ABSTRACT
Background: The cell-surface cysteine proteinases RgpA, RgpB (Arg-gingipain), and Kgp (Lys-gingipain) are major virulence factors of P. gingivalis, a keystone pathogen in the development of destructive periodontal disease. The gingipains function as proteinases and transpeptidases utilising small peptides such as glycylglycine as acceptor molecules. However, the characteristics of the gingipains from most P. gingivalis strains have not been determined.
Methods: We determined the phenotypes of a panel of P. gingivalis laboratory strains and global clinical isolates with respect to growth on blood agar plus whole-cell and vesicle-free culture supernatant (VFSN) Arg- and Lys-specific proteinase activities.
Results: The P. gingivalis isolates exhibited different growth characteristics and hydrolysis of haemoglobin in solid media. Whole-cell Arg-gingipain Vmax varied 5.8-fold and the whole cell Lys-gingipain Vmax varied 2.1-fold across the strains. Furthermore, the P. gingivalis strains showed more than 107-fold variance in soluble Arg-gingipain activity in VFSN and more than 371-fold variance in soluble Lys-gingipain activity in VFSN. Glycylglycine and cysteine stimulated Arg- and Lys-specific cleavage activities of all strains. The stimulation by cysteine was in addition to its redox effect consistent with both glycylglycine and cysteine promoting transpeptidation.
Conclusion: The global P. gingivalis clinical isolates exhibit different Arg- and Lys‑gingipain activities with substantial variability in the level of soluble proteinases released into the environment.
Introduction
Periodontal diseases are characterised by inflammatory responses to microbes within the plaque accreted to the tooth and surrounding epithelium. Overgrowth of supragingival plaque results in gingivitis (gingival inflammation). If this becomes chronic then an anaerobic periodontal pocket can develop through soft tissue swelling, and this environment can allow the emergence of periodontal pathogens. The resulting dysbiosis leads to destructive periodontal disease (periodontitis) involving degradation of the supporting tissues of the tooth including resorption of the alveolar bone [Citation1]. Porphyromonas gingivalis has been recognised as an important participant in the development of the pathology of periodontitis and has been described as a keystone pathogen [Citation2]. P. gingivalis expresses an array of virulence factors, including proteinases [Citation3]. It secretes serine and metalloproteases [Citation4,Citation5] plus cysteine proteases including gingipains, PrtT protease, periodontain, and Tpr protease [Citation3,Citation6–13]. Among the cysteine proteases, the Arg-specific gingipains RgpA and RgpB and Lys-specific gingipain Kgp are regarded as the major virulence factors [Citation14–16]. Gingipains help P. gingivalis in the processing of surface-associated proteins and hemagglutinins [Citation17], adherence, growth, development, and evasion of host defense [Citation16–18]. The activities of gingipains especially Lys-gingipain have been experimentally shown to be linked to the P. gingivalis binding to and degradation of haemoglobin and also to colony pigmentation on blood agar [Citation19–23].
Recently we demonstrated that gingipains also function as transpeptidases that utilize peptides such as glycylglycine as acceptor molecules [Citation24]. Transpeptidation using peptides derived from host proteins such as haemoglobin and serum albumin in vivo was suggested to potentially contribute to immune response dysregulation associated with P. gingivalis infection [Citation24].
Experimentally, strains of P. gingivalis vary in the manifestation of measured virulence traits. For example, following subcutaneous injection some strains produce spreading, ulcerating lesions at distant sites whereas others produce only small, localised abscesses [Citation25]. Collagenolytic activity varies between strains [Citation26] and platelet activation by P. gingivalis which is mediated by Rgp protease cleavage of protease-activated receptors [Citation27], has also been shown to be widely variable between clinical P. gingivalis isolates and the standard laboratory strains ATCC 33277 and ATCC 53977 (also known as strain W83) [Citation28]. The presence of proteinase-processed adhesin molecules, in particular Hgp44 domains, on the P. gingivalis cell surface has also been shown to be important for P. gingivalis to induce platelet aggregation in platelet-rich plasma [Citation29]. In the absence of RgpA, Kgp and the adhesin HagA P. gingivalis activation of platelets in platelet-rich plasma was not observed, whereas activation occurred with washed platelets [Citation29]. This observation was explained by reduction in effective RgpB exposure of the platelets through saturation of RgpB by plasma proteins [Citation29]. Following on from this concept, in vivo, a P. gingivalis strain such as 84–3 that releases abundant soluble gingipains may have more effect on platelet activation than one that releases fewer free proteinase molecules. Thus, the relationship between platelet activation and P. gingivalis strain protease activity requires further investigation.
The involvement of gingipains in the pathogenicity of P. gingivalis has made gingipains promising drug targets for the treatment of P. gingivalis-associated diseases including destructive periodontal disease [Citation30–32]. However, the characteristics of gingipain activities and cell distribution (that is whether the proteinases remain membrane associated or are released into the environment) has not been studied for a range of clinical isolates. Here we have determined the cell-associated and soluble (vesicle-free) gingipain proteinase activities of a panel of global P. gingivalis clinical isolates. We demonstrate widely varied relative proteinase activities between the isolates, particularly in the distribution between cell-associated and soluble forms released into the environment.
Materials and methods
P. gingivalis strains and growth conditions
The P. gingivalis strains Afr-5B1, ATCC 33277, ATCC 49417, A7A1-28, W50, YH522, 11A, 3A1, 3–3, 13–1, 15–9, 84–3, 381 and 7B TORR () were obtained from the collection of the Melbourne Dental School, The University of Melbourne [Citation33] and have previously been examined for ribotype, serotype plus recognition by patient sera and murine anti-P. gingivalis sera [Citation34]. P. gingivalis were cultured from frozen 50% (v/v) glycerol stock onto horse blood agar (HBA) plates [Blood Agar Base No 2 (Oxoid) supplemented with 10% (v/v) of lysed defibrinated horse blood (Equicell) and 1 µg/mL of menadione]. The primary broth culture of the P. gingivalis strains was prepared by inoculating several colonies into 20 mL of BHI broth [Brain Heart Infusion Broth (Oxoid) supplemented with 1 µg/mL menadione, 5 µg/mL haemin and 5 mg/mL L-cysteine hydrochloride]. The inoculated BHI broths were then incubated at 37°C in an anaerobic workstation for 24–48 h with an atmosphere of 10% CO2, 5% H2, and 85% N2. P. gingivalis were then sub-cultured by transferring enough volume into fresh BHI medium (20 mL) to give approximately 108 CFU/bottle. The cultures were incubated at 37°C for 18–24 h to obtain P. gingivalis at late log phase growth with an optical density at 650 nm (OD650) of 1.0. Four biological replicates were maintained for each P. gingivalis strain.
Table 1. Fold-change increase of the Arg- and Lys-gingipain activity of P. gingivalis whole cells following cysteine and glycylglycine addition to the assay
Plate colony phenotype
P. gingivalis strains initially grown on HBA were sub-cultured into BHI and incubated overnight at 37°C. Subsequently, cultures were diluted in fresh BHI to give approximately 2 × 103 cells/mL, and 50 µL spread onto fresh 20 mL HBA plates supplemented with 5 µg/mL menadione. The plates were incubated in the anaerobe chamber and colony growth was examined over time.
Harvesting of P. gingivalis
P. gingivalis cells were harvested from liquid culture by centrifugation at 8,000 g for 10 min at 4°C. The pellets (whole cells) were washed using an equal volume of Tris buffer (50 mM Tris, 150 mM NaCl and 5 mM CaCl2, pH 8.0) and re-centrifuged at 8,000 g for 10 min at 4°C. The cell pellet was resuspended in an equal volume of Tris buffer and aliquots, and stored at −80°C. The culture supernatant (~15 mL each) was transferred to 3 × 4.7 mL OptiSeal polypropylene centrifuge tubes (Beckman Coulter Inc., CA, USA) and subjected to ultracentrifugation at 90,000 rpm (438,545 g) in an Optima™ TLX ultracentrifuge (Beckman Coulter Inc., CA, USA) using a TLA-110 rotor (Beckman Coulter Inc., CA, USA) for 60 min at 4°C. The resulting vesicle free supernatants (VFSN) were carefully removed avoiding the vesicle pellet and stored as 1 mL aliquots at −80°C.
Proteolytic assay
Wholecell and VFSN Arg and Lys-gingipain proteolytic activities were determined using the synthetic chromogenic substrates Nα-benzoyl-L-Arg-4-nitroanilide (BApNA) and GlyProLys-4-nitroanilide (GPKNA) (Sigma-Aldrich) respectively, at 1 mM unless stated otherwise, in 200 µL volumes in 96 well flat-bottom plates. All assays were conducted in Tris buffer, with variation in L-cysteine (from a 2 M stock, pH 7.0), glycylglycine (from a 1.5 M stock, pH 7.0), and DTT (from a 2 M stock) as outlined in the text. The assay mixture for Lys-gingipain contained P. gingivalis whole cells at 5 × 107 cells per well or VFSN (5 to 40 µL), whilst the Arg-gingipains assay mixture had P. gingivalis whole cells at 5 × 106 cells per well or 40 µL of VFSN. Substrate cleavage activity was determined by measuring the release of p-nitroaniline by the change in absorbance at 405 nm, at 1 s intervals for up to 99 repeats, at 37°C using a Victor3 1420 Multilabel Counter (Perkin Elmer, MA, USA). The rate of product formation was calculated by dividing the change in absorbance over time by the extinction coefficient of 8,800 M1cm−1 and a path length of 0.6 cm.
Kinetic determinations
To calculate the Vmax and Km, the initial velocity of a series of enzymatic reactions in the presence of increasing concentrations of the substrate was determined. The curves were fitted individually by non-linear regression analysis to the Michaelis-Menten expression (v = Vmax [S]/(Km + [S]), where v is the rate of reaction, Vmax is the maximum rate, [S] is the concentration of substrate and Km is the half of Vmax [Citation35] using the program GraphPad Prism (GraphPad Software, Inc.). The rate of product formation was expressed as nM/s/5 x 106 cells or nM/s for VFSN.
Statistical analyses
Statistical analyses were done using Graph Pad Prism 7 software (GraphPad Software, Inc., La Jolla, CA). The data were analyzed with two-way ANOVA and Student’s t-test. Values are expressed as the ± standard error of the mean, standard deviation or at a 95% confidence interval. P < 0.05 was considered to indicate a significant difference. Regression analyses were done using Minitab® 19 (Minitab Pty Ltd).
Results
P. gingivalis strains reveal differing gingipain-dependent characteristics on agar plates
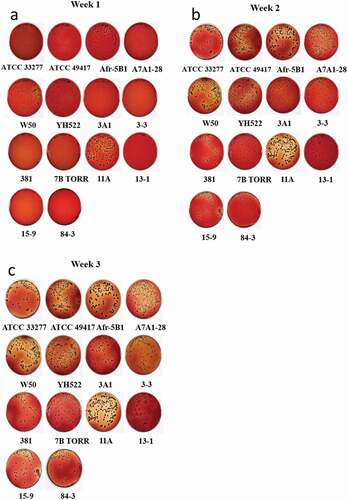
The 14 P. gingivalis strains to be assessed for gingipain activities were first compared for basic phenotype characteristics. After one week of incubation on HBA some variation in colony size and colour was evident between strains. Some had large, readily visible brown-black pigmented colonies (ATCC 33277, ATCC 49417, W50, Afr-5B1, 11A), small black-brown colonies (A7A1-28, 3A1, 3–3, 13–1) or pinhead-very small colonies (15–9, 84–3, 381). Some strains were seen to be degrading haemoglobin in the plates as shown by the loss of colour, although to varying extents (ATCC 33277, ATCC 49417, W50, Afr-5B1, A7A1-28, 381, 11A), whilst little haemoglobin degradation was in evidence for other strains. At week 2 colonies could be readily seen for all strains with haemoglobin degradation readily apparent for some strains and weak for others, e.g., 13–1, 3A1, 84–3, and 7B TORR. At week 3 haemoglobin degradation by the strain 13–1 was minimal despite the large size of the colonies ().
Both cysteine and glycylglycine influence whole-cell Arg-gingipain activities
The propensity for P. gingivalis isolates to cleave the substrate BApNA in the presence of cysteine and glycylglycine in the assays was systematically assessed. Pilot studies indicated that the use of 5 × 106 cells per assay in 96 well microtiter plate wells produced appropriate initial rates for data analysis. Relative to no cysteine addition, 20 mM of cysteine in the assays increased the rate of activity of Arg-gingipains of these 14 strains by 2.4 (Afr-5B1) to 7.4-fold (ATCC 33277) whilst adding cysteine to 200 mM, increased the rate of activity of Arg-gingipains significantly further (P < 0.001) by factors ranging from 5.7 (strain 3A1) to 15.6-fold (strain ATCC 49417) (). The Arg-gingipain activities of the whole cells of each strain were also stimulated by glycylglycine, producing a curve that approached, but did not reach a maximum even when 300 mM of glycylglycine was added. Relative to the absence of glycylglycine, the addition of 300 mM glycylglycine produced activity increases that overall were similar to 200 mM cysteine stimulation (P > 0.05), ranging from 7.3-fold (for strain 15–9) to 18-fold (for strain W50) (). The addition of 20 mM of cysteine plus 300 mM of glycylglycine resulted in an increase in the whole-cell Arg-gingipain activities from 21 to 59.7-fold across the strains (). However, increasing cysteine to 200 mM in an assay with 300 mM glycylglycine was less stimulatory of substrate hydrolysis than assays using only 20 mM cysteine ().
For a direct comparison of the magnitude of enzyme stimulation by either cysteine or glycylglycine, the assays were prepared using cysteine or glycylglycine at 200 mM. The results show that for some strains (13–1, 3–3, Afr-5B1 and A7A1-28) cysteine or glycylglycine were equally stimulatory, but for the other strains, glycylglycine had a more positive effect on Arg-gingipain activity than cysteine (P < 0.05) ().
Table 2. Comparison of whole-cell Arg-gingipain Vmax in the presence of 200 mM of cysteine (Cys) or glycylglycine (GlyGly) with 1 mM BApNA substrate
Both cysteine and glycylglycine influence whole-cell Kgp-gingipain activities
The propensity for P. gingivalis isolates to cleave the substrate GPKNA in the presence of cysteine and glycylglycine in the assays was also systematically assessed. Pilot studies indicated that the use of 5 × 107 cells per assay in 96 well microtiter plate wells produced appropriate initial rates for data analysis, 10-fold more cells than required for Arg-gingipain assay. The addition of 20 mM cysteine increased the Lys-gingipain activities by 1.5 to 4.1-fold whilst the addition of cysteine at 200 mM increased the Lys-gingipain activity by 1.9 (strain 13–1) to 5.6-fold (strain ATCC 49417) relative to no cysteine addition ().
Titration studies using P. gingivalis strains W50 and ATCC 33277 showed that the addition of 180 mM glycylglycine to the assay gave a maximum increase in Lys-specific gingipain activities (not shown). Strain 13–1 exhibited the lowest response to the dipeptide (2.1-fold) and strain ATCC 49417 had the highest (6.8-fold), which was not significantly different from the response to 200 mM cysteine (P > 0.05). In the presence of 20 mM of cysteine and 180 mM of glycylglycine, the overall Lys-specific gingipain activities of P. gingivalis increased by 4.1 (381 strain) to 12.1-fold (ATCC 49417) when compared to the activity in the absence of these compounds. However, in parallel with the Arg-gingipain results, in the presence of 200 mM of cysteine and 180 mM of glycylglycine, the rate of activity of the Lys-specific gingipain was slightly less than when using 20 mM cysteine (). Thus, similar to the observations of whole-cell Arg-gingipain activities, the different P. gingivalis strains exhibited varying Lys-gingipain activities with varying responses to the additives.
The kinetics of whole-cell-associated gingipains are strain-dependent
The enzyme kinetics of the Arg- and Lys-specific substrate cleavages by the P. gingivalis strains were then determined using 20 mM cysteine and glycylglycine (300 mM for Arg-specific activity or 180 mM for Lys-specific activity). The whole-cell Arg-gingipain Vmax varied by 5.8-fold, from 20 ± 0.3 nM/s/5 x 106 cells measured for strain 13–1 and 117 ± 1.3 nM/s/5 x106 cells measured for strain W50 ( and ). In contrast, the Vmax of whole-cell Lys-gingipains was more evenly distributed, varying among P. gingivalis strains by only approximately 2-fold, from 8 ± 0.2 nM/s/5 x 106 cells for the 13–1 strain and 17 ± 0.2 nM/s/5 x 106 cells for ATCC 49417 ( and ). The Vmax ratio for Arg/Lys whole-cell activities for the strains varied from 2.5 to 8.5-fold, with 11 of the 14 strains in the ratio range 5.0 to 7.3-fold (). The log10-transformed Arg- and Lys-gingipain activity data of the P. gingivalis whole cells could be fitted to a linear model with an R2 of 76.5% at a 95% prediction interval ()). Overall, strains that had relatively high Rgp activity also had high Kgp activity, and strains that had relatively low Rgp activity also had low Kgp activity. P. gingivalis W50 strain had the highest gingipain activity based on cumulative activities of Arg- and Lys-gingipains on wholecells, whereas, the 13–1 strain had the lowest.
Table 3. The Vmax of P. gingivalis whole-cell and VFSN Arg- and Lys-gingipains
Table 4. The comparison of Arg- and Lys- gingipain Vmax for whole cells (WC) and VFSN
Figure 2. (a) The kinetics curves for P. gingivalis whole-cell Arg-gingipain activities using BApNA as substrate. (b) The kinetics curves for P. gingivalis whole-cell Lys-gingipain activities using GPKNA as substrate
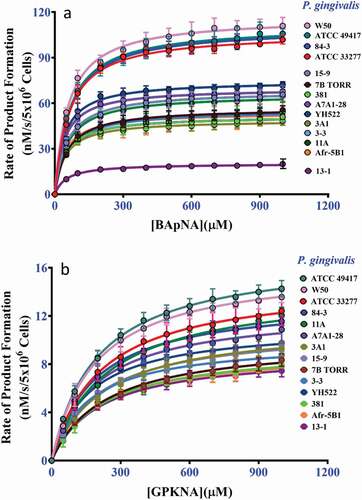
Figure 3. Correlation of the log10 transformed Arg-and Lys-gingipain activities of P. gingivalis (a) whole cells and (b) VFSN. Dashed lines indicate 95% PI with the model
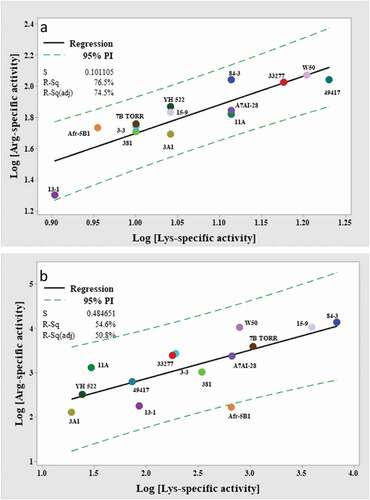
The whole-cell Arg-gingipain Km for BApNA was fairly similar from 40 ± 3 µM to 65 ± 6 µM (), whilst the range of whole-cell Lys-gingipain Km for GPKNA substrate was from 142 ± 8 µM to 234 ± 11 µM (). Strain A7AI-28 had the lowest Km for both BApNA and GPKNA whilst the highest Km for BApNA and GPKNA were shown by 84–3 and 11A strains respectively.
Table 5. The Km for substrates of whole-cell (WC) and VFSN Arg- and Lys-gingipains of P. gingivalis.
High concentration of cysteine negatively affected the overall Km of whole-cell Arg-gingipains for BApNA, however the decreased substrate affinity was, however, compensated by increased Vmax (). Comparing the Km of whole cells for BApNA determined using 20 mM cysteine and 300 mM glycylglycine () also indicated a negative effect of glycylglycine addition on substrate binding, however as shown by the data in , this was also compensated by increased Vmax.
Table 6. The calculated Km for BApNA and Vmax of P. gingivalis whole-cell Arg-gingipains with cysteine addition to the assay
The kinetics of gingipains in the VFSN are strain dependent
Remarkably, the Arg-gingipain VFSN Vmax varied by more than two orders of magnitude (up to 107-fold) across the P. gingivalis strains. The highest Vmax of 13.9 ± 0.153 µM/s was observed for strain 84–3 and the lowest Vmax was 0.13 ± 0.004 µM/s for strain 3A1. The VFSN Lys-gingipain Vmax also varied by more than two orders of magnitude (up to 371-fold) across the P. gingivalis strains. The highest Vmax of 6.7 ± 0.13 µM/s was observed for strain 84–3 and the lowest Vmax of 18 ± 4 nM/s was found for strain 3A1 (). P. gingivalis 84–3 was the highest soluble gingipain-producing strain based on cumulative activities of Arg- and Lys-gingipain in the VFSN, whereas the 3A1 strain produced the least amount of active soluble gingipains under these growth conditions.
A greater variation of the Arg/Lys Vmax ratios was observed for the VFSN fractions (average ratios 9.6 ± 11) compared with the whole cells (average of ratios 5.9 ± 1.4) with values ranging from 0.26 as a minimum (strain Afr-5B1) to 45 (strain 11A) as the maximum (). A linear model could be fitted to the log10-transformed Arg- and Lys-gingipain activity in the VFSN with the fitted equation [log10 (Arg-specific activity) = 1.58 + 0.643 log10 Lys-gingipain activity)], R2 54.6%. The plot suggested that strain Afr-5B1 was different from the others, with low Arg-gingipain activity in the VFSN given its Lys-gingipain activity ()). When strain Afr-5B1 was omitted from the data set, the fit of the model improved (R2 75.2%), with the revised equation remaining similar [log10 (Arg-gingipain activity) = 1.53 + 0.703 log10 (Lys-specific activity)]. Overall, the data indicated the strain Arg and Lys activities were positively correlated.
Across the P. gingivalis strains, the range of VFSN Arg-gingipain Km for BApNA was from 49 ± 8 µM to 70 ± 11 µM () which were mostly similar (P > 0.05) to Km of whole-cell Rgps. Slightly higher Km was determined for strains A7A1-28, 3–3, and Afr-5B1 for BApNA by Rgp enzymes in the VFSN than on whole cells (P < 0.05), but the values were not remarkably different. The VFSN Lys-gingipain Km for GPKNA, which ranged from 139 ± 8 µM to 228 ± 13 µM (), were not significantly different to whole-cell-located Lys-gingipain enzyme Km for GPKNA for each strain (P > 0.05) ().
Comparison of DTT and cysteine in the activation of whole-cell gingipain activity
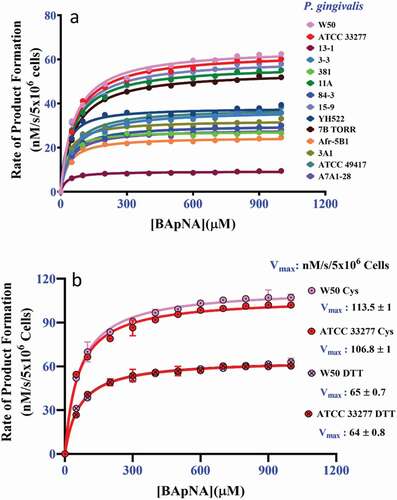
To observe whether the reducing compounds DTT and L-cysteine have a similar effect on gingipain activity the Vmax and Km of the Arg-gingipains were determined by replacing 20 mM cysteine with 20 mM of DTT in the presence of 300 mM glycylglycine. All strains exhibited higher Vmax for BApNA hydrolysis in the presence of 20 mM cysteine as a reducing agent than when using DTT, from a modest 1.1-fold to 3.7-fold more activity (P < 0.05) ( and ). The range of the whole-cell Arg-gingipain Km for BApNA when using DTT as a reducing agent was from 29 ± 5 µM to 67 ± 2 µM (). For some strains of P. gingivalis (15–9, W50, ATCC 33277, 7B TORR, 3–3, 11A, and A7A1-28) no significant difference in Km (P > 0.05) was observed between assays conducted with 20 mM cysteine or 20 mM DTT. Whereas P. gingivalis strains 84–3, ATCC 49417, 381, 13–1, Afr-5B1, 3A1, and YH522 exhibited significantly lower Km for BApNA in the presence of DTT (P < 0.05) than in the presence of cysteine ().
Table 7. The Km and Vmax of P. gingivalis whole-cell Arg-gingipains in the presence of 300 mM glycylglycine and 20 mM DTT or 20 mM L-cysteine
Figure 4. The kinetics of P. gingivalis whole-cell Arg-gingipains using BApNA substrate in the presence of 20 mM DTT and 300 mM glycylglycine. (a) The aggregate kinetic curves for 14 strains using 20 mM DTT as a reducing agent. (b) Control assays using strain W50 and ATCC 33277 cells in the presence of 300 mM glycylglycine and 20 mM DTT or 20 mM cysteine. The data shows DTT is less effective at stimulating the Arg-gingipain activity of these strains than cysteine. (P < 0.05)
Discussion
Growth and gingipain activities of P. gingivalis are not directly correlated
In this study we report gingipain characteristics of several global P. gingivalis clinical isolates. Previous studies have demonstrated that strains of bacterial species isolated from around the world can have a variable phenotype that impacts virulence [Citation36]. The phenotype variation can be due to the acquisition of virulence genes such as those coding for toxins, protein effector secretion systems, or antibiotic resistance [Citation37–40]. Single nucleotide changes can also affect the phenotype by introducing nonsense mutations, changing promoter function, and altering protein sequences [Citation41,Citation42]. The sequenced genomes of multiple P. gingivalis strains revealed that although the P. gingivalis genomes are prone to rearrangement [Citation33,Citation43] they exhibit low sequence divergence and low acquisition of other genes compared to other species [Citation33,Citation43–45]. The low sequence divergence includes high conservation within the catalytic domains of the gingipains [Citation33]. However, a genome sequence alone does not provide a comprehensive indicator of phenotype as it does not show the effect of small inter-strain variations, such as changes to codons or regulatory elements. The global P. gingivalis examined here exhibited different phenotypes as demonstrated by the growth rate on HBA and ability to utilize haemoglobin/haem in the HBA plates (clear agar pigment). The ability of P. gingivalis to cleave haemoglobin and assimilate haem has been attributed to the activity of the gingipains [Citation19,Citation46]. In this study, P. gingivalis strain 11A formed robust colonies after 7 days of incubation and indeed had robust glossy black-pigmented colonies after only 4 days of inoculation (data not shown), whereas, strain such as 84–3 produced colonies of pinpoint size after 7 days. Strains that grew poorly in BHI broth (13–1, 3A1, 84–3, and 7B TORR) degraded haemoglobin in the plate less rapidly than other strains signalling a potentially correlating phenotype.
Cysteine stimulates gingipain activities of P. gingivalis
Maintaining a reduced environment is necessary for optimum gingipain activity and a reducing agent such as cysteine is an important component of a gingipain proteolytic assay to protect the catalytic cysteine from oxidation. Glycylglycine can stimulate gingipain proteolytic activities in the presence of substrates, including BApNA, azocasein, and azocoll [Citation47]. The activity enhancement in the presence of glycylglycine was originally suggested by Potempa et al. [Citation9] to be due to the elimination of non-productive, high-affinity binding of the substrate arginine side chain. However, Zhang et al. [Citation24] subsequently showed that gingipains behave as transpeptidases, with it likely that the transfer reaction permits faster product release, thus enhancing the rate of substrate turnover. The stimulatory effect of cysteine beyond that required for active site cysteine reduction indicates it may also behave as a transpeptidation acceptor molecule. This was indicated by the higher whole-cell Arg-gingipain Vmax of strains ATCC 33277 and W50 in the presence of 20 mM cysteine relative to the whole-cell Arg-gingipain Vmax in the presence of 20 mM DTT (). This result is consistent with the previous report where it was shown that among the thiol-containing compounds, cysteine (0–50 mM) produced a 1.4-fold higher stimulatory effect than that of either DTT or β-mercaptoethanol in the presence of 100 mM glycylglycine [Citation47]. However, the addition of 200 mM cysteine to the assay had an adverse effect on the gingipain function. This could be due to the chelation of calcium (Ca2+), an ion that stabilizes gingipains [Citation47]. Relative to the assay containing 200 mM cysteine, the addition of 200 mM glycylglycine was found to only have a positive effect on Arg-gingipain activities produced by 10 of the 14 strains of P. gingivalis (). This may suggest that differences exist between the enzymes of these strains that are sufficient to affect substrate-acceptor molecule interactions during catalysis.
Cysteine and glycylglycine can have similar effects on the gingipains activities of P. gingivalis isolates
The calculated Km for BApNA of P. gingivalis whole cells Arg-gingipains in the presence of 20 mM cysteine ranged from 4 ± 1 to 14 ± 2 µM across the P. gingivalis strains tested (), consistent with a previous study where it was shown that the W50 strain and RgpB deficient mutant D7 Km were 5 and 5.6 µM, respectively [Citation48]. The Km for BApNA significantly increased when 200 mM cysteine was applied in the assay and still further when in the presence of both 300 mM glycylglycine plus 20 mM cysteine in the assay. This indicates a negative effect, potentially from molecular crowding. Previously it has been shown that the Km of rKgp and RgpB for their substrates increased by over 8 and 4.6-fold respectively when assayed in the presence of 200 mM glycylglycine compared to that of Km calculated in the absence of glycylglycine [Citation24]. But the activation of gingipains by glycylglycine is inversely correlated to the change of the Km value [Citation9,Citation24]. Similarly, our data show that cysteine and glycylglycine can have similar effects on the gingipains activities of a wide range of P. gingivalis isolates.
P. gingivalis strains may have different virulence potentials due to gingipain functions
A previous study of P. gingivalis W50 and its gingipain mutants in the animal lesion model has suggested that Lys-gingipain contributes more to virulence than Arg-gingipains [Citation49]. Similarly, in a periodontitis model, oral inoculation with P. gingivalis W50 and its gingipain mutants also indicated that Kgp was important for bone loss, with RgpB, but not RgpA, being similarly important in this model [Citation50]. In contrast, Wilenski et al. [Citation51] determined that RgpA is a major virulence factor as P. gingivalis that produced RgpA had less residual bone volume as shown by micro-computed tomography and suppressed phagocytosis by leucocytes in a subcutaneous chamber infection model [Citation51]. In the periodontitis model used by Pathirana et al. [Citation50], disease was induced by oral gavage with P. gingivalis suspended in media with reducing agents to maintain proteinase activities [Citation50]. In the experimental periodontitis model utilised by Wilenski et al. [Citation51] in addition to oral gavage, P. gingivalis were applied to the murine colorectal region, and the P. gingivalis were not suspended in a carrier medium containing reducing agent to maintain proteinase activities. The difference in treatment of P. gingivalis during murine inoculations could have had a significant effect on proteinase activities, host colonisation, induction of immune responses and disease outcomes.
Support for levels of gingipain activities, in particular Lys-gingipain activity, possibly influencing pathogenicity, is that P. gingivalis W50 and A7A1-28 (ATCC 53977) that exhibit higher levels of proteinase activity than strains 381 and ATCC 33277 (original strain designation 2561) () [Citation25] were previously demonstrated to be the more pathogenic in murine lesion models [Citation25,Citation52]. Furthermore, in a periodontitis model, mice challenged with A7A1-28 had less residual supportive bone volume present than those challenged with strain 381 [Citation53]. Overall, the balance of Lys and Arg-gingipain activities of strains could influence virulence outcome, potentially indicating why loss of RgpA in A7A1-28 had more impact on virulence of this strain than loss of RgpA from strain W50, and why A7A1-28 and W50 are more invasive than ATCC 33277 in murine and rat models [Citation54]. However, the explanation of gingipain production correlating with virulence is not clear cut. P. gingivalis W50 and ATCC 49417 were both found to be highly virulent in an animal lesion model [Citation25,Citation55,Citation56], but ATCC 49417 has similar Arg- and Lys-cleavage Vmax to ATCC 33277 and lower soluble gingipain activities. Furthermore, although strain ATCC 33277 is less virulent than other strains in some disease models [Citation43], which could potentially be attributed to secretion of less gingipain into the surrounding environment, in other models it shows greater virulence characteristics. For example, when added to gingival epithelial cells ATCC 33277 can completely inhibit IL-8 accumulation and degrade existing IL-8 in culture supernatants [Citation57] thus attenuating the inflammatory response. It is likely that the combined effect of multiple bacterial factors contributes to virulence, but an understanding of the effect of released gingipains on virulence is essential to understanding pathogenesis.
Conclusion
Global P. gingivalis clinical isolates exhibited different rates and distributions of Arg- and Lys-gingipain activities. Cysteine and glycylglycine were observed to enhance the Arg-and Lys-gingipain activities produced by the different P. gingivalis strains. The data suggested that different clinical isolates of P. gingivalis may exhibit different virulence characteristics based on the distribution and activities of their gingipains.
Authors contributions
NLH, KC, CAS and ECR conceived and designed the study. ECR obtained the funding. ASMM and CAS conducted experiments and analysed data. CAS and ASMM drafted the manuscript. NLH, KJC, and ECR revised the manuscript.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Sanz M, Beighton D, Curtis MA, et al. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J Clin Periodontol. 2017;44(Suppl 18):S5–S11.
- Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10(5):497–14.
- Mysak J, Podzimek S, Sommerova P, et al. Porphyromonas gingivalis: major periodontopathic pathogen overview. J Immunol Res. 2014;2014:476068.
- Nonaka M, Shoji M, Kadowaki T, et al. Analysis of a Lys-specific serine endopeptidase secreted via the type IX secretion system in Porphyromonas gingivalis. FEMS Microbiol Lett. 2014;354(1):60–68.
- Chen YY, Cross KJ, Paolini RA, et al. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J Biol Chem. 2002;277(26):23433–23440.
- Bhogal PS, Slakeski N, Reynolds EC. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143(7):2485–2495.
- Slakeski N, Bhogal PS, O’Brien-Simpson NM, et al. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology. 1998;144(6):1583–1592.
- Chen Z, Potempa J, Polanowski A, et al. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J Biol Chem. 1992;267:18896–18901.
- Potempa J, Mikolajczyk-Pawlinska J, Brassell D, et al. Comparative properties of two cysteine proteinases (Gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273(34):21648–21657.
- Pike R, McGraw W, Potempa J, et al. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411.
- Otogoto J, Kuramitsu HK. Isolation and characterization of the Porphyromonas gingivalis prtT gene, coding for protease activity. Infect Immun. 1993;61(1):117–123.
- Nelson D, Potempa J, Kordula T, et al. Purification and characterization of a novel cysteine proteinase (Periodontain) from Porphyromonas gingivalis. J Biol Chem. 1999;274(18):12245–12251.
- Bourgeau G, Lapointe H, Peloquin P, et al. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992;60(8):3186–3192.
- Hočevar K, Potempa J, Turk B. Host cell-surface proteins as substrates of gingipains, the main proteases of Porphyromonas gingivalis. Biol Chem. 2018;399(12):1353–1361.
- Potempa J, Sroka A, Imamura T, et al. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Peptide Sci. 2003;4(6):397–407.
- O’ Brien-Simpson NM, Veith PD, Dashper SG, et al. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Peptide Sci. 2003;4(6):409–426.
- Travis J, Pike R, Imamura T, et al. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32(1):120–125.
- Vincents B, Guentsch A, Kostolowska D, et al. Cleavage of IgG1 and IgG3 by gingipain K from Porphyromonas gingivalis may compromise host defense in progressive periodontitis. Faseb J. 2011;25(10):3741–3750.
- Dashper SG, Cross KJ, Slakeski N, et al. Hemoglobin hydrolysis and heme acquisition by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19(1):50–56.
- McKee AS, McDermid AS, Wait R, et al. Isolation of colonial variants of Bacteroides gingivalis W50 with a reduced virulence. J Med Microbiol. 1988;27(1):59–64.
- Smalley JW, Birss AJ, Kay HM, et al. The distribution of trypsin-like enzyme activity in cultures of a virulent and an a virulent strain of Bacteroides gingivalis W50. Oral Microbiol Immunol. 1989;4(3):178–181.
- Okamoto K, Nakayama K, Kadowaki T, et al. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273(33):21225–21231.
- Shi Y, Ratnayake DB, Okamoto K, et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J Biol Chem. 1999;274(25):17955–17960.
- Zhang L, Veith PD, Huq NL, et al. Porphyromonas gingivalis gingipains display transpeptidation activity. J Proteome Res. 2018;3(8):2803–2818.
- Neiders ME, Chen PB, Suido H, et al. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24(3):192–198. .
- Birkedal-Hansen H, Taylor RE, Zambon JJ, et al. Characterization of collagenolytic activity from strains of Bacteroides gingivalis. J Periodontal Res. 1988;23(4):258–264.
- Lourbakos A, Yuan Y, Jenkins AL, et al. Activation of protease-activated receptors by gingipains fromPorphyromonas gingivalis leads to platelet aggregation: a new trait in microbial pathogenicity. Blood. 2001;97(12):3790–3797.
- Jockel-Schneider Y, Kobsar A, Stellzig-Eisenhauer A, et al. Wild-type isolates of Porphyromonas gingivalis derived from periodontitis patients display major variability in platelet activation. J Clin Periodontol. 2018;45(6):693–700.
- Naito M, Sakai E, Shi Y, et al. Porphyromonas gingivalis-induced platelet aggregation in plasma depends on Hgp44 adhesin but not Rgp proteinase. Mol Microbiol. 2006;59(1):152–167.
- Grenier D, La VD. Proteases of Porphyromonas gingivalis as important virulence factors in periodontal disease and potential targets for plant-derived compounds: a review article. Curr Drug Targets. 2011;12(3):322–331.
- Olsen I, Potempa J. Strategies for the inhibition of gingipains for the potential treatment of periodontitis and associated systemic diseases. J Oral Microbiol. 2014;6(1):24800.
- Dominy SS, Lynch C, Ermini F, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5(1):eaau3333.
- Dashper SG, Mitchell HL, Seers CA, et al. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48.
- Slakeski N, Margetts M, Moore C, et al. Characterization and expression of a novel Porphyromonas gingivalis outer membrane protein, Omp28. Oral Microbiol Immunol. 2002;17(3):150–156.
- Michaelis L, Menten ML, Johnson KA, et al. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50(39):8264–8269.
- Siena E, Bodini M, Medini D. Interplay between virulence and variability factors as a potential driver of invasive meningococcal disease. Comput Struct Biotechnol J. 2018;16:61–69.
- Qiu X, Kulasekara BR, Lory S. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn. 2009;6:126–139.
- Flores AR, Galloway-Pena J, Sahasrabhojane P, et al. Sequence type 1 group B Streptococcus an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc Natl Acad Sci U S A. 2015;112(20):6431–6436.
- Fierer J, Guiney DG. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J Clin Invest. 2001;107(7):775–780.
- Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292(5519):1096–1099.
- Sakamoto Y, Suzuki Y, Iizuka I, et al. Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity. Sci Rep. 2015;5(1):11151.
- Liao Y, Brandt BW, Zhang M, et al. A single nucleotide change in the promoter mutp enhances fluoride resistance of Streptococcus mutans. Antimicrob Agents Chemother. 2016;60:7509–7512.
- Naito M, Hirakawa H, Yamashita A, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15(4):215–225.
- Kuwahara T, Yamashita A, Hirakawa H, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A. 2004;101(41):14919–14924.
- Cerdeno-Tarraga AM, Patrick S, Crossman LC, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307(5714):1463–1465.
- Smalley JW, Olczak T. Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol Oral Microbiol. 2017;32:1–23.
- Chen ZX, Potempa J, Polanowski A, et al. Stimulation of proteinase and amidase activities in Porphyromonas (Bacteroides) gingivalis by amino acids and dipeptides. Infect Immun. 1991;59(8):2846–2850.
- Rangarajan M, Aduse-Opoku J, Slaney JM, et al. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23(5):955–965.
- O’ Brien-Simpson NM, Paolini RA, Hoffmann B, et al. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect Immun. 2001;69(12):7527–7534.
- Pathirana RD, O’Brien-Simpson NM, Brammar GC, et al. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun. 2007;75(3):1436–1442.
- Wilensky A, Polak D, Houri-Haddad Y, et al. The role of RgpA in the pathogenicity of Porphyromonas gingivalis in the murine periodontitis model. J Clin Microbiol. 2013;40:924–932.
- van Steenbergen TJ, Kastelein P, Touw JJ, et al. Virulence of black-pigmented Bacteroides strains from periodontal pockets and other sites in experimentally induced skin lesions in mice. J Periodontal Res. 1982;17(1):41–49.
- Wilensky A, Gabet Y, Yumoto H, et al. Three-dimensional quantification of alveolar bone loss in Porphyromonas gingivalis-infected mice using micro-computed tomography. J Periodontol. 2005;76(8):1282–1286.
- Katz J, Ward DC, Michalek SM. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11(5):309–318.
- Grenier D, Mayrand D. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987;25(4):738–740.
- Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13(5):322–325.
- Darveau RP, Belton CM, Reife RA, et al. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66(4):1660–1665.