ABSTRACT
Background
Porphyromonas gingivalis and Treponema denticola are proteolytic periodontopathogens that co-localize in polymicrobial subgingival plaque biofilms, display in vitro growth symbiosis and synergistic virulence in animal models of disease. These symbioses are underpinned by a range of metabolic interactions including cooperative hydrolysis of glycine-containing peptides to produce free glycine, which T. denticola uses as a major energy and carbon source.
Objective
To characterize the P. gingivalis gene products essential for these interactions. Methods: The P. gingivalis transcriptome exposed to cell-free T. denticola conditioned medium was determined using RNA-seq. P. gingivalis proteases potentially involved in hydrolysis of glycine-containing peptides were identified using a bioinformatics approach.
Results
One hundred and thirty-twogenes displayed differential expression, with the pattern of gene expression consistent with succinate cross-feeding from T. denticola to P. gingivalis and metabolic shifts in the P. gingivalis folate-mediated one carbon superpathway. Interestingly, no P. gingivalis proteases were significantly up-regulated. Three P. gingivalis proteases were identified as candidates and inactivated to determine their role in the release of free glycine. P. gingivalis PG0753 and PG1788 but not PG1605 are involved in the hydrolysis of glycine-containing peptides, making free glycine available for T. denticola utilization.
Conclusion
Collectively these metabolic interactions help to partition resources and engage synergistic interactions between these two species.
Introduction
Periodontitis is a bacterial-induced chronic inflammatory disease characterized by destruction of the supporting tissue structures of the tooth. Porphyromonas gingivalis and Treponema denticola are known pathobionts and synergistic partners that co-localize in vivo, produce biofilms with increased biomass when grown together in vitro and increased pathogenicity during co-infection in animal models of disease [Citation1–Citation6]. Both species are asaccharolytic and primarily rely on amino acids as carbon and energy sources for growth. P. gingivalis possesses an extensive array of proteases for the degradation of host proteins in order to increase the accessibility of nutrients. Gingipains (RgpA, RgpB, Kgp) are the primary P. gingivalis proteases that initiate the process of degradation of host extracellular matrix and serum-derived proteins including haemoglobin [Citation7–Citation10]. Following the initial proteolytic degradation of complex proteins by gingipains or other proteases into oligopeptides, numerous endopeptidases work in concert for downstream digestion to produce tri- and dipeptides for P. gingivalis internalization and utilization [Citation7,Citation9,Citation11]. Peptidases of P. gingivalis that have been characterized include: dipeptidyl peptidase (DPP) III [Citation12], DPPIV [Citation13], DPP5 [Citation14], DPP7 [Citation15], DPP11 [Citation16], prolyl tripeptidyl peptidase A (PtpA) [Citation17], as well as acylpeptidyl oligopeptidase (AOP) [Citation18] and carboxypeptidase (CPG70) [Citation19,Citation20].
Besides acting as important macronutrient extracting factors for P. gingivalis, extracellularly localized peptidases are postulated to have a role in the release of small peptides and free amino acids for other oral bacterial species within the vicinity. A study using an oral polymicrobial inoculum grown in mucin has shown that the diversity and complementarity of enzymatic activities found in different bacterial species led to increasing accessibility of nutrients and proliferation of the microbial consortium [Citation21]. The efficiency and specificity of amino acid release and utilization by the oral subgingival polymicrobial community can indirectly govern their amino acid fermentation pathways and therefore influence cytotoxic end product production which exacerbates disease. In this regard, metabolic cooperation between the dysbiotic oral bacterial species presents as a virulence determinant for the initiation and progression of chronic periodontitis.
T. denticola has been previously shown to upregulate expression of glycine cleavage and reductase system encoding genes as well as a putative glycine transporter during co-culture with P. gingivalis [Citation22]. T. denticola preferentially utilizes free glycine as a major energy and carbon source. In addition, P. gingivalis was shown to produce free glycine in the presence of cell-free T. denticola conditioned media, by increasing the rate of hydrolysis of glycine-containing peptides [Citation22]. Together these data indicated that the increased release of free glycine by P. gingivalis was utilized by T. denticola as a carbon and energy source, which contributed to an observed increase in T. denticola biomass during co-culture [Citation22]. Interestingly, the biomass of P. gingivalis also increased during co-culture with T. denticola indicating mutualism between the two species [Citation22]. However, the microarray analysis of P. gingivalis and T. denticola in coculture identified only two putative P. gingivalis protease genes, PG0753 and PG0383, whose expression was upregulated, by 1.21 fold and 1.18 fold, respectively [Citation22].
In this study, RNA-seq was used to provide insights into the broad metabolic interactions between P. gingivalis and T. denticola. Three putative protease-encoding genes of P. gingivalis that were likely to contribute to cooperative free glycine release, PG0753, PG1605, and PG1788, were identified. Mutant strains were created and used to determine the contribution of these proteases to the release of free glycine by P. gingivalis.
Methods and materials
Bacterial strains and growth conditions
T. denticola ATCC 35405 and all P. gingivalis W50 strains were grown in Oral Bacterial Growth Medium (OBGM) as described previously [Citation23]. If required, 0.8% (w/v) agarose (Life Technologies, Thermo Fisher Scientific, Australia) was added prior to autoclaving for OBGM agar. All P. gingivalis and T. denticola bacterial strains were grown in an anaerobic chamber (MG500 anaerobic workstation; Don Whitley Scientific Pty. Ltd., NSW, Australia) with a gas composition of 5% H2, 10% CO2 in N2 at 37°C. P. gingivalis was also grown in T. denticola conditioned medium (TdCM), as described in Tan et al. [Citation22]. Briefly, TdCM was obtained by growth of T. denticola in OBGM under anaerobic conditions for 7 days until stationary phase. The culture supernatant collected after centrifugation (4,000 × g, 20 min, 4°C) was subjected to vacuum filtration through a 0.22 µm Steritop filter (Millipore), followed by a 0.1 µm filter (Sarstedt AG & Co., Germany). OBGM was mixed with TdCM in a 1:1 volume ratio with a final pH of 7.4, labelled as OB:CM and used in this study. When required erythromycin or ampicillin was added to the growth media of P. gingivalis mutant strains at a final concentration of 10 µg/mL and 5 µg/mL, respectively.
Escherichia coli strains were grown in Lysogeny Broth [LB: 1% (w/v) Tryptone (Oxoid), 0.5% (w/v) Yeast Extract (Oxoid) and 0.5% (w/v) sodium chloride (Chem-Supply)] or on LB agar plates made with LB containing 1.5% (w/v) Bacto agar (BD, DifcoTM). E. coli cultures were incubated aerobically at 37°C in a shaking incubator (NT INFORS Minitron, INFORS AG-CH 4103 Bottmingen, Infors AG, Switzerland), shaking at 180–220 rpm. Media for E. coli culture were supplemented with ampicillin at a final concentration of 100 µg/mL when required.
Differential gene expression
Log phase cells of P. gingivalis W50 (OD650 ~ 0.5) in OBGM or OB:CM were mixed with 0.2 vol of 5% (v/v) phenol in absolute ethanol at RT and centrifuged (8,000 × g, 15 min); the resulting cell pellets were snap frozen in liquid nitrogen and stored at −80°C for future use [Citation24,Citation25]. P. gingivalis cell lysis was performed by adding 1.4 mL TRIzolTM Reagent (Ambion, Invitrogen) to each frozen pellet consisting of ~1010 P. gingivalis cells. The mixture was transferred to 2 mL screw-capped tubes with pre-chilled glass beads (MP BioMatrix B) for homogenization in a Precellys24 homogeniser (Bertin Corp., USA) at 6,500 rpm for 23 s twice with a 5-s pause between cycles. Total RNA extraction was performed using TRIzolTM according to the manufacturer’s instructions. Extracted RNA was subjected to rigorous DNase I treatment according to the Turbo DNA-freeTM protocol (Ambion, Life Technologies), then the sample was purified using a NucleoSpin RNA II kit (Macherey-Nagel GmbH & Co. KG, Germany) as per the manufacturer’s instructions. To prevent degradation of RNA, Ribosafe RNase inhibitor (Bioline) was added to each RNA sample.
RNA integrity, quality score and quantification were examined using the DNA 5K/RNA Charge Variant Assay LabChip (PerkinElmer) on the LabChip GXII Touch 24 instrument (Perkin Elmer) according to the manufacturer’s instruction. The quality and quantity of RNA were reviewed using the LabChip GX Reviewer.
RNA library preparation using Epicentre ScriptSeq Complete Bacteria and NextSeq Illumina sequencing in Mid-Output mode and single-read 75 bp format was conducted at Micromon (Monash University). Data analysis was performed by the Monash Bioinformatics Platform (Monash University) using the P. gingivalis W83 NCBI reference sequence NC_002950.2 [Citation26]. Raw RNAseq FASTQ files were aligned with the BWA-MEM aligner and analyzed using the RNAsik pipeline to generate raw counts [Citation27]. Then, Degust was used for differential gene expression analysis and visualization [Citation28]. RNA-seq data have been submitted to the NCBI Short Read Archive with the Bioproject Accession Number PRJNA574726. MicrobesOnline [Citation29] was referred to for P. gingivalis W83 gene information including Clusters of Orthologous Groups (COGs) categorization, whilst the Kyoto Encyclopedia of Genes and Genome (KEGG) and BioCyc were employed for metabolic pathway analyses [Citation30,Citation31].
Predictive analysis of localization of P. gingivalis proteases
The P. gingivalis W83 annotated genome sequence (Accession number: NC_002950.2) was sourced from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). Amino acid sequences annotated as proteases were examined via the BLASTp suite and Conserved Domain Database (CDD) to identify predicted protein homologs and their conserved domains [Citation32,Citation33]. The MEROPS Peptidase Database Release 12.0 was used as the primary resource for compilation of P. gingivalis W83 proteases [Citation34,Citation35]. PSORTb and SignalP were employed for the prediction of protein cellular localization, signal peptide, and transmembrane topology [Citation36–Citation38].
Deletion of selected P. gingivalis protease-encoding genes
Isogenic P. gingivalis W50 protease mutants were constructed where the open reading frames of PG0753 and PG1788 were replaced with the ampicillin resistance gene cepA whilst PG1605 was replaced with the erythromycin resistance gene ermF. Briefly, recombination cassettes for the deletion of PG0753, PG1605, or PG1788 were constructed using splicing by overlap-extension (SOE) PCR [Citation39]. The primers used are shown in Supplementary Table S1. The upstream and downstream regions of the target genes were amplified by PCR from the chromosomal DNA of P. gingivalis W50 whilst the ermF gene was amplified from the shuttle vector pHS17 [Citation40] and the cepA gene was amplified from pEC474 [Citation41]. The three respective amplicons were sequentially fused in a step-wise manner into a single fragment using SOE PCR [Citation39], cloned into the pGEM®-T Easy vector and the fidelity of each recombination cassette was confirmed by DNA sequencing. Plasmids were linearized before transformation into P. gingivalis W50 by electroporation [Citation42]. P. gingivalis transformants were subjected to PCR to verify the homologous recombination event in the chromosome.
Whole-genome sequencing of ΔPG0753, ΔPG1605 and ΔPG1788 mutants
Chromosomal DNA of P. gingivalis ΔPG0753, ΔPG1605 and ΔPG1788 strains was isolated using a DNeasy Blood and Tissue kit (QIAGEN) as per the manufacturer’s instructions. Genome sequencing was performed using an Ion Torrent Personal Genome Machine (PGM; Thermo Fisher Scientific) according to the protocols of the manufacturer, unless otherwise stated. Briefly, 1 µg of P. gingivalis genomic DNA was fragmented to ~400 bp using a Covaris M220 Focused-ultrasonicator™ (TrendBio, Australia). A 1 µL aliquot of sheared DNA was visualized using a LabChip GXII Touch 24 Nucleic Acid Analyzer (PerkinElmer, USA) to ensure a peak fragment size of 400 bp. The DNA was end-repaired (Ion Xpress™ Plus Fragment Library Kit), purified (Agencourt™ AMPure™ XP Kit, Beckman Coulter), barcoded adaptors ligated to the DNA then nick-repaired (Ion Xpress™ Plus Fragment Library Kit; Ion Xpress™ Barcode Adapters Kit) and purified again (Agencourt™ AMPure™ XP Kit, Beckman Coulter). The labelled library was then size-selected using the Pippin Prep™ DNA Size Selection System (Sage Science), aiming for a target-peak size of ~480 bp collected over the specified range of 407 to 543 bp. Following sample purification (Agencourt™ AMPure™ XP Kit, Beckman Coulter) the concentration of the unamplified library was determined using qPCR (Ion Library TaqMan® Quantitation Kit). Barcoded libraries were pooled in equimolar amounts of 26 pM to ensure an equal representation of each barcoded library in the sequencing run. The library was then used to prepare enriched, template-positive Ion PGM™ Hi-Q™ Ion Sphere™ Particles (ISPs) using the Ion OneTouch™ 2 System (Ion PGM™ Hi-Q™ OT2 Kit). The recovered template-positive ISPs were enriched using the Ion OneTouch™ ES Instrument and Ion OneTouch™ ES Supplies Kit, then loaded onto an Ion 318™ Chip v2 BC and sequenced using the Ion PGM™ Hi-Q™ Sequencing Kit and Ion PGM™ Instrument.
The resulting sequencing reads were downloaded from the Torrent Server and the sequences were analyzed using Geneious 8.1.9 (Biomatters Ltd., New Zealand) with reference to P. gingivalis W83 genome reference sequences (NCBI Reference Sequences NC_002950.2 and CP025932). All sequencing data are accessible from NCBI’s Short Reads Archive using BioProject accession number PRJNA561669.
Glycine enzyme-linked immunosorbent assay (ELISA)
Two 1 mL aliquots of P. gingivalis cultures were simultaneously collected at various time points, t = 0, 20, 30, 40, and 50 h post inoculation. One sample was centrifuged (4,000x g, 10 min, 4°C) to remove P. gingivalis cells, snap-frozen with liquid nitrogen and stored at −80°C for later glycine quantification whilst the other was used for the measurement of OD650. Similarly, 100 µL and 60 µL sample aliquots were collected for glycine quantification and OD650 measurements using a 50 mm window quartz cuvette (Variance, Cary) respectively, from P. gingivalis grown in 2 mL cultures.
Quantification of total free glycine using a glycine enzyme-linked immunosorbent assay (ELISA) BAE-2100 (Labor Diagnostika Nord GmbH & Co. KG, Germany) kit was performed as per the manufacturer’s instructions.
Results
RNA extraction and sequencing
P. gingivalis W50 RNA was extracted from three biological replicate cultures during log phase growth in OBGM and OB:CM. All RNA samples were of high quality as determined by the LabChip GXII RNA High Sensitivity assay which showed distinctive peaks and baseline separation of the 5S (~0.12 kbp), 16S (~1.5 kbp) and 23S (~2.9 kbp) rRNAs, and a calculated RNA Quality Score >8 for each sample (Supplementary Table S2). All RNA samples were then used for library preparation and RNA sequencing.
Raw data processing and analysis of P. gingivalis W50 differential gene expression in OB:CM relative to OBGM was performed using the latest annotation of the P. gingivalis W83 reference sequence, as a complete sequence annotation of the P. gingivalis W50 genome was not available. The sequences of the genomes of P. gingivalis strain W83 and strain W50 have been shown to be nearly identical, differing by only 20 single nucleotide polymorphisms [Citation43]. Each processed RNA sample produced ~20 million sequencing reads with >90% mapped to gene features, except for the third biological sample of P. gingivalis W50 grown in OBGM (OB_3) that produced ~9 million reads, as well as the highest duplication and lowest assignment rates (Supplementary Table S3, Supplementary Figure S1). Nevertheless, OB_3 was included in the differential gene expression analysis as the counts per million (CPM) standardization of OB_3, which was used for the fold-change analysis, was similar to the other samples (Supplementary Table S3) and was deemed suitable for inclusion in the bioinformatics analysis. A high percentage of duplication for each sample was expected given the high number of sequencing reads and the small genome size of P. gingivalis of 2.343 Mbp (Supplementary Table S3), indicating deep sequencing for each library. The unassigned reads were composed of ~5% that were ambiguous and 4% with no feature indicating regions of the genome that had not been annotated, whilst less than 1% of the reads were unmapped to the reference sequence (Supplementary Fig. S1).
Differential gene expression
Of the 1915 genes predicted to encode proteins in the P. gingivalis genome, transcripts were detected from 1866 genes (97.4%) in this study. Forty-five transcripts were significantly upregulated during P. gingivalis growth in OB:CM, while 87 transcripts were significantly downregulated, based on a false discovery rate (FDR) cut-off of <0.05 and fold-change log2 > 0.585 (>1.5-fold) (Supplementary Table S4, Supplementary Figure S2). Of the 132 genes differentially regulated greater than 1.5-fold, 29 encode proteins with no known function (hypothetical proteins, 22%), the majority of which (20) were down-regulated during growth in OB:CM relative to OBGM (Supplementary Table S4). Clusters of Orthologous Group (COG) functional categories were used to assign functions to the remaining 103 differentially regulated gene products. Of those that had a functional group assigned, 32 were involved in metabolism (COG categories C, G, E, F, H, I, P, Q), confirming that exposure to T. denticola cell-free conditioned medium altered P. gingivalis metabolism.
The differentially expressed metabolism-related genes were enriched in pathways corresponding to cofactors, prosthetic groups, biosynthesis of electron carriers, vitamin biosynthesis, one-carbon compound assimilation, reductant biosynthesis, amino acid biosynthesis and melibiose degradation (Supplementary Table S4). Whilst there were many single intermediary genes in the metabolic pathways that showed differential expression, multiple enzyme-encoding genes from the one pathway provided a higher confidence in deducing potential metabolic and physiological changes exhibited by P. gingivalis during growth in OB:CM. For example, a number of metabolic enzyme-encoding genes that were differentially expressed corresponded to P. gingivalis succinate catabolism and one-carbon metabolism. These pathways were examined to gain insight into possible metabolic synergisms between P. gingivalis and T. denticola.
Succinate catabolism pathways
P. gingivalis showed significant upregulation of genes encoding enzymes in succinate catabolism pathways during growth in OB:CM relative to OBGM (). These genes are all transcribed from the one operon comprising PG0692 – PG0687 with genes encoding succinate semialdehyde reductase (PG0689; [Citation44]) and 4-hydroxybutyryl-CoA dehydratase (PG0692) significantly upregulated by 1.66- and 2.09-fold, respectively, whilst genes encoding succinyl CoA reductase (PG0687; [Citation45]) and butyryl CoA transferase (PG0690; [Citation46]) were upregulated by 1.41- and 1.36-fold, respectively.
Figure 1. Succinate catabolism pathways of P. gingivalis W50. The metabolic genes and pathway reactions of P. gingivalis W83 were sourced from KEGG with reference to Yoshida et al. [Citation44,Citation45] and Sato et al. [Citation46]. Gene products with their gene expression fold changes are listed in red for each gene that showed upregulation during P. gingivalis W50 growth in OB:CM relative to OBGM. Gene products listed in black were encoded by genes that did not show differential expression. The dotted arrow indicates that the gene coding for a 4-hydroxybutyrate CoA transferase in P. gingivalis is currently unknown.
![Figure 1. Succinate catabolism pathways of P. gingivalis W50. The metabolic genes and pathway reactions of P. gingivalis W83 were sourced from KEGG with reference to Yoshida et al. [Citation44,Citation45] and Sato et al. [Citation46]. Gene products with their gene expression fold changes are listed in red for each gene that showed upregulation during P. gingivalis W50 growth in OB:CM relative to OBGM. Gene products listed in black were encoded by genes that did not show differential expression. The dotted arrow indicates that the gene coding for a 4-hydroxybutyrate CoA transferase in P. gingivalis is currently unknown.](/cms/asset/c7bf090a-e31f-4ba7-b4d2-28c23903003c/zjom_a_1808750_f0001_oc.jpg)
One-carbon metabolism
Within two inter-related metabolic pathways that contribute to folate-mediated one-carbon metabolism, several genes had altered expression (). In the folate biosynthesis pathway, genes encoding formyltetrahydrofolate synthetase (fhs; PG1321), the bi-functional 5,10-methylene-tetrahydrofolate dehydrogenase/cyclohydrolase (folD; PG1116) and formate transporter (PG0209) were down-regulated by 2.71-, 1.87-, and 3.21-fold, respectively, in OB:CM compared with OBGM (). This could result in less tetrahydrofolate (THF) intermediates being produced, such as N10-formyltetrahydrofolate (N10-fTHF), 5,10-methenyl-tetrahydrofolate (5,10-CH+-THF) and 5,10-methylene-tetrahydrofolate (5,10-CH2-THF).
Figure 2. P. gingivalis folate-mediated one carbon superpathway. The red and cyan proteins (with corresponding colored arrows) are encoded by metabolic genes that showed significant (>1.5-fold, FDR <0.05) upregulation and downregulation respectively. The black proteins are encoded by genes that showed no change in expression during P. gingivalis W50 growth in OB:CM, relative to OBGM. Not shown is the formate transporter PG0209 whose gene was downregulated 3.21-fold. The dotted arrows indicate enzymatic steps that have not been expanded for the sake of brevity.
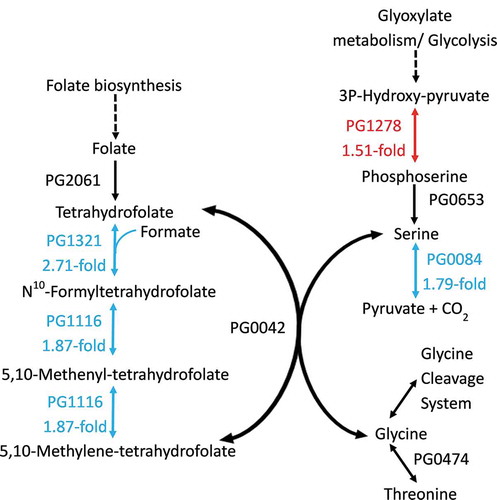
In the pathway for serine, glycine and threonine metabolism, the gene encoding phosphoserine aminotransferase (serC, PG1278) was up-regulated 1.51-fold, whilst the gene encoding the iron-sulfur-dependent L-serine dehydratase (sda, PG0084) was down-regulated 1.79-fold (). The overall effect on this metabolic pathway from the differential expression of these two gene products would be to build up cellular levels of serine, by increasing the levels of phosphoserine that can then be converted to serine and reducing the amount of serine that is converted to pyruvate and CO2. Using the serine and THF, serine hydroxymethyltransferase (GlyA, PG0042) can then catalyse the reversible conversion of serine and THF to glycine and 5,10-CH2-THF.
Proteases required for hydrolysis of glycine-containing peptides in OB:CM
Tan et al. [Citation22] showed that P. gingivalis W50 increased the rate of hydrolysis of glycine-containing peptides when grown in OB:CM compared with OBGM, contributing to an increased production of free extracellular glycine. However, RNA-seq examination of P. gingivalis W50 grown under these same conditions did not identify which proteases were potentially involved in this process. Although several protease-encoding genes such as rgpA (PG2024), rgpB (PG0506), kgp (PG1844), PG1788, PG0537, PG0418, and PG1548 were amongst the most highly expressed P. gingivalis transcripts, they were not differentially regulated; in fact, none of the P. gingivalis protease-encoding genes showed significant upregulation during growth in OB:CM compared with OBGM. Rather, expression of PG1542, that encodes PrtC, a Type I collagenase [Citation47,Citation48] was downregulated 2.3-fold in OB:CM (Supplementary Table S4). There were also significant reductions in the expression of genes encoding P. gingivalis ATP-dependent proteases PG0047 (2-fold), PG0620 (2-fold) and PG0010 (1.7-fold), and an annotated aminopeptidase P family protein (PG0889, 1.7-fold) (Supplementary Table S4). Therefore, a protein bioinformatics approach was taken to identify P. gingivalis peptidases that could be involved in peptide hydrolysis to release glycine.
Bioinformatic analyses of P. gingivalis putative peptidases
A search of the MEROPS Peptidase Database Release 12.0 resulted in identification of 64 putative peptidases and non-peptidase homologs in P. gingivalis W83 [Citation34]. Six non-peptidase homologs (PG2157, PG0634, PG0919, PG1768, PG0589, PG0328) were identified by the unacceptable replacement or absence of characteristic active site residues from their peptidase counterparts [Citation49] and these were excluded from further analyses. The NCBI database contained a further 10 unique peptidases not found in the MEROPS database including PrtT (PG1548) that is annotated as a T9SS C-terminal target domain-containing protein and Kgp (PG1844) that is annotated as a DUF2436 domain-containing protein; these were manually added to the P. gingivalis peptidase list. Based on this approach, a total of 68 unique potential proteases and peptidases were identified for further analyses (Supplementary Table S5).
Amino acid sequences of the selected peptidases were analyzed via PSORTb version 3.0, a bacterial protein subcellular localization prediction tool [Citation38] and the N-terminal signal peptide predictor program SignalP 4.0 [Citation36,Citation37]. Twenty-four P. gingivalis peptidases were predicted to be located in the cytoplasm, 17 in the cytoplasmic membrane, three peptidases in the periplasmic region, two in the outer membrane, one in the extracellular region and 21 proteases were of unknown location (Supplementary Table S5). Of the 43 proteases located outside the cytoplasm (Supplementary Table S5), 11 of these: RgpA, RgpB, Kgp, PorU, DPP5, periodontain, CPG70, DPP7, DPP11, PtpA, and PrtT, have been characterized previously and shown to have no specificity for glycine, whilst the remaining 32 predicted proteases have not been characterized.
In search of peptidase candidates that could potentially cleave glycine-containing peptides, PG1605 showed 35% overall sequence identity to Lactococcus lactis aminopeptidase C (PepC) which has been demonstrated to hydrolyze polyglycine and other glycine-containing peptides [Citation50], and so was selected as a target peptidase. Based on amino acid sequence analyses (not shown), both PG1605 and L. lactis PepC are members of the C1 family of clan CA cysteine peptidases. Other members of peptidase clan CA, such as ubiquitin-specific peptidases, bacteriocin-processing peptidases, and their homologs, cleave the peptide bonds of substrates that are C-terminal to double glycine residues (-Gly-Gly-) with high specificity. PG1788 was also predicted via sequence homology to be a C1 family cysteine peptidase in the MEROPS database and was thus selected as another potential target for investigation.
Of the identified putative proteases (Supplementary Table S5), microarray data from co-culture of P. gingivalis with T. denticola in a continuous culture system showed increased gene expression of PG0753 and PG0383, in comparison to monoculture [Citation22]. Therefore, the putative PrtQ collagenase (PG0753) that belongs to the U32 peptidase family was also selected for further analysis. PG0383, annotated as a regulated intramembrane proteolytic (RIP) metalloprotease RseP, was considered unlikely to be involved in free glycine release and was therefore not selected for further analysis. Although PG0753 was predicted to be located in the cytoplasm by PSORTb, it was selected for further investigation as the possibility of glycine-containing peptides being hydrolyzed intracellularly and the resultant free glycine being released into the environment could not be ruled out. Therefore, genes encoding PG0753, PG1605, and PG1788 were selected for inactivation. The resulting mutant strains were confirmed by PCR and whole-genome sequencing was performed to confirm the veracity of the mutants. All sequenced mutants had the appropriate target genes deleted from their genomes and strains ∆PG0753 and ∆PG1605 had no additional SNPs or mutations when compared to the laboratory wild type strain. However, two gene loci in ∆PG1788 each had a single nucleotide polymorphism that resulted in substitution of a highly conserved amino acid, as determined using COBALT (data not shown), which should not affect the resulting protein structure or function (Supplementary Table S6). The affected proteins were PG0867, a hypothetical protein, and PG1864, a TIR-domain containing protein.
Growth of P. gingivalis peptidase mutants
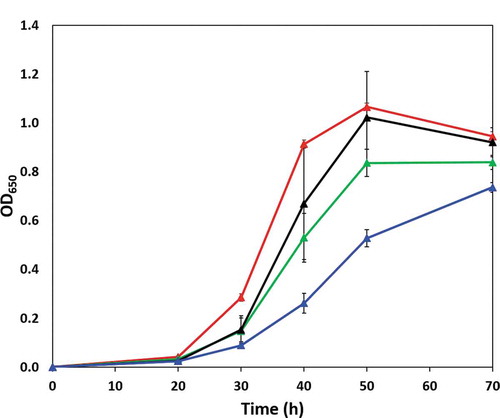
During growth in OBGM, P. gingivalis ΔPG0753 and ΔPG1605 peptidase mutants showed no growth differences relative to the wild type whilst ΔPG1788 showed an approximate 17% decrease in maximum cell density relative to the wild type (data not shown). In OB:CM, P. gingivalis wild type and the mutant strain ΔPG1605 displayed comparatively similar growth, with both strains achieving a maximum cell density equivalent to an OD650 of ~1.0 after 50 h of inoculation (). The growth of the ΔPG1788 strain was severely compromised, showing a significantly slower growth rate in OB:CM and reduced maximum cell density. Although ΔPG0753 grew better than the ΔPG1788 strain, it also showed a slower growth rate in comparison to the other strains ().
Figure 3. Growth of P. gingivalis wild type (red line) and ΔPG0753 (green line), ΔPG1605 (black line) and ΔPG1788 (blue line) strains in OB:CM. Bacterial strains were grown under anaerobic conditions at 37°C in static culture and the cell growth was determined by measurement of optical density at a wavelength of 650 nm (OD650). Data presented is the mean and standard deviation (n = 6) comprised of triplicates in two independent experiments.
Effects of P. gingivalis peptidase mutants on free glycine release
The free glycine concentration during P. gingivalis peptidase mutant and wild type strain growth at 30 to 40 h in OB:CM was determined. The rate of free glycine production as a function of P. gingivalis cell number was determined based on the change of free glycine concentration relative to the initial glycine concentration in OB:CM by P. gingivalis strains during the 10 h period of log phase growth (). P. gingivalis wild type demonstrated an approximately 25.1 ± 7.1 µmole increase in free glycine per 109 cells over the 10 h log growth phase in OB:CM (). Mutation of peptidase encoding genes resulted in changes in the free glycine production rate of these mutant strains. The ΔPG1605 strain, that had similar growth to the wild type in OB:CM, showed no difference in the rate of free glycine production (). There was a 16.3-fold reduction in the free glycine production by ΔPG0753 in OB:CM compared to wild type. Growth of ΔPG1788 strain in OB:CM from 30 to 40 h resulted in an overall negative rate of glycine production.
Table 1. Rate of free glycine production by P. gingivalis W50 wild type and mutant strains in OB:CM. The rate of free glycine production for each P. gingivalis W50 strain was determined from the slope of a linear regression line that indicated the change of free glycine against 109 P. gingivalis cells per h, as measured over a 10 h time period during the time interval of 30 to 40 h of growth.
Discussion
The close association of P. gingivalis and T. denticola in subgingival plaque and their increase in abundance prior to disease progression strongly supports a polymicrobial etiology for chronic periodontitis and cooperativity between the two species [Citation1]. When grown together, P. gingivalis and T. denticola share metabolites, resulting in an overall increased metabolic capability and increased fitness to survive competitive environments [Citation22,Citation51,Citation52]. In order to better understand the breadth of P. gingivalis metabolic pathways that respond to T. denticola, we initially determined the effect of T. denticola conditioned growth medium on the P. gingivalis transcriptome.
The high depth of coverage obtained from this RNA-seq analysis represents a comprehensive transcriptomic profile of P. gingivalis during growth in medium enriched with T. denticola produced effector molecules. The T. denticola conditioned medium used in this experiment resembled a condition where T. denticola was present in high abundance and pre-modified the environment. P. gingivalis showed significant upregulation of genes encoding enzymes in succinate utilization pathways during growth in OB:CM, relative to OBGM () suggesting the presence of succinate in the T. denticola conditioned medium. This supports an earlier study that inferred cross-feeding of succinate from T. denticola to P. gingivalis by detection of succinate in T. denticola conditioned medium and supplementation of succinate to increase P. gingivalis growth [Citation51]. These genes, however, were not detected previously as being differentially expressed in a microarray analysis of P. gingivalis W50 and T. denticola grown in OBGM co-culture versus monoculture [Citation22], possibly due to the comparatively lower sensitivity of microarrays.
Succinate catabolism leads to butyrate and propionate biosynthesis, short-chain fatty acids known to be toxic to mammalian cells [Citation53]. An increase of these highly cytotoxic products contributes to the exacerbation of disease [Citation54,Citation55]. Thus, the metabolic exchange of succinate between T. denticola and P. gingivalis could lead to increased cytotoxic end products and contribute to disruptive effects on host cell activities, as well as immune defense mechanisms.
As succinate is the catabolic end-product of many fermentative pathways, bacteria that utilize succinate during growth in complex polymicrobial biofilms could gain a selective advantage. For example, microbiota-derived succinate enabled Salmonella enterica serovar Typhimurium and Clostridium difficile to colonize and infect the gut of gnotobiotic mice [Citation56,Citation57]. Co-culture of T. denticola and P. gingivalis in a nutrient-rich medium in continuous culture resulted in a 54% increase in P. gingivalis cell density relative to monoculture growth in the same medium [Citation22]. This increase in P. gingivalis cell density was likely mediated by succinate cross-feeding. Other bacterial species that have close associations with P. gingivalis, such as Streptococcus gordonii, Fusobacterium nucleatum, Tannerella forsythia, Actinomyces species and A. actinomycetemcomitans [Citation58–Citation63] are also capable of producing succinate, suggesting that succinate could be an important carbon source for P. gingivalis in the oral polymicrobial biofilm. It has also been postulated that succinate could serve as a partial alternative to amino acid anaerobic respiration to rescue cell growth under heme-limited conditions [Citation25,Citation64]. Thus, the succinate catabolic pathway might enable metabolic interaction of P. gingivalis with other oral bacterial species that produce succinate, as well as increasing nutrient utilization efficiency in the polymicrobial community.
Increased gene expression of a serine biosynthesis enzyme and decreased gene expression of a putative serine degradation pathway suggest increased glycine synthesis from serine during P. gingivalis growth in OB:CM. This implies that P. gingivalis reduces its use of environmental glycine by altering its metabolism to the use of intracellular glycine produced from serine when co-cultured with T. denticola. In a similar experiment, Tan et al. [Citation22] reported that following growth of P. gingivalis in OB:CM for 48 h, the increase in free glycine was accompanied by a decrease in peptide-bound glycine, indicating that the increase in free glycine was due to increased hydrolysis of glycine-containing peptides in the medium, not from the de novo synthesis of glycine by P. gingivalis. T. denticola glycine utilization pathways and N10-fTHF-generating enzyme-encoding genes showed significant upregulation via microarrays when co-cultured with P. gingivalis [Citation22], whilst the P. gingivalis genes coding for PG1116 and PG1321 in the 5,10-CH2-THF generating pathway were significantly down-regulated, supporting the RNA-seq data. The reduced expression of genes encoding THF intermediates in P. gingivalis also implies a syntrophic exchange of folate intermediates from T. denticola to P. gingivalis. Together these processes help to partition resources and engage synergistic interactions between these two species.
The hydrolysis of peptides for the release of free glycine by P. gingivalis when grown in association with T. denticola and the use of that glycine as a major energy source by T. denticola is one of the mechanisms that underpin the synergistic relationships between these two species [Citation22]. Tan et al. [Citation22] hypothesized that cooperative proteolytic activities of P. gingivalis and T. denticola were mechanisms for the increased production of free glycine, which in turn enhanced T. denticola growth when the bacteria were co-cultured. Their microarray analysis found two P. gingivalis genes significantly upregulated by 1.2 fold that encoded putative proteases [Citation22]. The current RNA-seq analysis, however, offered no indication of the specific P. gingivalis protease-encoding genes responsible for the increase in free glycine, as there was no significant upregulation of any protease-encoding genes. The study by [Citation22], used coculture of T. denticola and P. gingivalis, thereby enabling physical contact between the species which may have altered gene expression. The [Citation22], study also utilised a comparative microarray analysis that can be subject to cross-hybridization bias. The current study determined the effect of cell-free conditioned T. denticola medium on P. gingivalis global gene expression using RNA-seq which is considered to be unbiased. The protease-encoding genes were generally highly expressed as determined by RNA-seq analysis, suggesting that the increased glycine hydrolysis by P. gingivalis grown in T. denticola conditioned medium was due to the different substrates present, i.e. the T. denticola produced peptides that could be further hydrolyzed by P. gingivalis. We therefore used a primarily bioinformatics approach to identify putative P. gingivalis proteases that may be involved in free glycine release. Three putative proteases were selected for study, PG0753, PG1605 and PG1788, and deletion mutants of each produced. PG0753 was predicted with high confidence to be localized in the cytoplasm, whereas PG1605 and PG1788 contain N-terminal signal peptides and are predicted to be localized non-cytoplasmically [Citation36]. PG1788 was found in the lumen of P. gingivalis OMVs, which indicated that it is contained in the cellular periplasm [Citation65]. The ΔPG1605 strain showed a similar rate of free glycine production to the wild type suggesting that the PG1605 protease was not involved in the release of free glycine from peptides. Deletion of either PG1788 or PG0753 (that encodes a putative PrtQ collagenase-like peptidase) resulted in a dramatic decrease in the rate of free glycine production compared with the wild type. These observations suggest that both PG1788 and PG0753 play roles in the cooperative hydrolysis of glycine-containing peptides and that the absence of these proteins has altered the process of peptide degradation and free glycine release. The putative collagenase PrtQ was not detected in the P. gingivalis OMV proteome [Citation65,Citation66] indicating that it likely has a cytoplasmic location. Together this suggests that the activities of PG1788 and PG0753 are complementary to T. denticola proteases, which in turn allows further processing of T. denticola generated glycine-containing peptides. These P. gingivalis peptidases are not necessarily specific for cleavage at glycine residues, although our bioinformatic analyses indicate that this is a probability. It is also likely that they perform hydrolytic functions that benefit P. gingivalis growth in the absence of T. denticola as seen by the slight inhibition of ΔPG1788 strain growth in OBGM. This could explain why there was a seemingly high constitutive expression of these peptidase encoding genes.
As P. gingivalis does not preferentially utilize glycine [Citation67], the production of exogenous glycine by P. gingivalis benefits T. denticola. These data support the concept that metabolic cooperativity between members of the periodontal polymicrobial community can increase the accessibility of nutrients in the environment and thus indirectly inflict destruction of the tooth supporting tissue structures. Type IV collagen is composed of the repetitive triplet sequence Gly–X–X in the collagenous domain, where X represents a variety of other amino acids. Type IV collagen is enriched in the gingival epithelial basement membranes [Citation68]. The involvement of PG0753 and PG1788 in the release of free glycine during P. gingivalis growth in OB:CM might extend their physiological roles to the degradation of proteinaceous substrates for T. denticola. This finding highlights the metabolic cooperativity between P. gingivalis and T. denticola resulting in an increased release of nutrients for their mutual growth in a competitive environment.
Supplemental Material
Download MS Word (207.3 KB)Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Byrne SJ, Dashper SG, Darby IB, et al. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 2009;24:469–11.
- Duran-Pinedo AE, Chen T, Teles R, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. Isme J. 2014;8:1659–1672.
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122.
- Jiao Y, Hasegawa M, Inohara N. The role of oral pathobionts in dysbiosis during periodontitis development. J Dent Res. 2014;93:539–546.
- Orth RKH, O’brien-Simpson NM, Dashper SG, et al. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. 2011;26:229–240.
- Wang J, Qi J, Zhao H, et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843.
- Dashper SG, Cross KJ, Slakeski N, et al. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19:50–56.
- Guo YH, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44.
- O’brien-Simpson NM, Veith PD, Dashper SG, et al. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Peptide Sci. 2003;4:409–426.
- Ruggiero S, Cosgarea R, Potempa J, et al. Cleavage of extracellular matrix in periodontitis: gingipains differentially affect cell adhesion activities of fibronectin and tenascin-C. Biochim Biophys Acta Mol Basis Dis. 2013;1832:517–526.
- Nemoto TK, Ohara-Nemoto Y. Exopeptidases and gingipains in Porphyromonas gingivalis as prerequisites for its amino acid metabolism. Jpn Dent Sci Rev. 2016;52:22–29.
- Hromić-Jahjefendić A, Jajčanin Jozić N, Kazazić S, et al. A novel Porphyromonas gingivalis enzyme: an atypical dipeptidyl peptidase III with an ARM repeat domain. PLoS One. 2017;12:1–27.
- Rea D, Lambeir A-M, Kumagai Y, et al. Expression, purification and preliminary crystallographic analysis of dipeptidyl peptidase IV from Porphyromonas gingivalis.. Acta Crystallographica: Section D. 2004;60:1871–1873.
- Ohara-Nemoto Y, Rouf SMA, Naito M, et al. Identification and characterization of prokaryotic dipeptidyl-peptidase 5 from Porphyromonas gingivalis. J Biol Chem. 2014;289:5436–5448.
- Banbula A, Yen J, Oleksy A, et al. Porphyromonas gingivalis DPP-7 represents a novel type of dipeptidylpeptidase. J Biol Chem. 2001;276:6299–6305.
- Ohara-Nemoto Y, Shimoyama Y, Kimura S, et al. Asp- and glu-specific novel dipeptidyl peptidase 11 of Porphyromonas gingivalis ensures utilization of proteinaceous energy sources. J Biol Chem. 2011;286:38115–38127.
- Banbula A, Mak P, Bugno M, et al. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis - A novel enzyme with possible pathological implications for the development of periodontitis. J Biol Chem. 1999;274:9246–9252.
- Nemoto TK, Ohara-Nemoto Y, Bezerra GA, et al. A Porphyromonas gingivalis periplasmic novel exopeptidase, acylpeptidyl oligopeptidase, releases n-acylated di- and tripeptides from oligopeptides. J Biol Chem. 2016;291:5913–5925.
- Chen YY, Cross KJ, Paolini RA, et al. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J Biol Chem. 2002;277:23433–23440.
- Masuda K, Yoshioka M, Hinode D, et al. Purification and characterization of arginine carboxypeptidase produced by Porphyromonas gingivalis. Infect Immun. 2002;70:1807–1815.
- Bradshaw DJ, Homer KA, Marsh PD, et al. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology. 1994;140:3407–3412.
- Tan KH, Seers CA, Dashper SG, et al. Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses. PLoS Pathog. 2014;10:e1003955.
- Veith PD, Dashper SG, O’brien-Simpson NM, et al. Major proteins and antigens of Treponema denticola. Biochim Biophys Acta. 2009;1794:1421–1432.
- Bhagwat AA, Phadke RP, Wheeler D, et al. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J Microbiol Methods. 2003;55:399–409.
- Dashper SG, Ang CS, Veith PD, et al. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J Bacteriol. 2009;191:1044–1055.
- Nelson KE, Fleischmann RD, Deboy RT, et al. Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601.
- Tsyganov K, Perry AJ, Archer SK, et al. RNAsik: A Pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J Open Source Software. 2018;3:583.
- Powell DR (2015). Degust: interactive RNA-seq analysis.
- Alm EJ, Huang KH, Price MN, et al. The MicrobesOnline Web site for comparative genomics. Genome Res. 2004;15:1015–1022.
- Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480.
- Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462.
- Marchler-Bauer A, Bo Y, Han L, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203.
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226.
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;191(D1):D343–D350.
- Rawlings ND, Barrett AJ, Thomas PD, et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2017;46:D624–D632.
- Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432.
- Petersen TN, Brunak S, Von Heijne G, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786.
- Yu NY, Wagner JR, Laird MR, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615.
- Ho SN, Hunt HD, Horton RM, et al. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59.
- Fletcher HM, Schenkein HA, Morgan RM, et al. Virulence of a Porphyromonas gingivalis W83 mutant defective in the PrtH gene. Infect Immun. 1995;63:1521–1528.
- Seers CA, Slakeski N, Veith PD, et al. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found. Porphyromonas Gingivalis J Bacteriol. 2006;188:6376–6386.
- Bélanger M, Rodrigues P, Progulske-Fox A. Genetic Manipulation of Porphyromonas gingivalis. Curr Protoc Microbiol. 2007;5:13C.12.11–13C.12.24.
- Dashper SG, Mitchell HL, Seers CA, et al. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48.
- Yoshida Y, Sato M, Nagano K, et al. Production of 4-hydroxybutyrate from succinate semialdehyde in butyrate biosynthesis in Porphyromonas gingivalis. Biochim Biophys Acta Gen Subj. 2015;1850:2582–2591.
- Yoshida Y, Sato M, Kezuka Y, et al. Acyl-CoA reductase PGN_0723 utilizes succinyl-CoA to generate succinate semialdehyde in a butyrate-producing pathway of Porphyromonas gingivalis. Arch Biochem Biophys. 2016;596:138–148.
- Sato M, Yoshida Y, Nagano K, et al. Three CoA transferases involved in the production of short chain fatty acids in Porphyromonas gingivalis. Front Microbiol. 2016;7:1146.
- Kato T, Takahashi N, Kuramitsu HK. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J Bacteriol. 1992;174:3889–3895.
- Takahashi N, Kato T, Kuramitsu HK. Isolation and preliminary characterization of the Porphyromonas gingivalis prtC gene expressing collagenase activity. FEMS Microbiol Lett. 1991;68:135–138.
- Rawlings ND, Morton FR. The MEROPS batch BLAST: A tool to detect peptidases and their non-peptidase homologues in a genome. Biochimie. 2008;90:243–259.
- Mistou MY, Gripon JC. Catalytic properties of the cysteine aminopeptidase PepC, a bacterial bleomycin hydrolase. Biochim Biophys Acta. 1998;1383:63–70.
- Grenier D. Nutritional interactions between 2 suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. 1992;60:5298–5301.
- Ng HM, Kin LX, Dashper SG, et al. Bacterial interactions in pathogenic subgingival plaque. Microb Pathog. 2016;94:60–69.
- Singer RE, Buckner BA. Butyrate and propionate: important components of toxic dental plaque extracts. Infect Immun. 1981;32:458–463.
- Kuniyasu O, Tomoko K-O. Effects of butyric acid on the periodontal tissue. Jpn Dent Sci Rev. 2009;45:75–82.
- Scragg MA, Cannon SJ, Williams DM. Comparative cytotoxic effects of short-chain fatty acids produced by periodontal pathogens on two cultured fibroblast lines. Microb Ecol Health Dis. 1994;7:83–90.
- Ferreyra JA, Wu KJ, Hryckowian AJ, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777.
- Spiga L, Winter MG, Zhu W, et al. An oxidative central metabolism enables salmonella to utilize microbiota-derived succinate. Cell Host Microbe. 2017;22:291–308.
- Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148:467–472.
- Kuboniwa M, Houser JR, Hendrickson EL, et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2017;2:1493–1499.
- Kuboniwa M, Tribble GD, James CE, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139.
- Maeda K, Tribble GD, Tucker CM, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164.
- Nagayama M, Sato M, Yamaguchi R, et al. Evaluation of co-aggregation among Streptococcus mitis, Fusobacterium nucleatum and Porphyromonas gingivalis. Lett Appl Microbiol. 2001;33:122–125.
- Simionato MR, Tucker CM, Kuboniwa M, et al. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun. 2006;74:6419–6428.
- Baughn AD, Malamy MH. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci U S A. 2002;99:4662–4667.
- Veith PD, Chen -Y-Y, Gorasia DG, et al. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. 2014;13:2420–2432.
- Veith PD, Luong C, Tan KH, et al. Outer membrane vesicle proteome of Porphyromonas gingivalis is differentially modulated relative to the outer membrane in response to heme availability. J Proteome Res. 2018;17:2377–2389.
- Takahashi N, Sato T, Yamada T. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J Bacteriol. 2000;182:4704–4710.
- Gultekin SE, Yucel OO, Senguven B, et al. Effect of periodontal inflammation on collagen IV, laminin 5, MMP-2, and MMP-9 expression in gingival tissues of diabetic rats. Austin Dent Sci. 2017;2:1013.
