ABSTRACT
Background
Periodontopathic bacteria Porphyromonas gingivalis in humans and Porphyromonas gulae in animals are phylogenetically close and commonly have FimA and Mfa1 fimbriae. However, little is known about how fimA and mfa1 are phylogenetically different between P. gingivalis and P. gulae. Here, we examined phylogenetic diversity in their fim and mfa gene clusters.
Methods
Twenty P. gulae strains were isolated from the periodontal pocket of 20 dogs. For their genomic information, along with 64 P. gingivalis and 11 P. gulae genomes, phylogenetic relationship between the genotypes of fimA and mfa1 was examined. Variability of amino acid sequences was examined in the three-dimensional structure of FimA. The distance between strains was calculated for fim and mfa genes.
Results
Some fimA genotypes in P. gulae were close to particular types in P. gingivalis. Two types of mfa1 were classified as 70-kDa and 53-kDa protein-coding mfa1. The variable amino acid positions were primarily at the outer part of FimA. The genes encoding the structural proteins and the main component were similarly distant from the reference strain in P. gingivalis, but not in P. gulae.
Conclusions
The differences in the gene clusters between P. gingivalis and P. gulae may result in their host specificity.
Introduction
The genus Porphyromonas contains Gram-negative anaerobic bacilli, and was formerly classified in the genus Bacteroides [Citation1]. Species in the genus Porphyromonas are prevalent in the oral cavity of mammals [Citation2–Citation5]. Among them, Porphyromonas gingivalis is most widely known as a periodontopathic bacterium in humans [Citation6]. Compared to other human oral bacteria, P. gingivalis has been extensively studied and characterized because it is one of the few oral bacteria that can be isolated and cultured, and produces various virulent factors such as proteases [Citation6]. P. gingivalis is classified as a member of the red complex species, which are highly detectable in deep periodontal pockets [Citation7]. In recent, P. gingivalis was called a keystone species, which has substantial effects on a bacterial community despite its low abundance [Citation8], and is therefore still influential in the etiology of periodontitis.
Porphyromonas gulae, on the other hand, is a species that is phylogenetically close to P. gingivalis and exists in animals such as dogs, cats, and monkeys [Citation9]. In the etiology of dog periodontitis, P. gulae has similar characteristics as those of P. gingivalis in being highly detectable at the periodontitis sites [Citation10] and in modulating the host immune system [Citation11]. P. gulae and P. gingivalis are highly similar in the nucleotide sequence of 16 S rRNA gene, but their genomes are homologous in only nearly one-half of the entire length [Citation9]. Despite the difference in nearly one-half of their genomes, P. gulae and P. gingivalis are highly close in the genus Porphyromonas, which was demonstrated by the examination of core genome-based phylogenetic relationships between various Porphyromonas species [Citation12]. Although the host specificity of P. gulae and P. gingivalis may result from genomic differences between them, little is known about how host specificity and genomic differences are linked.
P. gulae and P. gingivalis both have fimbriae on the cell surface. Fimbriae of P. gingivalis are the virulence factor for adhering to a host cell and tissue as the first step of colonization [Citation6]. In P. gingivalis, fimbriae are classified as FimA fimbriae and Mfa1 fimbriae [Citation13]. FimA is assembled into the polymer with the expression of accessory proteins FimBCDE [Citation14]. The genes encoding FimA and accessory proteins are located in tandem to form the fim gene cluster [Citation14]. Mfa1 fimbriae are similarly expressed by the mfa gene cluster, including mfa1 for the major subunit of Mfa1 fimbriae and mfa2345 for accessory proteins [Citation15]. In addition to the gene cluster, FimA assembly is also regulated in trans by the genes fimSR. FimSR form a two-component system and are encoded distant from the gene cluster [Citation16,Citation17].
The genotypes of fimA have been used for easily classifying P. gingivalis strains. Six genotypes of fimA (i.e., types I, Ib, II, III, IV, and V) have been classified, and are associated with the virulence of P. gingivalis [Citation18]. By contrast, the genotypes of Mfa1 fimbriae were unknown until the 53-kDa protein was revealed as a variant of the Mfa1 protein [Citation19]. Two genotypes of mfa1 are currently to be considered, as 70-kDa protein-coding mfa1 and 53-kDa protein-coding mfa1 [Citation19]. On the other hand, fimA in P. gulae strains were first classified as types A and B, independently of the P. gingivalis genotypes [Citation20], and type C fimA was then identified [Citation21]. However, the mfa1-based genotyping is impractical for P. gulae; therefore, the distribution of the mfa1 genotypes among P. gulae strains remains unknown. Moreover, the phylogenetic diversity of fim- and mfa-related genes, other than fimA and mfa1, has not been described.
Antigenicity in bacteria is diversified by mutations in the genes encoding surface proteins [Citation22]. The antigenicity of fimbriae between P. gingivalis and P. gulae may differ immunogenetically and in the style of host immunity evasion, and thus may cause a difference in the host specificity between the two species. We then hypothesized that the fimbrial gene clusters of P. gingivalis and P. gulae would be a genomic spot where the genetic differences would be detectable between the two species. In this study, we investigated how the fimA and mfa1 genotypes were distributed among strains. We also examined the relationship between strains in the nucleotide sequence similarity of fim- and mfa-related genes. We newly obtained P. gulae strains and their draft genome sequences to compare their fimbrial gene clusters with the genomic information of P. gingivalis and P. gulae in the public database.
Materials and methods
Sample collection
Twenty dogs with periodontitis were recruited for this study at the Fujita Animal Hospital (Saitama, Japan) from 2008 to 2010. All owners provided informed consent for participation. Under general anesthesia, a sterile paper point was inserted into the periodontal pocket for 20 seconds and was then transferred to an anaerobic transport medium [Citation23]. The sample was transported to the laboratory in Tokyo Medical and Dental University (Tokyo, Japan) and stored at −80°C until use. This study was approved by the Dental Research Ethics Committee of Tokyo Medical and Dental University (Tokyo, Japan; approval number 572).
Bacterial strains and culture conditions
Each sample was placed onto a trypticase soy agar plate containing 30 g/L trypticase soy broth (Becton-Dickinson, Franklin Lakes, NJ, USA), 5% defibrinated horse blood (Nippon Bio-Test Laboratories, Tokyo, Japan), 1 mg/mL yeast extract (Nacalai Tesque, Kyoto, Japan), 5 µg/mL hemin (Sigma-Aldrich, St. Louis, MO, USA), and 0.5 µg/mL menadione (Nacalai Tesque). The plate was anaerobically incubated at 37°C in 10% CO2, 10% H2, and 80% N2. To obtain a strain of P. gulae from each sample, a black-pigmented colony was selected on the plate and taxonomically identified using 16S rRNA gene sequencing with the ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA). The P. gulae strains were 20 in total and were named by connecting ‘FJ’ and distinct numbers ().
Table 1. Strains of P. gingivalis and P. gulae used in this study.
Determination of the draft genome sequences
Genomic DNA was extracted from the 20 P. gulae strains, and their draft genome sequences were determined and annotated, as described previously [Citation24]. The sequence reads were deposited in the DNA Data Bank of Japan under the accession number DRA006235. The complete or draft genome sequences of 64 P. gingivalis strains, 11 P. gulae strains, and P. asaccharolytica DSM 20707 were downloaded from the GenBank, and annotated with the same conditions used for the aforementioned 20 genomes. The number of genomes used in this study was 64 for P. gingivalis and 31 for P. gulae in total (). The genome of P. asaccharolytica DSM 20707 was used as an outgroup, as described in the next section.
Construction of a genome-based phylogenetic tree
With respect to the similarity of nucleotide and amino acid sequences, the protein-coding sequences (CDSs) were compared between genomes by using PGAP v1.02 with default parameters [Citation25]. Then, the CDSs that were located at the single genomic region and common among all genomes were identified. In each common CDS, the amino acid sequences were aligned by using MAFFT v7.245 [Citation26], and were examined using the Phi test in SplitsTree v4.11.3 to remove possible rearrangement regions inside the CDS [Citation27,Citation28]. After concatenating the amino acid sequences of all common CDSs, a phylogenetic tree was constructed with the maximum likelihood method under 100-times bootstrap iteration by using RAxML v8.2.4 [Citation29]. The Jones-Taylor-Thornton substitution model was used [Citation30], as suggested by ModelGenerator v851 [Citation31]. The tree was visualized by using Dendroscope v3.2.8 [Citation32].
Determination of the fimA and mfa1 genotypes
The CDSs of fimA and mfa1 were identified in the 64 P. gingivalis genomes and 31 P. gulae genomes. In addition to these data, the nucleotide sequences of fimA in other 34 P. gulae strains, previously determined for their genotyping [Citation21,Citation33], were downloaded from GenBank as a reference for the fimA genotypes of P. gulae (). The nucleotide sequences of these fimA CDSs were aligned by using MAFFT. A tree based on fimA was then constructed under the General Time Reversible model and 1,000-times bootstrap iteration by using RAxML, and was visualized by using Dendroscope. The tree based on the nucleotide sequences of the mfa1 CDSs was constructed and visualized in the same manner as for the tree based on fimA.
Table 2. Known fimA genotypes of P. gulae strains/isolates as a reference.
For the P. gingivalis and P. gulae strains that were previously unclassified by the fimA and/or mfa1 genotypes, the genotypes were determined, based on the phylogenetic relationship with other strains in the trees. In this study, the 70-kDa and 53-kDa protein-coding mfa1 were called ‘type 70’ and ‘type 53,’ respectively. The amino acid sequences of FimA and Mfa1 were aligned within each genotype by using MAFFT and were visualized by using WebLogo v3 [Citation34]. The amino acid sequence of P. gingivalis W83 FimA was downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank, and its crystal structure was visualized by using PyMOL v2.3.2 (http://www.pymol.org) to indicate conserved amino acid positions where only a single amino acid was observed among genotypes.
Calculation of pairwise distance from fim and mfa CDSs
The following CDSs were identified in the 64 P. gingivalis genomes and 31 P. gulae genomes: the CDSs in the fim gene cluster (fimABCDE) and mfa gene cluster (mfa12345), and the CDSs of the two-component system for regulating FimA fimbriation (fimSR). In each of these CDSs, the nucleotide sequences were aligned by using MAFFT, and the K80 pairwise distance from P. gingivalis ATCC 33277 was calculated by using R v3.5.2. The distance matrix was visualized as a heat map by using R.
Results
Phylogenetic relationship based on the fimA and mfa1 nucleotide sequences
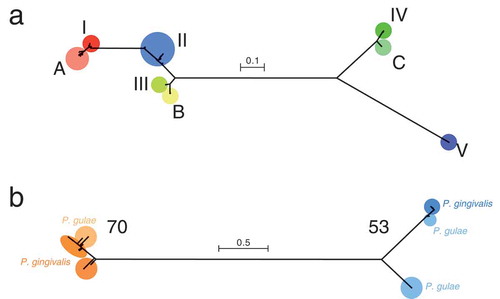
In the phylogenetic tree based on fimA, types I, II, III were distant from types IV and V (). Type Ib could not be distinguished from type I; we therefore did not distinguish between types I and Ib, and considered both of them as type I in this study. Types A, B, and C for P. gulae strains were close to types I, III, and IV, respectively, for P. gingivalis. On the other hand, the phylogenetic tree, based on mfa1, had whole branches that were nearly five times longer than those of the tree based on fimA (). Type 70 was considerably far from type 53, and P. gulae and P. gingivalis strains were mixed in the tree topology of each type. Based on the phylogenetic relationship, the fimA and mfa1 genotypes were determined (), whereas the fimA genotype of Co5 and the mfa1 genotypes of D34 and FJ81 were not classifiable because the corresponding CDSs could not be identified in these strains, possibly due to the limitation of data assembly.
Figure 1. Phylogenetic trees, based on the fimA and mfa1 nucleotide sequences. (a) The tree based on fimA is shown. Types I, II, III, IV, and V for P. gingivalis, and types A, B, and C for P. gulae are indicated by different colors. (B) The tree based on mfa1 is shown. Type 70 and type 53 are indicated by different colors. For each type, P. gingivalis and P. gulae are indicated by dark colors and light colors, respectively. The scale bar in each tree represents substitutions per nucleotide site.
Diversity in amino acid sequences of FimA and Mfa1
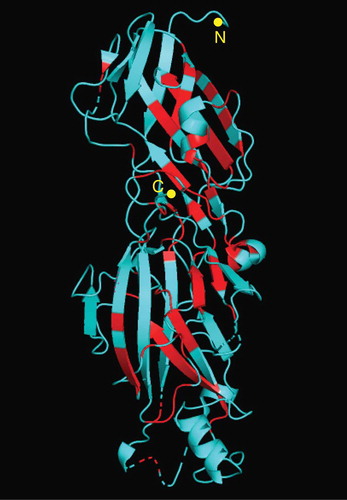
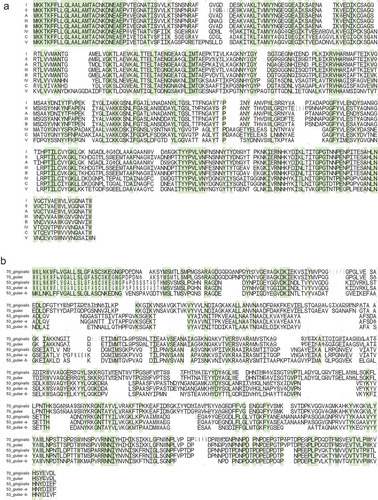
In each fimA and mfa1 genotype, the amino acid sequences of their encoding proteins were highly conserved although the variation in amino acids at a position within the genotype was observed throughout (). Most of the positions variable within the genotype seemed common among genotypes. The number of conserved positions, at which a single amino acid was exclusively observed among genotypes, was 143 in total 418 positions (34.2%) in FimA and 87 in total 607 positions (14.3%) in Mfa1. The N-terminal end of FimA and Mfa1 were highly conserved among genotypes. In the crystal structure of FimA, the conserved positions were primarily located at the inner part of the protein (), indicating that the positions variable among genotypes were primarily located at the outer part of FimA.
Figure 2. Variation in the amino acid sequences of FimA and Mfa1. The amino acid sequences of fimA (a) and mfa1 (b), as visualized using WebLogo, are shown. The alignment of the sequences is shown for each of eight fimA genotypes (a) and five mfa1 potential genotypes (b). Based on the phylogeny in ), the mfa1 genotypes 70 and 53 are divided into the clusters of each species. The cluster of P. gulae type 53 is further divided into putative subtype-a and subtype-b, which represent the upper and lower phylogroup, respectively, in ). The alignment is shown from the amino acid position 1 of N-terminal end to the last of C-terminal end, and for each 100 amino acids. In each genotype, the variation in amino acids at each position is indicated by the proportion of vertical length of characters. In particular strains, the absence of amino acids at a position is indicated by the width of characters. The positions where only a single amino acid exists among genotypes are colored.
Genome-based phylogeny and the fimA and mfa1 genotypes
In the phylogenetic tree based on 336 common CDSs, 31 P. gulae strains were clearly separated from 64 P. gingivalis strains (). When the fimA and mfa1 genotypes were considered in the phylogeny, the genotypes did not have a clear relationship with the tree topology. In P. gingivalis, fimA type II was distributed among the strains, and was mixed with other types such as types I, III, and IV, in the tree topology. Types 70 and 53 of mfa1 were also mixed throughout the P. gingivalis strains. These situations were similarly observed in P. gulae. A remarkable finding was that P. gulae FJ70 did not have any fimA genotypes for P. gulae but did had type II fimA for P. gingivalis.
Figure 4. Phylogenetic tree based on the amino acid sequences of common CDSs, the fimA and mfa1 genotypes, and heat map for K80 distances of fim and mfa CDSs from P. gingivalis ATCC 33277. The tree based on 336 common CDSs in 64 P. gingivalis strains and 31 P. gulae strains is shown. The outgroup P. asaccharolytica DSM 20707 is not shown. The scale bar represents substitutions per amino acid site. The names of strains are on the right side of the tree. The genotypes of fimA are indicated by colored circles on the right side of the names of strains. For each fim-related CDS, the K80 distance values are indicated by the color gradient in the heat map. Black boxes in the heat map indicate the absence of the corresponding CDSs. The mfa1 genotypes and the K80 distance values for each mfa-related CDS are shown on the far right.
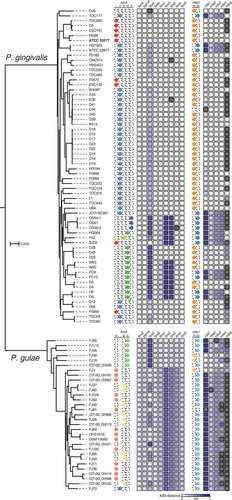
The distribution of fimA and mfa1 genotypes was reflected in the K80 distances in fimA and mfa1 (). In the heat map, strains with fimA types I and A had mostly white boxes for fimA, whereas the fimA boxes for types IV, V, and C were darkened. The mfa1 boxes were light for type 70 and darkened for type 53, although the boxes for type 70 showed a diversity in gradation, based on the distance from ATCC 33277.
Distances based on the nucleotide sequences of the fim and mfa CDSs
In P. gingivalis, the three CDSs fimX, pgmA, and fimB were nearly identical in their nucleotide sequences among the strains (). These CDSs in P. gulae were rather distant from P. gingivalis but were nearly identical among the P. gulae strains. Similar situations were observed for fimSR, although the P. gulae strains were divided into two groups, based on their distances from ATCC 33277. One of these two groups contained six strains (i.e., FJ55, FJ38, FJ19, COT-052_OH3439, FJ46, and FJ115), whereas the other group contained the remaining P. gulae strains. The two groups were separated by the genome-based phylogeny and by their distances from ATCC 33277, based on fim-related CDSs.
The distances based on three fimA-related CDSs (i.e., fimCDE) appeared to be associated with the fimA-based distances in most P. gingivalis strains (). P. gingivalis SJD2 was exceptionally far from ATCC 33277 when using fimCDE-based distances, and nearly identical to ATCC 33277 when using fimA-based distances. By contrast, the fimCDE-based distances in P. gulae showed a rather opposite relationship to the fimA-based distance. P. gulae strains with a low distance of fimCDE from ATCC 33277 (i.e., the aforementioned group consisting of six strains) were far from ATCC 33277 in the fimA-based distance. On the other hand, the mfa234-based distances were associated with the mfa1-based distance in P. gingivalis and in P. gulae. Possibly because of insufficient assembly of genomes, the CDSs encoding mfa5 could not be identified in 40 of 64 P. gingivalis strains and in 27 of 31 P. gulae strains.
Discussion
The relationship between the fimA genotypes and the observable phenotypes of P. gingivalis was reported nearly two decades ago. P. gingivalis strains with type II or IV were virulent, whereas strains with type I or III were mostly avirulent [Citation35]. In particular, type II FimA was known to be highly virulent, compared to the other types with regard to adhesion to and invasion into host cells [Citation36] and causing subcutaneous abscess in mice [Citation37]. The fimA genotypes and phenotypes of P. gulae were also associated with each other, as shown in mouse abscess models; the P. gulae strains with type B have been reported as more virulent than type A [Citation20], and strains with type C were more virulent than strains with types A and B [Citation21]. On the other hand, in this study, we demonstrated that the fimA types for P. gulae were phylogenetically close to certain fimA types for P. gingivalis (). We observed that types A, B, and C in P. gulae were close to types I, III, IV, respectively, in P. gingivalis. A close relationship was also observed in the alignment of the amino acid sequences (). The signal peptide of FimA was highly conserved, whereas the N-terminal extension, the region cleaved by the gingipain at the arginine residue first appearing in the N-terminal end [Citation15,Citation38], was variable at most positions among the genotypes. The close relationship was partially consistent with the aforementioned relationship that the type II and IV strains in P. gingivalis and the type B and C strains in P. gulae were virulent whereas the type I and III strains in P. gingivalis and the type A strain in P. gulae were less virulent. The relationship and differences in fimA in the phylogeny among genotypes may be explained by localizing the variable positions, primarily at the outer part of FimA (). The inner part of FimA may have been highly conserved to maintain the basic structure of protein, whereas the outer part may have allowed the substitution of amino acids to diversify the antigenicity of the fimbriae. Although no novel fimA type was observed in P. gulae, other than the three fimA types A, B, and C, only P. gulae FJ70 had the fimA type II that was considered to be unique to P. gingivalis. This may be a variant of type B, based on the close relationship among types II, III, and B in the fimA-based phylogeny () and in the similarity in amino acid sequences (). It may also be a novel type for P. gulae that has not been described previously. This exceptional type will be further examined in the future by collecting the corresponding P. gulae strains, with a possibility of their infection from dogs to humans, and vice versa.
With regard to the mfa1 genotypes, we demonstrated that types 70 and 53 were the major types and prevalent among the P. gulae and P. gingivalis strains. Type 70 seemed a major mfa1 genotype for P. gingivalis, whereas most P. gulae strains had type 53 (). In a previous study, the relationship between the fimA and mfa genotypes was weakly observed in P. gingivalis, such as type II strains harboring type 70 mfa1 rather than type 53, and mfa1 was absent in the type V strains [Citation13]. These previous findings were consistent with our observation that most of type II strains were type 70 in the mfa1 genotypes, but were not consistent with the presence of mfa1 in P. gingivalis strains with type V fimA. The topology of the mfa1-based tree suggested that each type, especially type 53, may be further classified into subtypes or distinct types (). This concept will be considered with the phenotypic differences in P. gulae strains between the potential subtypes. Although two mfa1 genotypes may possibly be further subtyped, a clear separation between the two genotypes was remarkable for detecting them as major mfa1 genotypes. A signal peptide of Mfa1 at the N-terminal end was highly conserved and most positions in the N-terminal extension [Citation39] were variable among genotypes in similar manner as FimA. However, the positions conserved among all genotypes were fewer for Mfa1 than for FimA (), despite the length of Mfa1 being longer than that of FimA. The difference in the extent of amino acid variation between FimA and Mfa1 may have resulted in the higher number of fimA genotypes than that of mfa1 genotypes. Identifying the crystal structure of Mfa1 will help in understand how the variation of amino acids occurs in the three-dimensional structure of protein and how this variation has a role in the function of fimbriae.
The fimA-related proteins FimCDE are accessory components that bind to the FimA polymer as a part of the fimbrial structure [Citation40–Citation42], and seem to be functionally different from other fimA-related proteins. The two-component system proteins FimSR regulate the transcriptional expression of the fim gene cluster [Citation17,Citation43], and FimB regulates fimbriation as a terminator [Citation44]. The functions of FimX and PgmA are not fully characterized [Citation42], although PgmA is suggested to be the usher [Citation14]. The relationship between fimA and fimCDE in P. gingivalis with respect to the distances from ATCC 33277 () possibly reflected the functional difference between FimCDE and the other fimA-related proteins. In P. gingivalis, fimCDE may have phylogenetically evolved together with fimA, whereas the other fimA-related CDSs may have retained their gene structure to keep regulatory or supportive functions for fimbriation. The mfa-related CDSs mfa345 were similarly associated with mfa1 with respect to the distances from ATCC 33277 (). Mfa345 binds to the Mfa1 polymer as a part of the fimbrial structure, similar to FimCDE [Citation15,Citation39]. mfa2 also showed a weak relationship with mfa1 with respect to distance, although Mfa2 contributes to the regulation of fimbrial length and is not included in the actual fimbrial structure [Citation15,Citation45]. The structural and regulatory CDSs of Mfa1 fimbriae in P. gingivalis may have evolved in a similar manner as FimA fimbriae.
However, fimA and fimCDE in P. gulae did not show a clear relationship with each other with respect to their distances from ATCC 33277 (), despite that mfa1 and mfa345 showed a similar relationship to P. gingivalis. The distances, based on fimCDE and fimSR, seemed to reflect the phylogenetic distance from P. gingivalis, whereas the distances based on fimA were irrelevant to the phylogeny between P. gulae and P. gingivalis. The combination of fimA distant from P. gingivalis and fimA-related CDSs close to P. gingivalis, and vice versa, may characterize P. gulae as a species independent from P. gingivalis and lead to its unique habitats segregated from P. gingivalis. In P. gingivalis, homologous recombination was suggested to shape the genetic diversity among the strains [Citation46–Citation48]. Chromosomes in other P. gingivalis cells are potential sources of the recombination partner, transferred by conjugation [Citation49,Citation50]. Natural competence is also important for recombination by introducing extracellular DNA, which is released from P. gingivalis cells [Citation47,Citation51]. Although it has been still unknown whether these mechanisms are also valid in P. gulae, homologous recombination that would occur within P. gingivalis or P. gulae and would occur between P. gingivalis and P. gulae across the hosts, may be a possible reason for the phylogenetic differentiation of fimbrial genes between P. gingivalis and P. gulae, thereby resulting in the difference in host specificity.
Conclusions
We demonstrated the relationship of the fimA genotypes between P. gingivalis and P. gulae, and the two mfa1 genotypes that were clearly separated from each other. In addition, we observed that fimA and fimCDE in P. gingivalis were similarly distant from the reference strain, whereas the distance of fimA was inversely related to the distance of fimCDE in P. gulae. A genomic region of a clustered regularly interspaced short palindromic repeat (CRISPR) array generally has the function of acquired immunity [Citation52], whereas the CRISPR arrays in P. gingivalis were suggested to regulate homologous recombination of the genome with the DNA introduced from nonself P. gingivalis cells [Citation24]. The function of arrays in P. gulae has not been described but may have similar role as the arrays in P. gingivalis, considering their phylogenetic closeness. Future studies will elucidate how the CRISPR arrays in P. gulae are involved in genetic diversification and in the differentiation of the fim and mfa gene clusters.
Data availability
The sequence reads obtained in this study are available in the DNA Data Bank of Japan under the accession number DRA006235.
Acknowledgments
Supercomputing resource was provided by the Human Genome Centre at the Institute of Medical Science (the University of Tokyo, Tokyo, Japan; http://sc.hgc.jp/shirokane.html). We thank Keiichi Fujita (Fujita Animal Hospital, Saitama, Japan) for sample collection.
Disclosure statement
The authors declare that they have no competing interests.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Shah HN , Collins MD. Proposal for reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new genus, Porphyromonas . Int J Syst Bacteriol. 1988;38(1):128–11.
- Finegold SM , Barnes EM . Report of the ICSB taxonomic subcommittee on Gram-negative anaerobic rods. Int J Syst Evol Microbiol. 1977;27:388–391.
- TJM VS , van Winkelhoff AJ , Mayrand D , et al. Bacteroides endodontalis sp. nov., an asaccharolytic black-pigmented Bacteroides species from infected dental root canals. Int J Syst Evol Microbiol. 1984;34:118–120.
- Collins MD , Love DN , Karjalainen J , et al. Phylogenetic analysis of members of the genus Porphyromonas and description of Porphyromonas cangingivalis sp. nov. and Porphyromonas cansulci sp. nov. Int J Syst Bacteriol. 1994;44:674–679.
- Coykendall AL , Kaczmarek FS , Slots J . Genetic heterogeneity in Bacteroides asaccharolyticus (Holdeman and Moore 1970) Finegold and Barnes 1977 (Approved Lists, 1980) and proposal of Bacteroides gingivalis sp. nov. and Bacteroides macacae (Slots and Genco) comb. nov. Int J Syst Evol Microbiol. 1980;30:559–564.
- Lamont RJ , Jenkinson HF . Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev. 1998;62:1244–1263.
- Socransky SS , Haffajee AD , Cugini MA , et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144.
- Hajishengallis G , Darveau RP , Curtis MA . The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725.
- Fournier D , Mouton C , Lapierre P , et al. Porphyromonas gulae sp. nov., an anaerobic, gram-negative coccobacillus from the gingival sulcus of various animal hosts. Int J Syst Evol Microbiol. 2001;51:1179–1189.
- Senhorinho GN , Nakano V , Liu C , et al. Detection of Porphyromonas gulae from subgingival biofilms of dogs with and without periodontitis. Anaerobe. 2011;17:257–258.
- Lenzo JC , O’Brien-Simpson NM , Orth RK , et al. Porphyromonas gulae has virulence and immunological characteristics similar to those of the human periodontal pathogen Porphyromonas gingivalis . Infect Immun. 2016;84(9):2575–2585.
- O’Flynn C , Deusch O , Darling AE , et al. Comparative genomics of the genus Porphyromonas identifies adaptations for heme synthesis within the prevalent canine oral species Porphyromonas cangingivalis . Genome Biol Evol. 2015;7:3397–3413.
- Nagano K , Hasegawa Y , Iijima Y , et al. Distribution of Porphyromonas gingivalis fimA and mfa1 fimbrial genotypes in subgingival plaques. Peer J. 2018;6:e5581.
- Nagano K , Abiko Y , Yoshida Y , et al. Porphyromonas gingivalis FimA fimbriae: roles of the fim gene cluster in the fimbrial assembly and antigenic heterogeneity among fimA genotypes. J Oral Biosci. 2012;54(3):160–163.
- Kloppsteck P , Hall M , Hasegawa Y , et al. Structure of the fimbrial protein Mfa4 from Porphyromonas gingivalis in its precursor form: implications for a donor-strand complementation mechanism. Sci Rep. 2016;6:22945.
- Hayashi J , Nishikawa K , Hirano R , et al. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis . Microbiol Immunol. 2000;44(4):279–282.
- Nishikawa K , Yoshimura F , Duncan MJ . A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol. 2004;54(2):546–560.
- Enersen M , Nakano K , Amano A . Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013;5.
- Nagano K , Hasegawa Y , Yoshida Y , et al. A major fimbrilin variant of Mfa1 fimbriae in Porphyromonas gingivalis . J Dent Res. 2015;94:1143–1148.
- Nomura R , Shirai M , Kato Y , et al. Diversity of fimbrillin among Porphyromonas gulae clinical isolates from Japanese dogs. J Vet Med Sci. 2012;74:885–891.
- Yamasaki Y , Nomura R , Nakano K , et al. Distribution and molecular characterization of Porphyromonas gulae carrying a new fimA genotype. Vet Microbiol. 2012;161:196–205.
- Foley J . Mini-review: strategies for variation and evolution of bacterial antigens. Comput Struct Biotechnol J. 2015;13:407–416.
- Takeuchi Y , Umeda M , Ishizuka M , et al. Prevalence of periodontopathic bacteria in aggressive periodontitis patients in a Japanese population. J Periodontol. 2003;74:1460–1469.
- Watanabe T , Nozawa T , Aikawa C , et al. CRISPR regulation of intraspecies diversification by limiting IS transposition and intercellular recombination. Genome Biol Evol. 2013;5:1099–1114.
- Zhao Y , Wu J , Yang J , et al. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418.
- Katoh K , Standley DM . MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780.
- Bruen TC , Philippe H , Bryant D . A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681.
- Huson DH , Bryant D . Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267.
- Stamatakis A . RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313.
- Jones DT , Taylor WR , Thornton JM . The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282.
- Keane TM , Creevey CJ , Pentony MM , et al. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29.
- Huson DH , Scornavacca C . Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067.
- Iwashita N , Nomura R , Shirai M , et al. Identification and molecular characterization of Porphyromonas gulae fimA types among cat isolates. Vet Microbiol. 2019;229:100–109.
- Crooks GE , Hon G , Chandonia JM , et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190.
- Amano A , Nakagawa I , Okahashi N , et al. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–142.
- Nakagawa I , Amano A , Kuboniwa M , et al. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70:277–285.
- Nakano K , Kuboniwa M , Nakagawa I , et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004;19:205–209.
- Hasegawa Y , Murakami Y . Porphyromonas gingivalis fimbriae: recent developments describing the function and localization of mfa1 gene cluster proteins. J Oral Biosci. 2014;56:86–90.
- Hall M , Hasegawa Y , Yoshimura F , et al. Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis . Sci Rep. 2018;8:1793.
- Yoshimura F , Takahashi Y , Hibi E , et al. Proteins with molecular masses of 50 and 80 kilodaltons encoded by genes downstream from the fimbrilin gene (fimA) are components associated with fimbriae in the oral anaerobe Porphyromonas gingivalis . Infect Immun. 1993;61:5181–5189.
- Nishiyama S , Murakami Y , Nagata H , et al. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology. 2007;153:1916–1925.
- Nagano K , Hasegawa Y , Abiko Y , et al. Porphyromonas gingivalis FimA fimbriae: fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PLoS One. 2012;7:e43722.
- Nishikawa K , Duncan MJ . Histidine kinase-mediated production and autoassembly of Porphyromonas gingivalis fimbriae. J Bacteriol. 2010;192:1975–1987.
- Nagano K , Hasegawa Y , Murakami Y , et al. FimB regulates FimA fimbriation in Porphyromonas gingivalis . J Dent Res. 2010;89:903–908.
- Hasegawa Y , Iwami J , Sato K , et al. Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology. 2009;155:3333–3347.
- Frandsen EV , Poulsen K , Curtis MA , et al. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;69:4479–4485.
- Tribble GD , Kerr JE , Wang BY . Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607–620.
- Dashper SG , Mitchell HL , Seers CA , et al. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48.
- Tribble GD , Lamont GJ , Progulske-Fox A , et al. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis . J Bacteriol. 2007;189:6382–6388.
- Naito M , Sato K , Shoji M , et al. Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: determination of the integration site and the genes essential for conjugal transfer. Microbiology. 2011;157:2022–2032.
- Tribble GD , Rigney TW , Dao DH , et al. Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis . MBio. 2012;3:e00231–11.
- Marraffini LA , Sontheimer EJ . CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190.