ABSTRACT
In this study, 181 healthy individuals, including 29 couples, were analysed regarding oral yeast colonization using a culture-based approach. Results showed that 39% of the individuals were yeast carriers, 89% being colonized with Candida albicans, 5% with C. guilliermondi, 3% with C. lusitaniae and 3% with C. parapsilosis.
Sixty-two percent of the couples had at least one member colonized. Colonization and CFU counts were higher in the couples´ group. Eighty percent of the volunteers were colonized with C. albicans strains with only one CAI genotype, while two but similar CAI genotypes inhabited the oral cavity of the remaining 20% individuals. The same CAI genotypes were found in 66.6% of the couples when both were colonized.
Our results indicate that the intimacy among couples increases the probability of heavy cross-colonization, which is potentiated when one member of the couple is a smoker.
Introduction
Advances in metagenomics allowed a more complete understanding of the different microorganisms that inhabit on or within human tissues and fluids. The number and type of these microbes vary with age, diet and personal hygiene habits, being collectively referred to as the normal microbiota of the human body. In particular, the oral cavity is colonized by a highly diverse set of microbes from different species, including bacteria, protozoa and fungi, and also by viruses [Citation1,Citation2]. They are always present in metabolic inter-dependent and highly organized polymicrobial communities on the surface of teeth, gums and tongue. The bacterial microbiome makes up over 99% of the total microbial counts and is coined as the core microbiome, while the remaining, less abundant and more diverse microbiota forms the ‘rare biosphere’ [Citation3]. The mycobiome represents a significant proportion of the rare biosphere. In culture-independent studies, 85 fungal genera were reported in healthy hosts, with Candida, Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium and Cryptococcus being most predominant [Citation4]. In culture-dependent studies from salivary samples, the most predominant moulds were Penicillium spp., Aspergillus spp. and Cladosporium, while the most abundant yeasts were Candida spp. and Rhodotorula spp [Citation5]. In these studies, Candida species were the most prevalent and existed commensally in the oral cavity of up to 70% of healthy individuals [Citation6,Citation7]. In fact, Candida species like C. albicans, C. dubliniensis, C. parapsilosis and C. glabrata belong to the normal microbiota of the skin and mucosal surfaces of healthy individuals, not only in the oral cavity, but also in the gastrointestinal tract and the vagina [Citation8].
Despite its relatively low abundance, the impact of the mycobiome in human health and disease is well established [Citation9, Nguyen 2015]. Reduced host defence or inadequate clearance promotes Candida opportunistic infection, leading to a spectrum of oral mucosal disorders, from simple to chronic candidosis [Citation10]. In severely immunocompromised patients, such as those under chemotherapy, patients with AIDS or with endocrinal or blood diseases, the commensal fungi can proliferate, penetrate the bloodstream and disseminate throughout the human body causing life-threatening infections [Citation8,Citation11,Citation12].
The increasing number of infections caused by Candida spp. and the appearance of strains resistant to conventional treatment, led to the development of new approaches to effectively identify different Candida spp., among which microsatellite typing was used [Citation13,Citation14]. The C. albicans microsatellite CAI was first described in 2003 and has proved to be an efficient method for strain differentiation with a discriminatory power of 0.97 [Citation13].
Most studies on the prevalence of C. albicans in the human oral cavity have been performed in individuals with different pathologies or belonging to risk groups. It is widely described that C. albicans predominates among Candida spp. in the oral cavity, but little is known about its genotypic diversity, stability and transmissibility between individuals. Hence, the two main goals of the present study were to determine yeast colonization in the oral cavity of healthy individuals and to evaluate patterns of C. albicans transmission between couples, and their stability over time. Moreover, correlations with gender and smoking habits were also studied.
Material and methods
Participants and sampling
One hundred and eighty-one healthy individuals volunteered to participate in this study, including students and staff of the University of Minho. The study had been approved by the Ethics Committee for Research in the Life and Health Sciences of the University. All the participants signed an informed consent, and data regarding food habits, oral hygiene and smoking habits were registered. The ages ranged from 18 to 60 years, with 116 females and 65 males. Exclusion criteria included antibiotic or steroid therapy within the 3 months preceding the collection. To maintain the anonymity of the donors a number was assigned to each. Members of a couple received the same number, being distinguished only by the use of the letters ‘a’ (female) and ‘b’ (male). To assess individual variation of oral yeast colonization over time, a follow-up of 10 participants was carried out approximately 1 year after the first sampling. The selection criteria for the follow-up participants included identification of C. albicans in the first sampling and continued participant availability.
A saliva sample (approximately 1 mL) from each participant was collected between 10 a.m. and 12 a.m., to avoid variation in salivary flow rates, and directly plated and spread on Yeast extract glucose chloramphenicol agar (YGC) with 50 mg/L chloramphenicol to inhibit bacterial growth. The plates were incubated at 37°C for 5 days. The cultures were examined every day, and the number of fungal colonies was recorded in colony-forming units per millilitre (CFU/mL).
Phenotypic and molecular identification of the isolates
The yeast colonies (a maximum of 50 per individual) were sub-cultured on CHROMagar Candida to facilitate the presumptive identification of Candida yeast species. Following this preliminary phenotypic characterization, isolates identified on the CHROMagar as C. albicans and the non-albicans species were further analysed through the use of molecular techniques. From each individual, a maximum of eight C. albicans colonies were randomly selected for CAI analysis, and one non-albicans colony of each different colour was chosen for ITS species identification. Bacteria isolates were not identified in this study.
DNA extraction, amplification, and sequencing conditions
Before DNA isolation, cells were cultivated overnight on Sabouraud medium at 30°C and DNA was extracted according to [Citation15]. Isolates presumptively identified as C. albicans were genotyped with the CAI species-specific microsatellite marker. CAI locus was amplified with primers 5´-ATG CCA TTG AGT GGA ATT GG-3´ (forward) and 5´-AGT GGC TTG TGT TGG GTT TT-3´ (reverse), according to [Citation13]. PCR products were analysed by capillary electrophoresis in an ABI 310 genetic analyser (AB Applied Biosystems) and fragment sizes were determined automatically using the GeneScan 3.1 Analysis software. Alleles were designated according to the number of trinucleotide repeats [Citation13].
Molecular identification of non-albicans isolates was performed by sequencing the internal transcribed spacer (ITS) regions of ribosomal RNA genes. Sequence analysis was carried out using primers ITS1 (5ʹ TCCGTAGGTGAACCTGCGG 3ʹ) and ITS4 (5ʹ TCCTCCGCTTATTGATATGC 3ʹ), according to [Citation16], and PCR products purified using the commercial Kit GenElute® PCR Clean-up (SIGMA). Sequences were edited with the Sequencer version 4.9 software package (Genes Codes Corporation), aligned with MEGA-X software [Citation17] and compared by BLAST with sequences available from NCBI GenBank and ISHAM-barcoding database.
Statistical analyses
Comparisons between categorical variables were performed using the Chi-square statistical test with Yates correction to compare between different conditions. Variables were compared using a 2-tailed t-test in order to evaluate significance associated with higher or lower CFUs values. A p value <0.05 was considered as significant. To evaluate if the statistically significant difference was correlated with helpful information in decision-making, the Cohen´s d value was calculated [Citation18]. A d value below 0.1 indicates trivial effect, a value between 0.1 and 0.3 indicates a small effect, between 0.3 and 0.5 a moderate effect, and a value higher than 0.5 indicates a large difference effect.
The C. albicans CAI genotypes were compared according to their Shriver´s genetic distance, using PowerMarker v3.25 software [Citation19]. A distance tree was constructed applying clustering with the unweighted pair group method with arithmetic means, using the MEGA-X software [Citation17]. Comparison between groups was tested with the log-likelihood (G) statistic, using CAI genotypes [Citation20]. Markov chain (MC) algorithm (1,000 iterations) was used to estimate, without bias, the exact p-value of this test [Citation21]. These calculations were performed with Genepop sotware, version 4.2 [Citation22].
Results
Yeast colonization in the oral cavity of healthy individuals
As previously reported, 181 healthy donors participated in this study, including students and staff of the University of Minho, among which 29 were couples. Results showed that 39% (70 out of 181) of the individuals carried yeasts in the oral cavity. The number of CFUs found in saliva samples was variable, with eight individuals showing a countless number of colonies (>300 CFUs) in the saliva, while others presented only one or two colonies. The majority of the individuals (55.7%) presented between one to 20 colonies ().
Figure 1. Distribution of the total individuals (A), males and females (B), and smokers and non-smokers (C) according to classes of yeast CFUs/mL in the saliva.
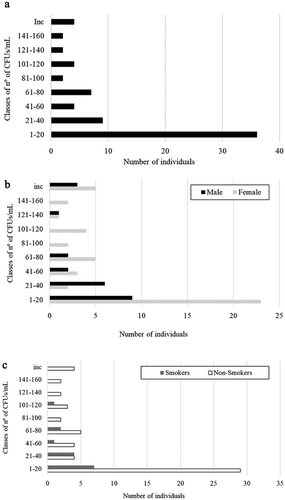
From the population studied, 116 were females and 65 were males. Although the number of males and females per classes of CFUs was different (), the percentage of colonized individuals did not differ significantly according to gender (40.5% of females and 35.4% of males were carriers), nor the level of colonization (number of CFUs) (not significant; p < 0.05). For comparative purposes, individuals with countless number of colonies were assigned as having 300 CFUs.
Eighteen percent (n = 34) of the participants were smokers. When correlating colonization with smoking habits (), the results showed that the number of colonized non-smokers was higher (n = 55) than colonized smokers (n = 15), but the differences were not statistically significant (not significant for p < 0.05).
To assess whether the intimacy results in cross-transmission of microorganisms, 29 couples were analysed. Interestingly, 11 couples (37.9%) showed no yeast colonization, nine (31.0%) presented colonization in only one member (five males and four females), and nine (31.0%) in both members of the couples.
Results were particularly significant when comparing colonized and non-colonized individuals between the couples and the remaining population (Yates chi-square is 16.09, p = 0.00006). Also, in the couples´ group, the average CFU number was 31.7, while in the remaining single individuals it was 13.9 CFUs (t-value is 2.15, p = 0.03, significant at p < 0.05, Cohen´s d = 0.36). CFU counts were also higher in the couples´ group with smoking habits (CFUs average = 26.3) than in single individuals (CFUs average = 6.1) with smoking habits (t-value is 2.23, p = 0.03, significant at p < 0.05, Cohen´s d = 1.23), contrary to CFUs number in non-smokers between the two groups (t-value is 1.73, p = 0.08, not significant at p < 0.05). These results indicate that the probability of colonization, together with high amount of yeast cells in the oral cavity, is moderately higher within the couples, but is largely potentiated by smoking habits. Colonization was likely not associated with hygiene habits or diet, because all the participants declared to follow regular oral hygiene habits and balanced diet.
Identification of yeasts isolated from the oral cavity of healthy individuals
Differentiation of yeast isolates was performed on CHROMagar Candida medium. In this medium, C. albicans isolates formed yellow-green to blue-green colonies, while the remaining species presented a wide range of colours, from white to dark purple. However, since the colours may differ according to the strain, sequencing of the ribosomal internal transcribed spacer region was performed to identify non-albicans isolates, while C. albicans isolates were amplified with a CAI species-specific CAI marker [Citation13]. The results confirmed that from the 70 samples positive for yeast species, 84.2% (n = 59) presented only C. albicans alone, 5.7% (n = 4) a mixed culture of C. albicans and other yeast species, 5.7% (n = 4) non-albicans species only, 2.8% (n = 2) C. albicans and bacteria, and in only one individual (1.4%) C. albicans co-inhabited the oral cavity with other yeast species and bacteria. Regardless of the presence of chloramphenicol in the culture medium, bacteria isolates were identified in five individuals, and from those, three also presented yeast colonization.
Candida guilliermondi, C. lusitaniae, C. parapsilosis, and Rhodotorula mucilaginosa were among the other yeast species identified (). The results also revealed that only one individual was colonized with R. mucilaginosa instead of Candida spp. None of the volunteers were colonized with more than two yeast species.
Table 1. Number and percentage of individuals with different colonization patterns in the oral cavity of the study subjects.
Genotyping of C. albicans isolates using CAI microsatellite
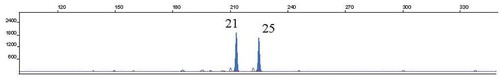
As indicated above, 66 individuals were colonized with C. albicans strains which were genotyped using the CAI microsatellite marker. To identify different clones within the sample, a maximum of eight different colonies were genotyped. Results showed that a total of 39 distinct C. albicans genotypes were identified in isolates from the oral cavity of the 66 C. albicans colonized volunteers. From the 39 different genotypes identified, 13 were shared by more than one individual. The most frequent genotype found was 21–25 (), present in 11 participants, followed by the genotype 25–25, shared by eight individuals (Supplementary data).
Figure 2. GeneScan profile of the most frequent CAI genotype (21–25). Electropherogram of PCR products obtained from a strain isolated from participant S019, showing allele 21 (213 base pairs) and allele 25 (225 base pairs).
Fifty-three participants (80%) were colonized by strains with only one CAI genotype and 13 (20%) with two genotypes. The different genotypes isolated from the same individual were probably due to rearrangements at the CAI locus, such as slippage (in seven individuals) or loss of heterozygosity (LOH, in five individuals). The slippage is observed when one or two microsatellite repeated units are added or reduced in one or both alleles, as observed in strains isolated from individual S148, while the LOH is observed when one of the alleles is missing, as shown in strains isolated from the individual S031 (). Considering the internal structure of this microsatellite, a block of 10 repeat units distinguished alleles of group II (16 to 29) from group III (>30) [Citation23]. This transition followed by rearrangements could be observed in strains isolated from the volunteer S142 between alleles 27 (group II) and 38 (Group III) (). In couples sharing C. albicans colonization, 55.6% (S035, S046, S071, S095 and S128) showed isolates with the same CAI genotype, 33.3% (S017, S036 and S054) presented isolates with different CAI genotypes, and only one couple (11.1%, S010) presented a mix of genotypes derived from slippage events, such as microvariations ().
Table 2. Individuals with different C. albicans CAI genotypes.
Table 3. Candida albicans CAI genotypes and species identified in the couples.
No significant difference was observed in CAI genotypes or alleles between couples in which both members were colonized, and couples in which only one member was colonized (p > 0.05). Interestingly, in couples colonized with species other than C. albicans, only the S054 couple also shared the non-albicans species, C. guillermondii. Couples S015 and S095 did not share the non-albicans species, C. parapsilosis or C. lusitaniae ().
shows a dendogram correlating all CAI genotypes. For the dendrogram construction, in case of more than one CAI genotype present, only the most frequent was used. Results showed that there is no correlation between CAI genotypes and gender or smoking habits (p > 0.05).
Stability of C. albicans genotypes over time
To evaluate whether C. albicans colonization is stable over time, 10 C. albicans carriers were selected for a second sampling, approximately 1 year after the first analysis. The collected isolates were again identified and C. albicans CAI genotypes compared with the first sampling. Results showed that the CAI genotypes of the colonizing strains were stable over 1 year (). Indeed, in seven individuals, the CAI genotype was the same as in the first analysis and, in subject S032, only one of the genotypes identified in the first analysis persisted after 1 year. Curiously, only individuals of the couple S054, lost yeast colonization after 1 year.
Table 4. Comparison of the genotypes found in the first and in the second sampling performed approximately one year later.
Discussion
The impact of yeast colonization on human health and disease is well established [Citation9,Citation24]. In this study, culture-dependent identification methods were used to evaluate oral yeast colonization in healthy individuals. Thirty-nine percent of the volunteers were colonized by yeasts, a number within the broad range reported in the literature: from 18.5% [Citation25] to 92.5% [Citation5]. This large range may be due to diverse potential confounding factors, including age, ethnicity, presence of neglected co-infection, as well as microbiological culture variables, including type of sample collected, culture media, time or temperature of incubation.
As observed in other studies [Citation5,Citation25], gender and oral hygiene habits (tooth brushing frequency) did not affect the percentage of yeast carriage or CFU counts. However, [Citation26], in a crowdsourced population study reported that youth oral microbiomes are affected by gender and adult oral microbiomes by oral health habits (flossing frequency).
The relationship between smoking habits and oral yeast colonization has been a matter of debate. Some studies have shown that oral Candida prevalence is higher in smokers than in non-smokers [Citation7], while other recent reports were inconclusive [Citation27]. In the present work, smoking did not appear to increase yeast oral colonization or CFU counts in single volunteers, however, higher colonization and CFU counts were correlated with smoking in couples. The transferring of microorganisms through kissing, together with the described immunosuppressive nature of tobacco [Citation28], may have contributed to this correlation. However, selection mechanisms resulting from a shared lifestyle, environment, or genetic factors from the host could also contribute. Three out of four participants with countless number of colonies (>300 CFUs) were within the couple non-smoking group.
In the present study, we observed that C. albicans was by far the most prevalent species (94.2%) among yeast carriers, which is also widely reported in the literature [Citation29–32]. Indeed, in 84.2% of the volunteers, C. albicans was isolated alone and in 10% in a mixed culture, including chloramphenicol resistant bacteria. Curiously, C. albicans was identified together with C. guillermondii and C. lusitaniae, but not with C. parapsilosis or R. mucilaginosa. A lower competitiveness between C. guillermondii and C. lusitaniae with C. albicans may account for this result. Candida tropicalis and C. glabrata are frequently identified in oral samples, however, in our survey, these species were not found. Indeed, it is described that C. glabrata is innately resistant to antifungal agents [Citation33], so the selection of volunteers that were not being treated with any antimicrobial compound may explain this result. The composition of the studied population (mostly young healthy adults) may also have influence on the oral population because functions of the innate immune system are downregulated in aged healthy volunteers compared with healthy young volunteers [Citation34].
Another species frequently identified in oral samples is C. dubliniensis, but this species was also not isolated in our study. However, to the best of our knowledge, C. dubliniensis has not been identified yet in oral samples in Portugal [Citation35,Citation36].
Since the mode of reproduction of C. albicans is essentially clonal, it is described that variations in biological properties may be traced by only one variable marker [Citation37]. However, due to evidence that some recombination may also occur in C. albicans, analysis of loci associated with known biological properties would provide more accurate data [Citation37,Citation38]. CAI microsatellite fulfils these criteria since it is located within the coding region of an important transcription factor, RLM1 [Citation23], and has been linked to some known biological properties [Citation23,Citation39,Citation40], thus, justifying the use of only one marker. Genotyping of isolates from the 66 C. albicans colonized volunteers with the CAI marker identified a total of 22 different alleles and 39 distinct genotypes. Different studies have correlated strains presenting CAI alleles with 30 or more CAA/CAG repetitions with resistance to stress agents, virulence in the mouse model, and severity of human vulvovaginal candidiasis [Citation23,Citation39,Citation40]. However, in this study no correlation was observed between strains with those CAI alleles and CFU counts. Similarly, no correlation was observed between any CAI genotypes and smoking habits or gender, possibly because only healthy participants were involved in the present study.
Results showed that the majority of the volunteers were colonized with the same CAI genotype, but some were colonized by two distinct genotypes that could result from a microevolutionary step, such as an LOH or slippage event [Citation41]. This is a curious observation since this microsatellite is so polymorphic [Citation13], yet at the oral surface of healthy participants, the local variability is, apparently, very reduced.
In hospitalized patients without oral infections, up to nine different C. albicans genotypes in the same individual were identified by using two molecular markers (Hist3 and CAI) [Citation42]. Analysing only the CAI marker in the profile of isolates from [Citation42] study, 74% of those patients (n = 17) had only one CAI genotype in the different clones, while 22% of the patients presented two CAI genotypes (n = 5). In the five cases with two CAI genotypes, only one could be explained by LOH, a simple mutational step, while the other four cases consisted of different CAI genotypes.
The results of the present study corroborated these findings, suggesting that in healthy individuals, only one C. albicans CAI genotype or microvariations of the same CAI genotype is predominant in the oral cavity. However, the difference between the two studies may indicate that hospitalized patients are more immunocompromised, disturbing the host-fungus interaction, enabling different CAI clones to inhabit the oral cavity. In CIn immunocompromised individuals the host-microorganism relationship changes from commensal to opportunistic and may eventually lead to the appearance of oral diseases [Citation4,Citation43,Citation44].
In the couples group, it was interesting to observe that 37.9% were not colonized, 31.0% showed colonization in only one member of the couple, and only 31% in both members of the couple, indicating that transmission of C. albicans strains between couples is not a dominant phenotype, but the intimacy among couples increased the probability of heavy cross-colonization. It has been reported that a massive transmission of microbes may occur through kissing [Citation45], so, related individuals are likely to share the oral microbiome [Citation26]. However, when couples share C. albicans strains, the same or similar CAI genotype is predominant, suggesting an exogenous origin followed by CAI stabilization due to constraint sharing of microorganisms.
One important observation of the present study was that C. albicans strains are stable over time in the oral cavity, despite its continuous exposure to the external environment. In all individuals the same CAI genotypes persisted after 1 year, except one case with two CAI genotypes in which only the CAI genotype with a high molecular weight allele persisted. In agreement with this, it has been described that high molecular weight CAI alleles could confer higher adaptability to certain niches, such as mucosal surfaces, enabling specific clones to persist [Citation23Citation40Citation46], Carvalho-Pereira et al. 2018. Our results also indicate that the cross-colonization between the two members of the couple is potentiated when the mates are smokers.
Overall, our study presents interesting new findings regarding yeast colonization in healthy individuals and, as regards C. albicans strains, the stability of the present genotypes over time. Nevertheless, we acknowledge the existence of some limitations regarding the number of couples included in the study and the need for further information about the yeast isolates, for instance, their antifungal susceptibility profiles.
Supplemental Material
Download MS Excel (15.9 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28(8):405–9.
- Zaura E, Nicu EA, Krom BP, et al. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:1–8.
- Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103(32):12115–12120.
- Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6(1):1–8.
- Monteiro-da-Silva F, Araujo R, Sampaio-Maia B. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol. 2014;52(5):498–505.
- Arslan SG, Akpolat N, Kama JD, et al. One-year follow-up of the effect of fixed orthodontic treatment on colonization by oral. Candida J Oral Pathol Med. 2008;37(1):26–29.
- Mun M, Yap T, Alnuaimi AD, et al. Oral candidal carriage in asymptomatic patients. Aust Dent J. 2016;61(2):190–195.
- Mavor AL, Thewes S, Hube B. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets. 2005;6(8):863–874.
- Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;335(6082):647.
- Williams D, Lewis M. Pathogenesis and treatment of oral candidosis. J Oral Microbiol. 2011;3(1):5771.
- Bodey GP, Anaissie E, Gutterman J, et al. Role of granulocyte-macrophage colony-stimulating factor as adjuvant therapy for fungal infection in patients with cancer. Clinl Infect Dis. 1993;17(4):705–707.
- Voss A, le Noble JL, Verduyn Lunel FM, et al. Candidemia in intensive care unit patients: risk factors for mortality. Infection. 1997;25(1):8–11.
- Sampaio P, Gusmão L, Alves C, et al. Highly polymorphic microsatellite for identification of Candida albicans strains. J Clin Microbiol. 2003;41(2):552–557.
- Sampaio P, Gusmão L, Correia A, et al. New microsatellite multiplex PCR for Candida albicans strain typing reveals microevolutionary changes. J Clin Microbiol. 2005;43(8):3869–3876.
- Kaiser, C., Michaelis, S., Mitchell, A., & Laboratory, C. S. H. (1994). Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. New York, United States: Cold Spring Harbor Laboratory Press.
- White T, Bruns T, Lee S, et al. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Pcr Protocols: a Guide to Methods and Applications. Vol. 31. Academic Press Inc, San Diego, USA; 1990. p. 315–322.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;(2018)(35):1547–1549.
- Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. New Jersey: Lawrence Erlbaum; 1988.
- Liu K, Muse SV. PowerMaker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2120–2129.
- Goudet JM, Raymond M, Meeüs T, et al. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940.
- Guo SW, Thompson EA. Performing the exact test for Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372.
- Raymond M, Rousset F. GENEPOP: population genetics software for exact tests and ecumenicism. J Heredity. 1995;86: 248–249.
- Sampaio P, Nogueira E, Loureiro AS, et al. Increased number of glutamine repeats in the C-terminal of Candida albicans Rlm1p enhances the resistance to stress agents. Antonie Van Leeuwenhoek. 2009;96(4):395–404.
- Nguyen LDN, Viscogliosi E, Delhaes L. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol. 2015;6:1–8.
- Gupta B, Gupta S, Chaudhary M, et al. Oral candida prevalence and species specificity in leprosy. Dis Mon. 2019;65(11):100920.
- Burcham ZM, Garneau NL, Comstock SS, et al. Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci Rep. 2020;10(1):2133.
- Rivera RE, Zuluaga A, Arango K, et al. Characterization of oral yeasts isolated from healthy individuals attended in different Colombian dental clinics. J Biomed Res. 2019;33(5):333–342.
- Arcavicv L, Benowitz NL. Cigarette smoking and infection. Arch Intern Med. 2004;164(20):2206–2216.
- Colombo AL, Nucci M, Park BJ, et al. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol. 2006;44(8):2816–2823.
- Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect. 2014;20(Suppl 6):5–10.
- Nucci M, Queiroz-Telles F, Alvarado-Matute T, et al. Epidemiology of candidemia in Latin America: a laboratory-based survey. PloS One. 2013;8(3):1–7.
- Tortorano AM, Kibbler C, Peman J, et al. Candidaemia in Europe: epidemiology and resistance. Int J Antimicrob Agents. 2006;27(5):359–366.
- Sardi JCO, Scorzoni L, Bernardi T, et al. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62:10–24.
- Oouchi M, Hasebe A, Hata H, et al. Age-related alteration of expression and function of TLRs and NK activity in oral candidiasis. Oral Dis. 2015;21(5):645–651.
- Martins M, Henriques M, Ribeiro AP, et al. Oral Candida carriage of patients attending a dental clinic in Braga, Portugal. Rev Iberoam Micol. 2010;27(3):119–124.
- Pinto E, Ribeiro IC, Ferreira NJ, et al. Correlation between enzyme production, germ tube formation and susceptibility to fluconazole in Candida species isolated from patients with denture-related stomatitis and control individuals. J Oral Pathol Med. 2008;37(10):587–592.
- Taylor JW, Geiser DM, Burt A, et al. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12(1):126–146.
- Gräser Y, Volovsek M, Arrington J, et al. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci U S A. 1996;93(22):12473–12477.
- Carvalho-Pereira J, Oliveira MME, Costa-Barbosa A, et al. High variability within Candida albicans transcription factor RLM1: isolates from vulvovaginal infections show a clear bias toward high molecular weight alleles. Med Mycol. 2018;56(5):649–651.
- Li J, Fan S-R, Liu X-P, et al. Biased genotype distributions of Candida albicans strains associated with vulvovaginal candidosis and candidal balanoposthitis in China. Clinl Infect Dis. 2008;47(9):1119–1125.
- Sampaio P, Correia A, Gusmão L, et al. Sequence analysis reveals complex mutational processes for allele length variation at two polymorphic microsatellite loci in Candida albicans. In: Méndez-Vilas, editor. Communicating Current Research and Educational Topics and Trends in Applied Microbiology. Volume II. A. 2007. p. 926–935. Formatex. Badajoz.
- Shimizu K, Hattori H, Adachi H, et al. Microsatellite-based genotyping of Candida albicans isolated from patients with superficial candidiasis. Med Mycol J. 2011;52(2):129–138.
- Aslani N, Janbabaei G, Abastabar M, et al. Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect Dis. 2018;18:24.
- Kitada K, de Toledo A, Oho T. Increase in detectable opportunistic bacteria in the oral cavity of orthodontic patients. Int J Dent Hyg. 2009;7(2):121–125.
- Kort R, Caspers M, van de Graaf A, et al. Shaping the oral microbiota through intimate kissing. Microbiome. 2014;2:41.
- Wu C-X, Cheng J, Wang -Y-Y, et al. microRNA expression profiling of macrophage line Raw264.7 infected by Candida Albicans. Shock. 2017;47(4):520–530.

