progression, recurrence, as well as in peri-implantitis [Citation5,Citation30].
P. gingivalis can be possibly shared between parents and children. This raises another important question: does an early inoculation increase the chances of a permanent colonization and development of periodontitis later on in children? Early studies using culture probably underestimated the prevalence of P. gingivalis in young subjects, seldom detecting it before puberty [Citation31,Citation32]. Later on, studies relying upon more sensitive techniques, such as DNA-based technologies, demonstrated the presence of P. gingivalis in a large fraction of young subjects and showed it to be equally common in children of all ages [Citation19]. Indeed, P. gingivalis can be detected in children aged of 20 days as of 18 years [Citation33–Citation35]. An epidemiological study, using DNA probe checkerboard assay, found that 71% of the 18- to 48-month-old children were infected with at least one periodontal pathogen [Citation36]. P. gingivalis detection rates was estimated at 68.8% in children. A study evaluating the influence of mother’s periodontal clinical status on the prevalence of periodontal pathogens in newborns (aged of 3 months) showed that P. gingivalis was the most prevalent pathogen followed by others periodontal bacteria (Prevotella intermedia, Tannerella forshythia, Campylobacter rectus, Aggregatibacter actinomycetemcomitans) [Citation37]. They concluded that the maternal clinical periodontal status is a significant indicator of the oral microbiota composition in the newborn children.
If the colonization in young children occurs, it appears to be composed by the same P. gingivalis strain of the parent(s) only transitory, whereas it becomes more stable during teenage years, possibly as deeper pockets develop [Citation33]. Indeed, the presence of P. gingivalis is favored by the presence of deep probing depths, and thus it is likely that transmission from a proband infected individual to a periodontally healthy one is possible in terms of transient presence in the oral cavities, but in the absence of a permanent niche, such as a deep pocket, it may no longer survive.
It is noteworthy that the exposure and chance of colonization of P. gingivalis may vary in relation to the proband status, the severity of the periodontal disease, and the administration of a successful periodontal treatment. Moreover, bacterium-specific virulence factors may facilitate the familial sharing of P. gingivalis. Particularly, the fimbriae A (FimA), a specific component of the cell surface [Citation38], seems to play a strategic role in the colonization and invasion of the periodontal tissues [Citation39,Citation40]. The FimA gene has been classified into six types (I to V and Ib) [Citation41]. A higher rate of type II FimA was detected in couples who shared the same strains of P. gingivalis [Citation9,Citation23,Citation42], being this strain mainly associated with severe periodontitis [Citation43]. Conversely, the type IV FimA was not found in couples with identical PFGE patterns [Citation23].
The present systematic review has some limitations. Retrieved data are heterogeneous: different methods for bacterial sampling, detection, and quantification were applied, requiring subgroup analyses. Findings are based upon a low-to-moderate level of evidence coming mainly from retrospective, cross-sectional studies or case series with a small sample size, which hamper any conclusion on the specific colonization patterns or on the eventual deterioration of periodontal health after the occurrence of P. gingivalis sharing within the family. Moreover, it should be stressed that a shared detection of pathogens (or colonization concordance) in cohabiting family members does not necessarily prove the transmission of these pathogens. Finally, the majority of the available studies were published in the ‘90. Due to relevant technical progress in microbiology in the last decade, the presented findings should be replicated and updated in lights of more recent knowledge about the diversity and richness of the oral microbiota [Citation44].
Conclusion
The present systematic review and meta-analysis supports the co-occurring detection of P. gingivalis within family members. Due to the role of P. gingivalis in the etiology of periodontitis, the likelihood of sharing P. gingivalis should be considered in the assessment of patient’s periodontal risk.
Author contributions
M.B.: study conception, literature search, data extraction, data analysis, manuscript drafting, and manuscript final version approval. H.R.: study conception, critical appraisal, manuscript revisions, and manuscript final version approval. V.M.: critical appraisal, manuscript revisions, and manuscript final version approval. F.M.: critical appraisal, manuscript revisions, manuscript final version approval. P.B: study conception, critical appraisal, manuscript revisions, and manuscript final version approval. M.C.C.: Reviewer and supervisor, study conception, data analysis, manuscript revisions, and manuscript final version approval.
Supplemental Material
Download MS Word (18.2 KB)Acknowledgments
The authors would like to thank the entire team of the European Postgraduate Program in Periodontology and Implant Dentistry of the University of Paris for their support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 World workshop on the classification of periodontal and peri-implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):S173–S182.
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–13.
- Palmer RJ. Composition and development of oral bacterial communities. Periodontol 2000. 2014;64:20–39.
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44.
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725.
- Van Winkelhoff AJ, Boutaga K. Transmission of periodontal bacteria and models of infection. J Clin Periodontol. 2005;32(Suppl 6):16–27.
- Kilian M, Chapple IL, Hannig M, et al. The oral microbiome - an update for oral healthcare professionals. Br Dent J. 2016;221:657–666.
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188.
- Feng X, Zhu L, Xu L, et al. Distribution of 8 periodontal microorganisms in family members of Chinese patients with aggressive periodontitis. Arch Oral Biol. 2015;60:400–407.
- Monteiro MDF, Casati MZ, Taiete T, et al. Periodontal clinical and microbiological characteristics in healthy versus generalized aggressive periodontitis families. J Clin Periodontol. 2015;42:914–921.
- Monteiro MF, Casati MZ, Taiete T, et al. Salivary carriage of periodontal pathogens in generalized aggressive periodontitis families. Int J Paediatr Dent. 2014;24:113–121.
- Park O-J, Min K-M, Choe S-J, et al. Use of insertion sequence element IS1126 in a genotyping and transmission study of Porphyromonas gingivalis. J Clin Microbiol. 2004;42:535–541.
- Asikainen S, Chen C, Slots J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol Immunol. 1996;11:387–394.
- Petit MD, van Steenbergen TJ, Scholte LM, et al. Epidemiology and transmission of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans among children and their family members. A report of 4 surveys. J Clin Periodontol. 1993;20:641–650.
- Petit MD, van Steenbergen TJ, Timmerman MF, et al. Prevalence of periodontitis and suspected periodontal pathogens in families of adult periodontitis patients. J Clin Periodontol. 1994;21:76–85.
- Al Yahfoufi Z. Prevalence of periodontal destruction and putative periodontal pathogens in the same lebanese family. J Contemp Dent Pract. 2017;18:970–976.
- Belcheva MD, Kiselova-Yaneva A, Krasteva A. Transmission of Porphyromonas gingivalis from caregivers to children. J Imab. 2012;18:157–162.
- Okada M, Hayashi F, Soda Y, et al. Intra-familial distribution of nine putative periodontopathogens in dental plaque samples analyzed by PCR. J Oral Sci. 2004;46:149–156.
- Tuite-McDonnell M, Griffen AL, Moeschberger ML, et al. Concordance of Porphyromonas gingivalis colonization in families. J Clin Microbiol. 1997;35:455–461.
- Asikainen S, Chen C. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol 2000. 1999;20:65–81.
- Song SJ, Lauber C, Costello EK, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458.
- Stahringer SS, Clemente JC, Corley RP, et al. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 2012;22:2146–2152.
- Asano H, Ishihara K, Nakagawa T, et al. Relationship between transmission of Porphyromonas gingivalis and fimA type in spouses. J Periodontol. 2003;74:1355–1360.
- von Troil-lindén B, Torkko H, Alaluusua S, et al. Salivary levels of suspected periodontal pathogens in relation to periodontal status and treatment. J Dent Res. 1995;74:1789–1795.
- Socransky SS, Haffajee AD, Ximenez-Fyvie LA, et al. Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontol 2000. 1999;20:341–362.
- Sanz M, Lau L, Herrera D, et al. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a review. J Clin Periodontol. 2004;31:1034–1047.
- Teles R, Teles F, Frias-Lopez J, et al. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000. 2013;62:95–162.
- Socransky SS, Haffajee AD, Smith GL, et al. Difficulties encountered in the search for the etiologic agents of destructive periodontal diseases. J Clin Periodontol. 1987;14:588–593.
- Von Troil-Lindén B, Torkko H, Alaluusua S, et al. Periodontal findings in spouses. A clinical, radiographic and microbiological study. J Clin Periodontol. 1995;22:93–99.
- Zhuang L-F, Watt RM, Mattheos N, et al. Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clin Oral Implants Res. 2016;27:13–21.
- Könönen E, Asikainen S, Jousimies-Somer H. The early colonization of gram-negative anaerobic bacteria in edentulous infants. Oral Microbiol Immunol. 1992;7:28–31.
- Könönen E, Kanervo A, Takala A, et al. Establishment of oral anaerobes during the first year of life. J Dent Res. 1999;78:1634–1639.
- Lamell CW, Griffen AL, McClellan DL, et al. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J Clin Microbiol. 2000;38:1196–1199.
- Mättö J, Saarela M, Alaluusua S, et al. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J Clin Microbiol. 1998;36:157–160.
- McClellan DL, Griffen AL, Leys EJ. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1996;34:2017–2019.
- Yang EY, Tanner ACR, Milgrom P, et al. Periodontal pathogen detection in gingiva/tooth and tongue flora samples from 18- to 48-month-old children and periodontal status of their mothers. Oral Microbiol Immunol. 2002;17:55–59.
- Aquino DR, Franco GCN, Cortelli JR, et al. The impact of the maternal’s periodontal status on the detection of periodontal pathogens in newborn children. Revista Odonto Ciência. 2010;25:333–338.
- Travis J, Potempa J, Maeda H. Are bacterial proteinases pathogenic factors? Trends Microbiol. 1995;3:405–407.
- Amano A, Kuboniwa M, Nakagawa I, et al. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–1668.
- Nakagawa I, Amano A, Kuboniwa M, et al. Functional differences among FimA variants of porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70:277–285.
- Amano A, Nakagawa I, Okahashi N, et al. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–142.
- Martelli FS, Mengoni A, Martelli M, et al. Comparison of periodontal microbiological patterns in Italian spouses. Ig Sanita Pubbl. 2012;68:589–599.
- Olsen I, Chen T, Tribble GD. Genetic exchange and reassignment in Porphyromonas gingivalis. J Oral Microbiol. 2018;10:1457373.
- Schwarzberg K, Le R, Bharti B, et al. The personal human oral microbiome obscures the effects of treatment on periodontal disease. PloS One. 2014;9:e86708.