ABSTRACT
Introduction: The oral cavity harbors an abundant and diverse microbial community (i.e. the microbiome), whose composition and roles in health and disease have been the focus of intense research. Down syndrome (DS) is associated with particular characteristics in the oral cavity, and with a lower incidence of caries and higher incidence of periodontitis and gingivitis compared to control populations. However, the overall composition of the oral microbiome in DS and how it varies with diverse factors like host age or the pH within the mouth are still poorly understood. Methods: Using a Citizen-Science approach in collaboration with DS associations in Spain, we performed 16S rRNA metabarcoding and high-throughput sequencing, combined with culture and proteomics-based identification of fungi to survey the bacterial and fungal oral microbiome in 27 DS persons (age range 7–55) and control samples matched by geographical distribution, age range, and gender.
Results: We found that DS is associated with low salivary pH and less diverse oral microbiomes, which were characterized by lower levels of Alloprevotella, Atopobium, Candidatus Saccharimonas, and higher amounts of Kingella, Staphylococcus, Gemella, Cardiobacterium, Rothia, Actinobacillus, and greater prevalence of Candida.
Conclusion: Altogether, our study provides a first global snapshot of the oral microbiome in DS. Future studies are required to establish whether the observed differences are related to differential pathology in the oral cavity in DS.
Introduction
Down syndrome (DS), also known as trisomy of chromosome 21, is the most common genetic cause of mental disability worldwide [Citation1]. The estimated incidence of DS is between 1 in 1,000 and 1 in 1,100 live births worldwide, according to the World Health Organization. DS is generally caused by maternal nondisjunction errors during meiosis, which results in chromosome 21 trisomy in all cells of the body, wherein advanced maternal age is the main risk factor [Citation2]. DS is characterized by variability in cognitive development and distinct physical features causing unique health conditions, including congenital heart disease, immune system alterations, premature dementia, Alzheimer’s disease, and many other symptoms related to premature aging. Indeed, current clinical and experimental findings support the concept that DS may be considered a premature aging disorder [Citation3]. Consistent with this, DS presents with premature immune system senescence, increased plasmatic levels of inflammatory markers resembling the chronic increase in proinflammatory status observed during aging, as well as oxidative stress due to mitochondrial dysfunction [Citation3]. Furthermore, consistent with premature aging, two markers of biological age, DNA methylation and quantification of circulating N-glycan species (GlycoAgeTest), show differences in DS compared with control individuals [Citation4,Citation5].
The composition of the gut microbiome has been suggested to be a powerful marker for distinguishing between biological and chronological age [Citation6–9], and, therefore, the characterization of the gut microbiome in DS populations may be of interest [Citation10]. Development of new technologies and the application of metagenomic analyses have enabled the characterization of the human microbiome at different body sites [Citation11,Citation12]. The profiling of the gut microbiome of 17 individuals with DS and matching controls showed a similar structure of the gut microbiome, with alterations in only two genera: Parasporobacterium and Sutterella [Citation13].
The microbiome of the oral cavity has been related to several diseases, including not only common oral diseases such as gingivitis or periodontitis, but also systemic ones [Citation14]. The study of the oral microbiome in DS is of particular interest due to the many specific features of the oral cavity associated with this syndrome. These include, among others, different saliva composition, poor occlusal correlation, high frenum insertion, early mucogingival problems and advanced tongue position. In addition, due to genetic abnormalities in their immune system and environmental factors, DS patients are more susceptible to infections. In particular, it has been largely reported that persons with DS have an increased prevalence of periodontal disease [Citation15–20]. First observations pointed to an excessive inflammatory response of the gums followed by manifestations of aggressive and/or early-onset periodontitis, characterized by high plaque formation and increased presence of pathogenic species (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Eikenella corrodens, Tannerella forsythia, Fusobacterium nucleatum, Treponema denticola, and Campylobacter rectus) when compared with age-matched control groups and age-matched groups with intellectual disability [Citation21]. The early periodontal colonizers in DS are facilitated by a combination of lower salivary flow rate, limited antibody production in the saliva, and neutrophils with impaired chemotaxis which prevents the immune cells from reaching target pathogens [Citation22]. Several studies have focused on identifying factors influencing the onset of aggressive variants of oral diseases that often affect youths and adults with DS, such as destructive forms of periodontitis [Citation23,Citation24]. More recently, some authors have suggested that the composition of oral biofilms, independently of the immunological alterations in DS, has a critical role in periodontal development [Citation25]. Although recent studies have failed to find a specific combination of pathogens causing periodontitis in DS patients, other than the ones causing periodontitis in the general population, it appears that these might be established earlier in young adult DS, which is associated with early-onset and more aggressive forms of the disease [Citation26]. Despite the impaired immune responses and comparatively poor oral health in DS, a lower or similar incidence of dental caries in DS has been seen compared to non-DS [Citation27,Citation28]. This is potentially due to the relatively late eruption of teeth in DS, microdontia, more missing teeth and greater dental spacing [Citation29,Citation30], and so it would also be interesting to explore whether specific oral microbiotas are also related to this relatively lower incidence of caries in DS. Most previous studies have been based on identification by conventional PCR of already described individual species. Hence, we still lack a comprehensive understanding of the global oral microbiome composition in DS.
With the aim of shedding light on the composition of the oral microbiota in DS, and how it differs from similar non-affected populations, we used 16S rRNA metabarcoding coupled to culture and proteomics-based identification of fungi to characterize the bacterial and fungal components of the oral microbiome of 27 DS volunteers and their relatives, and compared it with non-affected volunteers. To this end, we collected and processed saliva samples from different locations across Spain in the context of the second edition of the citizen-science project ‘Saca la lengua’ (SLL2) (www.sacalalengua.org [Citation31],). We expected to find differing abundances of organisms that may explain the greater incidence of periodontitis and lower incidence of dental caries in DS. Furthermore, considering the various signs of premature aging across the body in DS, we attempted to analyze potential signs within the oral microbiome. Overall, our study provides a first global overview of the species present in the oral cavity of DS. Future studies would benefit from an increment of sample size and the characterization of relevant variables for the study, such as the presence of comorbidities that may be shaping differences in the composition of the microbiome of the oral cavity.
Materials and methods
Sample collection
The target population of this study was individuals with DS and their relatives, which were contacted with the collaborations of local associations of DS families. We collected 27 oral rinse samples from individuals with DS (ages 7–33) in the context of the second edition of the ‘Stick out Your Tongue’ citizen science project (SLL2, see http://www.sacalalengua.org [Citation31],), and in close collaboration with DS family associations in Spain. Sample collection was coupled to science communication activities with DS individuals and their relatives, aiming to raise awareness about the microbiome, its role in health and disease, and its potential particularities in DS. The SLL2 project questionnaire about health and lifestyle was adapted with the help of DS associations and was answered jointly by DS participants and their relatives [see metadata file at the following github link: https://github.com/Gabaldonlab/ngs_public/blob/master/SLL2/SLL2.metadata.xlsx]. There were 20 relatives that participated, 18 of which were parents, and two of which were siblings. The siblings were 10 and 28 years old (their DS siblings were 7 and 22 years old, respectively), while the parents ranged in age from 44 to 77 years old.
All participants signed an informed consent form allowing the use of their saliva samples for microbiological research. For participants under the age of 18, the consent form was also signed by one of the parents or a legal guardian. This project was approved by the ethics committee of the Barcelona Biomedical Research Park (PRBB). Samples were collected from January to November 2017. Participants were asked not to ingest any food or beverage (except water) for 1 h before collecting the sample. All donors received clear indications about the sample collection procedure in person, and the collection of the samples was carried out with the assistance of a researcher involved in the project, following a demonstration. All participants responded to a uniform questionnaire (see below), which was adapted for DS in collaboration with DS partner associations, so that DS participants could decide for themselves whether or not to participate, and be able to answer most questions on their own. Before collection of the oral rinse, the pH of the saliva was measured using pH test strips (MColorpHast, Merck, range 5.0–10.0; 0.5 accuracy units), the accuracy of which has been previously validated [Citation31]. Saliva samples were collected using a mouthwash as described earlier [Citation31]. In brief, the protocol is as follows: participants rinsed their mouth with 15 mL of sterile phosphate-buffered saline (PBS) solution, for 1 min. Then, they returned the liquid into a 50 mL tube. The samples were then centrifuged at 4,500 g for 12 min at room temperature (r.t.) in an Eppendorf 5430 centrifuge equipped with an Eppendorf F-35-6-30 rotor. The supernatant was discarded and the pellets were resuspended with the remaining PBS, transferred to 1.5 ml tubes and centrifuged at 4,500 g for an additional 5 min at r.t. using an Eppendorf FA-45-24-11-HS rotor. Supernatants were discarded, and pellets were frozen and stored at − 80°C until further analysis.
DNA extraction and sequencing
Sample DNA was extracted using the ZR-96 Fungal/Bacterial DNA kit (Zymo research Ref D6006), following the manufacturer’s instructions. The extraction tubes were agitated twice in a 96-well plate using Tissue lyser II (Qiagen) at 30 Hz/s for 5 min at 4°C. We included as controls of library preparation and MiSeq sequencing processes two DNA samples derived from bacterial mock communities from the BEI Resources of the Human Microbiome Project: each sample comprised genomic DNA of ribosomal operons from 20 bacterial species. The ‘HM-782D’ community contained an even number of ribosomal DNA per species (100,000 operons per species). The ‘HM-783D’ community contained a variable number of operons, ranging from 1,000 to 1,000,000 per species.
Obtained DNA was diluted to 12.5 ng/μl and used to amplify the variable regions known as V3–V4 of the 16S ribosomal RNA gene, using a pool of modified universal primers in a limited cycle PCR:
V3-V4-Forward
(5′- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′)
V3-V4-Reverse
(5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′)
Low sequence diversity or unbalanced base composition in template DNA can negatively affect the sequence output, quality, and error rate due to problems in cluster identification in MiSeq sequencing. To prevent this unbalanced base composition with all libraries having the same initial sequence, we performed the first PCR with a mix of four forward and four reverse primers, shifting the sequencing phases by adding a varying number of bases (from 0 to 3 N bases) as spacers. The PCR was carried out using KAPA HiFi HotStart ReadyMix (Roche) in a total volume of 10-μl with 0.2-μM of the primers. Cycling conditions were: 3 min at 95°C followed by 20 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final step of 5 min at 72°C. PCR products were purified using AMPure XP beads (Beckman Coulter), with a 0.9 x ratio and eluted with 32 μl of Buffer EB (Qiagen), then 30 μl of the eluate were transferred to a fresh 96-well plate.
Next, for the purpose of multiplex sequencing, we performed a second PCR in which full-length Nextera adapters (Illumina) with barcodes were added to the overhangs of the primers used in the first PCR. Thus, products of the second PCR step were sequencing ready libraries with an insert size of approximately 450 bp. Specifically, 5 μl of the first PCR was used as a template for a second PCR with Nextera XT v2 adaptor primers (Illumina) in a final volume of 50 μl using the same mix and conditions as the first PCR, but limited to eight cycles. Twenty-five μl of the final product of this second PCR was used for purification and normalization with the SequalPrep normalization kit (Invitrogen), using the manufacturer’s protocol. Libraries were eluted in a volume of 20 μl and pooled for sequencing. Pools were quantified by qPCR using the Kapa library quantification kit for Illumina (Kapa Biosystems) on an ABI 7900HT real-time cycler (Applied Biosystems). Libraries were sequenced on an Illumina MiSeq with 2 × 300 bp reads using v3 chemistry with a loading concentration of 15 pM. To increase the diversity of the sequenced sample, 10% of PhIX control libraries were spiked. Negative controls with the same conditions and reagents but with sterile water instead of samples were made for the buffer, DNA extraction, and PCR amplification steps. The controls provided no visible band or quantifiable amount of DNA by gel visualization or Bioanalyzer, whereas all samples resulted in clearly visible bands after 20 cycles. Twelve such controls were subjected to library preparation and sequenced. Expectedly, these sequenced non-template controls systematically yielded very few reads (a range of 155–1,005 reads per sample), in contrast to an average of ~64,000 reads/sample in sample-derived libraries.
Fungal composition analysis
Attempts to identify fungi through ITS amplification failed in many samples that nevertheless were positive for a culture-based approach. This likely related to a low presence of fungi in oral rinse samples and the difficulty to break the fungal cell wall to access the DNA in comparison to bacteria. We therefore used traditionally culture-based methods to enrich the possible fungal species present in the samples and identify them. First, we optimized the experimental procedures for fungal composition detection with a small subset of control oral rinse samples, testing the following growing conditions: i) starting with fresh versus frozen oral rinse samples, ii) using of different antibiotics concentrations, iii) growth temperature (25°C, 30°C, 37°C), iv) plating method (spread plate method versus pour plate method), and iv) duration of incubation (2, 4 or 7 days). We observed no significant differences in the number of grown colonies when starting with fresh versus frozen samples or when using different incubation temperatures, while the number of grown colonies when using the spread plate method was somewhat higher as compared to the pour plate method, and longer incubation times resulted in a higher number of grown colonies (data now shown). Thus, the preliminary results obtained helped us to design the following working protocol to carry out the analysis of the oral fungal composition with the study samples. First, we resuspended the frozen pellets in 100 μl of sterile PBS, taking 10 μl of those to be diluted in 40 μl of PBS (the remaining 90 μl were used for DNA extraction), from which we plated 30 μl (6% of the original sample) onto a YPD sterile plate with chloramphenicol and ampicillin (100 μg/ml each). After 7 days of incubation at 30°C, we counted the number of colonies, their phenotypes, and the presence of bacteria. A total of 10 colonies were randomly selected per sample (or all plate colonies if there were fewer than 10) and were re-grown under the same conditions in a fresh plate for 24 h. We used MALDI-TOF analysis for fungal identification and, in the instance of inconclusive results after two or three rounds of MALDI-TOF, we performed colony PCR to amplify the Internal Transcribed Spacer (ITS) hypervariable region of the ribosomal gene 5.8S (fungal marker) and further Sanger sequencing. Data are summarised in a table ().
Table 1. Analysis of colonies grown onto YPD + antibiotics plates. The table summarises the number (n) and frequency (%) of samples which formed colonies for a) yeasts, – with b) indicating the mean and the range of the number of colonies for the yeast positive samples – c) molds, d) bacteria, and e-q) identified fungal species as determined by MALDI-TOF per group: subjects with DS (Down Syndrome) and matched control individuals
MALDI-TOF analysis was performed with a MALDI Biotyper in the Centre for Omics Sciences (COS) in EureCat (Centre Tecnològic de Catalunya, Reus, Spain). Total proteins of colonies were extracted in our laboratory following their standard protocols and then samples were sent to COS for the analysis. In brief, we picked fresh grown colonies into 1.5 ml tubes, added 300 μl of milliQ water and mixed thoroughly. Next, we added 900 μl of 100% ethanol to the sample, mixed thoroughly again, and centrifuged the tubes for 2 min at 13,000 rpm in a benchtop centrifuge to remove the supernatant (the centrifugation step was repeated twice). To increase the efficiency of identification, we dried pellets for some minutes at room temperature. After that, we resuspended the pellet in 70% aqueous formic acid (covering the pellet), added an equal volume of 100% acetonitrile, mixed carefully, and centrifuged for 2 min at 13,000 rpm in a benchtop centrifuge. Finally, we transferred the supernatant with the fungal protein extract into a fresh tube. Samples were frozen at −20°C and sent to COS for analysis in a MALDI Biotyper (Bruker Daltonik MALDI Biotyper). Samples were deposited in duplicate on a Polished Steel Target Plate (Bruker), coated with the matrix α-Cyano-4-hydroxycinnamic (HCCA) and analyzed with MALDI-TOF/TOF (MALDI ultrafleXtreme, Bruker, Germany). Spectrum identification was performed using the real-time classification software MALDI Biotyper (Bruker, Germany). Thus, for each colony sample we obtained a score with the following meaning according to the manufacturer’s recommended score identification: i) 2.3–3.0, highly probable species identification; ii) 2.0–2.299, secure genus identification, probable species identification; iii) 1.7–1.999, probable genus identification; iv) < 1.7, not reliable information. Samples identified with scores lower than 2.0 were manually re-analyzed, and its spectrum was classified using the Biotyper Offline Classification software. We considered that the results were consistent with species identification when the best match had a score >2.0 and the second best match was at least >1.7 with at least the same genus as the first one. The rest of the samples were re-grown and the whole process was repeated. If after two more experiments with MALDI-TOF analysis the identification of the fungal species was still inconclusive, we performed ITS amplification and sequencing.
For ITS-Sequencing, we performed a colony PCR from fresh colonies (replated 12–24 h before) with DongSheng Biotech (DSBio) Taq mix (#2012) and 20 pmol of each primer (ITS1: 5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ; NL4: 5ʹ-GGTCCGTGTTTCAAGACGG-3ʹ) in a total volume of 40 μl, and the following PCR program: 5 min at 94°C, then 30 cycles of 30 s at 94°C, 30 s at 55°C, 1.5 min at 72°C, and a final extension of 5 min at 72°C. PCR products were purified with the QIAquick PCR Purification Kit (Qiagen), following the manufacturer’s instructions. Samples were eluted in 33 ul of elution buffer, and sent for Sanger sequencing to Eurofins Genomics sequencing service, following the SUPREMERUN recommendations. The resulting sequences were evaluated with the webtool Blast [Citation32]. Only samples with a high score (>95%) of identity were considered as correctly identified. Additionally, for some samples, we were able to identify the predominant species responsible for bacterial growth on the YPD plates with antibiotics.
Pre-processing of 16S rRNA sequence reads and taxonomy assignment
Sequence reads from fastq files were filtered using the ‘dada2’ R package (version 1.10.1) [Citation33] to produce counts of amplicon sequence variants (ASVs). Low-quality reads were first removed by applying the filterAndTrim function with the following parameters: forward and reverse reads were trimmed to lengths of 275 and 230 nucleotides, respectively (truncLen = c(275,230)); the leading 10 nucleotides were trimmed in both reads (trimLeft = c(10,10)); reads with maximum expected errors greater than five in both reads were discarded (maxEE = c(5,5)); all other parameters used the default values. The remainder of the pipeline followed the suggestions in the tutorial from the authors of the tool ([Citation34], https://benjjneb.github.io/dada2/tutorial.html). Taxonomy was assigned using the dada2-formatted database of SILVA version 132 [Citation35]. A phylogenetic tree for use in UniFrac distance calculations was generated by following a protocol [Citation36] that uses the ‘DECIPHER’ (version 2.10.2) [Citation37] and ‘phangorn’ (version 2.5.5) [Citation38] R packages. After processing reads with the dada2 pipeline, only those samples with at least 5,000 reads were retained. At the end of this process there were a total of 1,648 samples, though only a portion of those were used for the analyses of this study, as described below in the section ‘Statistical analyses’.
For analyses regarding the abundances of taxa, a centered log-ratio transformation was applied to the ASV counts. Zeros were first replaced with the ‘count zero multiplicative’ method in the cmultRepl function from the ‘zCompositions’ R package (version 1.3.4) [Citation39]. Then centered log ratios were calculated using the codaSeq.clr function from the ‘CoDaSeq’ R package (version 0.99.5) [Citation40,Citation41].
Diversity measures
Alpha diversity measures were calculated using the estimate_richness function from the ‘phyloseq’ R package (version 1.30.0) [Citation42]. For beta diversity measures, both the weighted and unweighted UniFrac distances, which weight dissimilarity between samples by phylogenetic distances between taxa, were calculated using the UniFrac function from the ‘phyloseq’ package. The weighted UniFrac distances gives additional weight based on taxa abundances. Bray-Curtis and Jaccard distances were calculated using the vegdist function from the ‘vegan’ R package (version 2.5–6) [Citation43]. As the Jaccard distance is based on the presence or absence of taxa, the decostand function, also from the ‘vegan’ package, was applied to the ASV counts table, using the method ‘pa’ for presence/absence, before the vegdist function was applied. The Aitchison distance was calculated using the aDist function from the ‘robCompositions’ R package (version 2.2.1) [Citation44].
Statistical analyses
When running statistical tests, we first randomly selected representative-matched non-DS samples as controls 100 times to ensure consistency in the results. These same 100 sub-samples were used for each of the relevant tests, and were matched for geographical location, age, and gender by the following process. Of the 1,621 non-DS samples in the SLL2 dataset, we removed those samples with any other chronic disorder, leaving 1,335 samples. The DS samples came from four autonomous communities in Spain (Andalucia, Catalunya, Galicia, and the Basque Country), so from the non-DS controls, we first randomly selected two times the proportion of DS samples from each of those locations (i.e. 2 x 3/27, 14/27, 1/27, and 9/27, respectively). To ensure a comparable age range, we determined rough age brackets of youth (under 20), adult (20–60), and senior (60 and over), and randomly selected from the geographically matched samples the same proportions of each age group from DS samples (i.e. 12/27 youths, 15/27 adults, 0/27 seniors). Among the DS samples, there were 12 females and 15 males. Thus, a given sub-sampling was finally rejected and reselected if the proportions of males and females were not similar to that of DS samples (i.e. (12±2)/27 females and (15±2)/27 males). Among all of the 100 sub-samplings, a total of 332 samples were used as matched controls.
For each of these sub-samples, a number of statistical tests were run with the DS and matched controls together. First, we performed a permutational multivariate analysis of variance (permanova) based on each of the five distance metrics mentioned above using the adonis function from the ‘vegan’ package. The model included the following fixed effects: DS/non-DS, gender, age, and population of the city/town from which the sample came (as a generalized proxy of both location and lifestyle).
Then, to determine differential abundances of taxa and variation in other variables like alpha diversity and pH, we performed a linear model using the function lm from the base R package ‘stats’ (version 3.6.3) [Citation45], again using the same fixed effects as for the permanova test. The abundance values used for these tests were the centered log ratios of the ASV counts, as described above. The Anova function from the ‘car’ R package (version 3.0–7) [Citation46] was used to calculate type-II anova tables, from which p-values were taken for each fixed effect in the model. These p-values were corrected for multiple testing with the p.adjust function from the ‘stats’ package, using the ‘fdr’ method.
Results
Increased abundance of periodontal pathogens in DS
A number of genera consistently showed significant differences in abundance between DS and controls among the 100 sub-samples (, see Methods section for explanation of this process) and all of these organisms have potential implications in the pathogenesis of periodontitis and dental caries, as will be explored in the discussion section. The genera found at higher abundance in DS included Kingella, Gemella, Cardiobacterium, Staphylococcus, Rothia, and Actinobacillus (, , Supplementary Figure 1). In addition, reads that could not be classified at even the phylum level were found at greater abundance in DS. While the relevance of these to DS is questionable, this observation suggests that rare and perhaps understudied organisms were more abundant in these DS samples. On the other hand, the genera Alloprevotella (and its phylum Bacteroidetes), Atopobium, and Candidatus_Saccharimonas (and its phylum Patescibacteria) were found at lower abundance in DS (, , Supplementary Figure 1). There were no taxa from the genus to the phylum level that differed significantly between the 20 relatives of DS individuals and the same sub-samplings of matched controls.
Table 2. Significance of differentially abundant taxa and other variables between DS and matched controls. Columns indicate, in this order, the taxonomic level or the type of variable considered, the organism name or the variable name, the tendency of the difference in DS (↗: higher in DS, ↘: lower in DS, permanova results are not directional), the mean adjusted p-value of the statistical comparison between DS and matched controls, and the numbers of matched controls sub-samples for which the test is significant (out of the 100 matched control groups that were sampled from the total SLL2 dataset). Rows are ordered by mean adjusted p-value within each variable group
Figure 1. Down Syndrome differs in factors affecting oral health. (a) Mean relative abundances of 15 of the most abundant genera in DS samples and matched controls. The remaining genera are grouped together and colored in white. (b) The two most significantly differentially abundant genera are shown (Kingella and Alloprevotella), as well as the salivary pH and alpha diversity as calculated by the Shannon diversity index
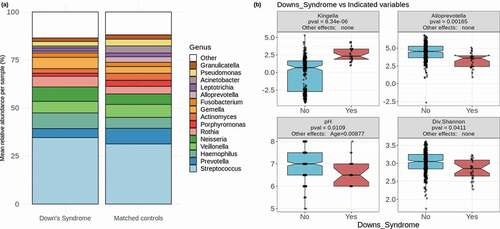
Lower pH and species diversity in DS
DS samples consistently had lower pH and lower alpha diversity than the matched controls, as calculated by the Shannon diversity index (). There was no significant difference in either alpha diversity or pH between the 20 relatives of DS individuals and the same sub-samplings of matched controls. Overall microbiome composition differed significantly in all 100 sub-samples between DS and matched controls based on a permanova test on each of the five distance metrics mentioned in the methods section, each of which calculates the distance between given samples using different criteria ().
Opportunistic pathogenic Candida species were more prevalent in DS
We assessed the presence of yeast in the oral microbiome through a culture-based approach coupled with proteomics identification (, see Materials and methods). This information was available for 26 of the 27 DS samples, and all of the matched control samples. Of the 26 DS samples, four were positive for Candida parapsilosis, while just two of the 332 matched controls used in all sub-samples were positive for C. parapsilosis (p = 0.00082 for multinomial log-linear model including all DS and matched control samples, mean adjusted p among 100 sub-samples = 0.0282, significant in 79 sub-sample tests, ). Candida dubliniensis was also found at a greater proportion in DS samples: four out of 26 as compared to five out of 332 matched controls (p = 0.00082), though this association was not consistently significant among 100 sub-samples (mean adjusted p = 0.0897, significant in two sub-sample tests, ). Yeasts in general were also proportionately more prevalent among DS samples, found in 14 of 26 samples as compared to 85 of 332 (p = 0.0161), though again this association was not consistently significant (mean adjusted p = 0.119, significant in 18 sub-sample tests, ). There was no significant difference in the prevalence of any yeast between the 20 relatives of DS individuals and the same sub-samplings of matched controls.
No evidence for premature aging in the oral microbiome of DS
Given the above-mentioned notion that the microbiome composition might reflect premature aging in DS, we looked for potential signatures of this process in the oral microbiome following two main approaches: first we assessed whether DS samples were more similar to those from non-DS older individuals than to those of non-DS younger individuals, and second, we checked whether changes across age were more drastic within DS samples than within non-DS samples. For the first approach we grouped all samples with no chronic disorders from the full SLL2 dataset, which included 1,335 samples ranging in subject age from 7 to 85 years old. These were placed into four age groups: child (under 13 years old), teen (13 to 19 years old), adult (20–59 years old), and seniors (60 years or older). We then compared the overall composition of DS samples to the samples in each of these age groups using five different distance metrics (Aitchison, weighted and unweighted UniFrac, Bray-Curtis, and Jaccard) (). Overall, we observed no apparent ‘premature aging’ effect in the DS samples in the sense that they would be more similar to samples from individuals older than themselves. Generally, DS samples showed the lowest difference with children, suggesting this is the most similar group to DS samples, while teens were typically the most distant, depending on the metric used. But none of the metrics showed that DS samples were most similar to either adults or seniors, which would be the expected result in the case of premature aging of the oral microbiome in DS. For the sake of clarity, in DS we had one child (age 7), 11 teens, and 15 adults (the maximum age was 33). We also did the same comparison including only the adult DS samples, as well as only the child and teen samples together, but in both instances the results were essentially the same as when including all 27 DS samples (Supplementary Figure 2a-b). In a similar approach, we grouped the non-DS samples into smaller age bins of 10 years, with the exception of 10–20 years old, which was made into two groups (10–15 and 15–20) since the majority of samples were teenagers of age 15–16, but again the same conclusions could be drawn (Supplementary Figure 2 c).
Figure 2. Comparisons across age. (a) Distributions of distances between DS samples and non-DS samples of each age group based on five distance metrics. Red stars represent p-values for Kruskal-Wallis tests to compare the means of each age group within the comparison of a given distance metric. The representation of p-values are as follows: 0 ‘****’ 0.0001 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘ ’ Not significant. (b) Scatterplot of age differences between pairs of samples vs Aitchison distance between those samples. Red points represent DS samples compared to other DS samples, blue points represent non-DS samples compared to other non-DS samples, with corresponding lines of the best fit
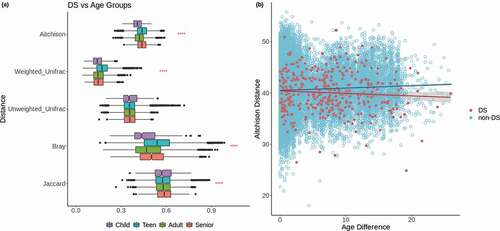
For the second approach, in order to determine whether changes in the microbiome across age were more extreme in DS, we looked for correlations between the difference in age between any pair of samples and the compositional distance between those samples, as calculated by the Aitchison distance. In this case, a premature aging effect might be inferred from a stronger positive correlation in DS samples between age difference and compositional distance than that seen in non-DS controls. However, as with the first approach, we did not see evidence of this effect in the DS samples, which actually showed a slightly negative correlation in these values which was not statistically significant (Pearson r = −0.065, p = 0.08) (). We compared this to non-DS samples of all ages (n = 1,335), as well as just those matched controls up to a maximum age of 33 (n = 262), in order to see the effect on the full range of ages, and just within the same age range as that of the DS samples, respectively. The full age range showed a statistically significant positive correlation between age difference and compositional distance (Pearson r = 0.101, p = 0). The matched controls up to age 33 showed a similar trend, though with a weaker correlation than that seen in the full age range (Pearson r = 0.0499, p = 4.9e-39). So, while there is evidence that there is a greater difference in overall composition as age difference increases between two non-DS samples, there is no evidence to suggest that this difference is more extreme in DS samples as a result of faster aging. In fact, this trend does not appear to occur at all in DS samples, though this may be the result of the relatively limited number of DS samples compared to non-DS samples. There were no differences between DS and matched control samples in the trajectory of abundances of any particular genus across age. However, there were differences for reads unclassified at the phylum level, which show the opposite trend between DS and non-DS, with a decrease across age in non-DS samples and an increase in DS samples (Supplementary Figure 3).
Discussion
Our study provides a first snapshot of the oral microbiome in DS and how it compares to non-DS individuals. The results indicate a significant shift in the overall composition of the oral microbiome between DS and non-DS individuals, with significantly lower pH and species diversity, as well as a larger presence of periodontal pathogens and Candida in DS. Although many DS symptoms relate to premature aging, we did not find such a signature in the composition of the oral microbiome.
Regarding the differences in microbiome composition between DS and non-DS individuals, the five distance metrics for which we ran the permanova test each measures the distance between samples using different criteria, and all five of these calculations showed statistically significant differences between the sample groups. The Bray-Curtis dissimilarity is most heavily influenced by dominant taxa in the samples, while the Jaccard index is based on the presence or absence of taxa, and thus primarily measures differences in rare taxa. The weighted and unweighted UniFrac distances use similar considerations to those of Bray-Curtis and Jaccard, respectively, but add additional weights based on phylogenetic distances between taxa. The Aitchison distance is based on centered log-ratio values, and thus is robust to changes in variation of abundances despite potentially different relative abundances of taxa that result from the compositional nature of the data. These results indicate strong differences between DS and non-DS across all levels of the oral microbiome, from rare taxa to the most dominant.
We do not have data on the incidence of any oral diseases in our samples, such as periodontitis or dental caries, as they are a subset of a larger exploratory study without an initial focus on oral disease (SLL2 – from ‘Saca La Lengua’ in Spanish, see http://www.sacalalengua.org[Citation31]). Nevertheless, our results do provide a platform from which to reflect on findings in the literature on oral health in both the general population and in DS. We found significant differences between DS and matched control samples in the abundances of a number of key genera of bacteria that are complicit in the pathogenesis of some oral diseases, particularly periodontitis. This follows with the increased incidence of periodontal disease in DS [Citation15–20], which has been posited to result from a number of factors, including diminished salivary flow leading to a reduced immune response in the oral cavity [Citation22,Citation47–49], as well as difficulties in dental treatment [Citation29,Citation50,Citation51].
However, we find a less straightforward connection to dental caries, wherein differential abundances of particular taxa suggest a non-caries environment, while the lower alpha diversity and low salivary pH suggest the potential occurrence of caries in DS samples. The literature has generally shown either lower incidence of caries in DS or no significant difference compared to non-DS [Citation27,Citation28]. This has been explained by the relatively late eruption of teeth in DS, microdontia, more missing teeth and greater dental spacing [Citation29,Citation30]. Many of the studies that, in the context of the differential abundances in our data, would suggest a low incidence of dental caries in our DS samples, were conducted with samples from young children [Citation52–58], and so are likely to represent early stages of cariogenesis. It may be that DS has a low incidence of dental caries because, despite typically worse oral hygiene, the unique dentition does not allow for optimal growth of many of the early plaque colonizers that initiate caries, while promoting the growth of organisms associated with the lack of caries. Nonetheless, the combination of poor oral health with the increase in acidogenic organisms in our DS samples and a low alpha diversity still may suggest a potentially cariogenic environment in DS.
The genera Kingella and Cardiobacterium, more abundant in DS samples in this study, form part of the HACEK group (made up of Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, and Kingella), which are the primary pathogens in infective endocarditis [Citation59]. This is a disease strongly linked to periodontal disease [Citation60–62], and one which can also be linked to individuals with DS [Citation63]. Increases in Kingella [Citation64–67] and Cardiobacterium [Citation68,Citation69] in the oral cavity have been associated with the pathogenesis of periodontitis, regardless of their involvement in endocarditis. On the other hand, Kingella has been found at lower abundance in samples with dental caries than in healthy controls [Citation52–54], though it is acidogenic [Citation67], a trait which may promote caries. Thus, the interpretation of the connection between Kingella and caries in the context of DS may be a bit muddled, but our data do follow the trend in the literature of low caries incidence in DS.
The other HACEK genera of Haemophilus, Aggregatibacter, and Eikenella were not differentially abundant in our dataset, but the genus Actinobacillus was more abundant in DS. This may be interesting in that Aggregatibacter is a relatively recently described genus made up of two former organisms of Haemophilus and the former Actinobacillus actinomycetemcomitans [Citation70], the latter of which (now labeled Aggregatibacter actinomycetemcomitans) is implicated in the pathogenesis of an aggressive form of periodontitis, acting to prime a site for growth of other periodontal pathogens [Citation71]. Thus, while Aggregatibacter itself is not differentially abundant in these DS samples, the increase of the closely related Actinobacillus may represent a similar effect upon oral health in DS.
The literature is less certain on the roles of some other organisms in periodontitis. DS samples also showed higher abundance of the genus Staphylococcus. The species Staphylococcus aureus is of particular interest, as it has been found to be both associated with periodontal health [Citation72] and periodontal disease [Citation73–75]. Another study found that there was no difference in abundance between healthy and diseased sites of individuals with periodontitis, and suggests that Staphylococcus is not involved directly in the pathogenesis, but rather acts synergistically to promote the growth of other pathogens [Citation76], a notion corroborated in another study saying that virulence factors of S. aureus may work to form biofilms along with other periodontal pathogens [Citation77]. In addition, S. aureus is the world’s most prominent pathogen of infective endocarditis [Citation78], a disease which may be linked to bacteremia resulting from dental procedures or even tooth brushing in individuals with poor periodontal health [Citation79–81]. The conjunction of DS with increased prevalence of periodontal disease and infective endocarditis illuminates the potential roles of Staphylococcus and the organisms of the HACEK group which we have seen at increased abundance in DS samples in this study.
Gemella and Rothia, also found at higher abundances in DS here, have had similar links to these DS-associated health concerns, as seen with Staphylococcus. Both Gemella [Citation82] and Rothia [Citation83,Citation84] have been associated with periodontal health. Conversely, Gemella [Citation85] and Rothia [Citation86] have been linked to periodontal disease in other studies. Furthermore, both Gemella [Citation87,Citation88] and Rothia [Citation89,Citation90] have been shown to occasionally cause endocarditis. One study [Citation91] showed that the species Gemella sanguinis, Gemella haemolysans, and Rothia dentocariosa were prevalent in infected sites of individuals with chronic periodontitis. They remained prevalent in infected sites of individuals that did not respond well to treatment. G. sanguinis even increased in these sites, but the species G. haemolysans and R. dentocariosa actually increased in individuals that responded well to treatment (the species associated with therapeutic success also included Cardiobacterium hominis and Kingella oralis, from two other genera that were higher in DS samples in our study) [Citation91]. It is difficult to conclude from this whether these species would then be associated with periodontal health, or if rather they were involved in particular stages of periodontal disease and, after treatment which worsened conditions for other organisms, were then primed to thrive in the now healthier host environment. For example, it has been posited that R. dentocariosa and K. oralis, among other organisms, are important for biofilm formation that may be used by other pathogens, due to the ability to form strong adhesive interactions [Citation65]. R. dentocariosa has also been shown to induce the production of tumor necrosis factor α (TNF-α) in the oral cavity [Citation92], leading to periodontal inflammation. Therefore, these species may be important in the initial pathogenesis of periodontitis, and then after a successful treatment that removes other competitors that have taken advantage of their biofilms, they are able to prosper. Regardless, the generally poorer oral health in DS may create the conditions necessary at least for the initial stages of periodontal disease, allowing for increased abundances of those organisms we have seen in this study. The increased Rothia in DS here also follows the results of the literature regarding dental caries, which show that Rothia is typically associated with low caries incidence [Citation56,Citation93], potentially due in part to nitrate reduction and the production of enterobactin, which can reduce the growth of some acidogenic and cariogenic organisms [Citation94]. Gemella has been conflictingly associated both with dental caries [Citation58] and with relative health [Citation52]. However, due its high degree of autoaggregation, which can allow for the initiation of plaque [Citation95], the species Gemella morbillorum may only be prominent in the initial stages of cariogenesis.
Apart from these relatively well-studied organisms, there was also an increased abundance in DS samples of reads that could not be classified at the phylum level. That would suggest that there is an increase in some rare and/or understudied bacteria, for which a 16S rRNA gene sequence is not available. No conclusive argument can be made as to the impact or cause of this increase in DS samples without knowing what they are, but for future studies it may be worthwhile to take note of any rare organism with an increased presence in DS samples. In this regard, whole-genome shotgun studies of the oral microbiome in DS will be helpful.
DS samples had lower abundances of the genera Alloprevotella (and its phylum Bacteroidetes as a whole), Atopobium, and Candidatus_Saccharimonas (as well as its phylum Patescibacteria). As with some of the other genera mentioned already, Alloprevotella and Atopobium have been shown in some instances to be associated with periodontitis [Citation68,Citation96,Citation97], and in others to not be significantly different between periodontitis and health [Citation98,Citation99]. Respectively, these studies showed that, while neither was different between periodontitis and health, subgingival Alloprevotella was associated with the early stages of rheumatoid arthritis, and Atopobium was significantly associated with gingival squamous cell carcinoma. However, Alloprevotella has been shown to be involved in nitrate reduction in saliva as an initial step in the circulation of nitrate between the saliva and digestive tract [Citation57]. This nitrate/nitrite cycle is part of the host defense reaction against periodontal disease [Citation100], since salivary glands boost the immune response as a reaction to periodontitis [Citation101]. Thus, the lower abundance of Alloprevotella in DS samples could be a result of the decreased salivary flow in DS, which may inhibit the overall nitrate availability to organisms like Alloprevotella, diminishing its ability both to thrive and to contribute to the host immune response in the pathogenesis of periodontitis. One study on the production of nitric oxide (NO) in the airways of cystic fibrosis patients, a process which also creates an anti-inflammatory effect, found that increasing dietary nitrate led to greater exhaled NO as compared to placebo treatments [Citation102]. The NO is produced by the same pathway of nitrate reduction by oral and airway bacteria, so increasing the nitrate intake in DS individuals may help to promote the growth of Alloprevotella and other nitrate reducers to boost the immune response, though this would require further study in the context of DS. Both Alloprevotella [Citation55–57,Citation103] and Atopobium [Citation52,Citation56,Citation104] have been shown to be associated with increased incidence of dental caries, so their low abundances in DS here again support the low caries in DS seen in the literature. The lower abundance of the genus Candidatus_Saccharimonas, of the fairly recently described Patescibacteria phylum [Citation105], does not seem to show the same connection from the literature, as this genus [Citation57] and its family, Saccharimonadaceae [Citation106], were shown to be found at lower abundances in individuals with dental caries than in healthy controls. This is the only taxon in this study that, on its own, would suggest greater incidence of dental caries in DS.
The alpha diversity was lower in DS samples than matched controls in this study, meaning the compositions of the DS samples were dominated more by particular organisms than those of the controls. The lower diversity is actually more reminiscent of the pathogenesis of dental caries than that of periodontitis. It has been shown that alpha diversity is lower in severe early childhood caries [Citation107,Citation108] and that it decreases as dental caries progresses over time [Citation109]. An explanation that has been posited for this was called the ‘ecological plaque hypothesis’ [Citation110], which suggests that acidogenic bacteria lower the pH and the diversity is reduced as other species intolerant of the acidic environment are inhibited. Conversely, alpha diversity has been shown to be higher in periodontitis [Citation111,Citation112] and increases with increasing severity of the disease [Citation113], as well as with particular periodontitis indicators, like periodontal pocket depth (PPD), bleeding on probing (BOP), and mean plaque index [Citation114]. As we do not have data related to specific aspects of the dental health in the samples of this study, we can only speak to the trend in our data that suggests generally worse oral health in DS samples than in matched controls, whether the characteristics be reminiscent of the pathogenesis of periodontitis or dental caries. Furthermore, the low pH that was seen in DS samples here, is important to cariogenesis according to the ecological plaque hypothesis, and has also been seen in periodontitis [Citation115,Citation116]. Salivary pH has also been shown to be lower in DS than in non-DS [Citation117,Citation118].
Yeasts were generally found to be more prevalent in DS, consistent with earlier reports [Citation119]. Candida species that most prominently differed with respect to matched controls were C. parapsilosis and, to a less significant extent, C. dubliniensis. Both of these species are opportunistic pathogens in the oral cavity that have been associated with periodontitis [Citation120–123] and dental caries [Citation124–126]. One study, however, from the Basque Country in Spain, actually found lower levels of both C. parapsilosis and C. dubliniensis in periodontitis as compared to controls [Citation127]. They suggested that this may be due to geographical differences with the other studies, as the pathogenesis of some fungal infections has been linked to geographical location [Citation128,Citation129]. Nevertheless, location cannot explain the differences from that study to ours, as all of our samples came from Spain, and nine of the 27 came specifically from the Basque Country. Thus, despite the findings of potential geographical aetiologies for fungal infections, DS may have a stronger impact on the growth of these Candida species (assuming at least a periodontitis-like environment in our DS samples, for the sake of comparison, which the evidence does suggest). For instance, studies have found negative correlations between salivary flow rates and the number of Candida colony forming units (CFUs) [Citation130,Citation131], so the low salivary flow in DS may promote oral Candida growth. Oral C. dubliniensis has generally been associated with immunocompromised individuals, and the hindered immune responses in the oral cavity in DS have already been touched upon here. C. parapsilosis has been shown to be acidogenic and able to induce salivary proteolysis via the production of secreted aspartyl proteinases [Citation132] and to demineralize tooth enamel [Citation133]. Thus, the fungal component of our data suggests an environment suitable to both periodontitis and cariogenesis in DS.
Between the 20 relatives of DS individuals and the matched controls used with the analyses of the DS samples, there were no differences in either abundances of particular bacterial taxa, alpha diversity, pH, or the prevalence of yeast species. Of these 20 relatives, 18 were parents of DS individuals and two were siblings. One of the sibling pairs were of the ages 7 and 10 years old, and the other pair was 22 and 28 years old. So, while we do not have information regarding the living situations of the participants of the study, we assume that most of the relatives live in the same households as the DS participants. Thus, the relevance of not finding any differences between relatives and the matched controls is that we can say with greater confidence that the differences found in DS samples are related to this disorder, and are likely not trends that are shared within a household or due to particular lifestyle factors that are specific to individual households or families.
Despite findings that have shown various effects of premature aging in DS [Citation3–5], there was no such effect apparent from the oral microbiome in DS in this study. DS samples were actually most similar to those of non-DS children and least similar to those of non-DS teens. Furthermore, while non-DS samples showed significantly greater differences in overall composition as age differences between samples increased, the DS samples did not show any trend. This may have been due to a lack of statistical power compared to the tests in the non-DS samples, or may suggest that there is even an ‘anti-aging’ effect on the oral microbiomes of DS individuals, in that the evolution of the composition of the oral microbiome occurs at a slower rate in DS than in healthy populations (though this claim would require more extensive analysis with a larger sample size and a longitudinal study design). A hypothesis for such a phenomenon might be that the oral microbiome composition in DS individuals is less dynamic than in non-DS individuals due to the limited salivary flow, unique dentition, and hindered immune responses in the oral cavity, creating a relatively static environment. If so, this idea may even help to explain the comparative lack of cariogenesis and some cariogenic organisms in DS, despite an apparently cariogenic environment. The confluence of factors may promote the formation of periodontal biofilms that can be maintained, but may be less favorable to the dental plaques that erode the hard tissues of teeth in dental caries.
Conclusions
This study has shown that there are significant differences in the overall composition of the oral microbiome between DS and non-DS individuals from across Spain. Differences in the abundances of particular organisms, both bacterial and fungal, follow what has been seen in the literature for increased incidence of periodontitis and decreased incidence of dental caries, and both of these tendencies have typically been seen in DS individuals. Nonetheless, the low pH and alpha diversity seen in DS samples do present a potentially cariogenic environment, but it may be that the unique dentition typically seen in DS impedes the early colonizers of dental plaques. Although there are various physiological signs of premature aging in DS, we have found no evidence of this phenomenon within the oral microbiome. There was nothing to suggest either that the oral microbiome in DS is more similar to older non-DS individuals, or that changes in the microbiome across age were more extreme in DS. Rather, there appears to be a relatively static microbial environment, potentially due to the limited salivary flow, unique dentition, and poor immune responses, though this hypothesis would require further exploration in a larger longitudinal study. Taken together, these results provide a glimpse into the distinctive oral microbiome in Down Syndrome and allow for a deeper understanding of the oral health trends therein.
Disclosure of interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author contributions
Conceptualization, T.G. and J.R.W.; formal analysis, J.R.W.; validation, J.R.W.; visualization J.R.W., T.G.; methodology, E.S., S.I.G., E.K., L.C. L.A.B., N.A.S., M.A.T., A.P.S., A.B., C.C., J.P., T.G., M.A.; data curation, L.C., J.P.; writing original draft, J.R.W., S.I., E.S., T.G.; supervision, T.G.; project administration, T.G.; funding acquisition, T.G.; resources T.G., J.P., J.H. All authors have read and agreed to the published version of the manuscript.
Availability of data and materials
The fastq files for the samples used for the analyses in this study were uploaded to the Sequence Read Archive (SRA) with the BioProject accession number PRJNA667,146 and can be found here: http://www.ncbi.nlm.nih.gov/bioproject/667146.
Supplemental Material
Download MS Word (412.6 KB)Acknowledgments
We are thankful to all citizens that participated in the second edition of the “Saca La Lengua” project by contributing samples and sharing ideas. In particular, for the work described here we are extremely thankful to the DS associations around Spain that actively participated in the project, including: Down España and the local associations Down Lleida, Down Bilbao, Down Vigo, and Down Malaga. Only with their efforts are studies like this possible. The authors acknowledge the CRG Genomics Core Facility, CRG Bioinformatics Core Facility, CRG biomolecular screening and protein technologies unit, CRG communication and public relationships department, and UCT ICTS High-Performance Computing unit for providing access to the computing facilities. CRG authors acknowledge the Spanish Ministry for Economy, Industry and Competitiveness (MEIC) for the EMBL partnership, and Centro de Excelencia Severo Ochoa.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lukowski AF, Milojevich HM, Eales L. Cognitive functioning in children with down syndrome: current knowledge and future directions. Adv Child Dev Behav. 2019;56:257–17.
- Sherman SL, Allen EG, Bean LH, et al. Epidemiology of Down syndrome. Ment Retard Dev Disabil Res Rev. 2007;13:221–227.
- Franceschi C, Garagnani P, Morsiani C, et al. The continuum of aging and age-related diseases: common mechanisms but different rates. Front Med. 2018;5:61.
- Horvath S, Garagnani P, Bacalini MG, et al. Accelerated epigenetic aging in down syndrome. Aging Cell. 2015;14(3):491–495..
- Borelli V, Vanhooren V, Lonardi E, et al. Plasma N-glycome signature of down syndrome. J Proteome Res. 2015;14:4232–4245.
- Rampelli S, Soverini M, D’Amico F, et al. Shotgun metagenomics of gut microbiota in humans with up to extreme longevity and the increasing role of xenobiotic degradation. mSystems. 2020;5. DOI:https://doi.org/10.1128/mSystems.00124-20.
- Liu A, Lv H, Wang H, et al. Aging Increases the Severity of Colitis and the Related Changes to the Gut Barrier and Gut Microbiota in Humans and Mice. J Gerontol A Biol Sci Med Sci. 2020;75:1284–1292.
- Maffei VJ, Kim S, Blanchard E 4th, et al. Biological aging and the human gut microbiota. J Gerontol A Biol Sci Med Sci. 2017;72:1474–1482.
- Biagi E, Franceschi C, Rampelli S, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26:1480–1485.
- Thevaranjan N, Puchta A, Schulz C, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2018;23(4):570.
- Peterson J, Garges S, Giovanni M, et al.; NIH HMP Working Group. The NIH human microbiome project. Genome Res. 2009;19(12):2317–2323..
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214.
- Biagi E, Candela M, Centanni M, et al. Gut microbiome in down syndrome. PLoS One. 2014;9(11):e112023.
- Willis JR, Gabaldón T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms. 2020;8(2):308.
- Cichon P, Crawford L, Grimm W-D. Early-onset periodontitis associated with down’s syndrome–a clinical interventional study. Ann Periodontol. 1998;3:370–380.
- Khocht A, Yaskell T, Janal M, et al. Subgingival microbiota in adult down syndrome periodontitis. J Periodontal Res. 2012;47:500–507.
- Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J Periodontol. 2008;79(8s):1560–1568.
- Barr-Agholme M, Dahllöf G, Linder L, et al. Actinobacillus actinomycetemcomitans, capnocytophaga and porphyromonas gingivalis in subgingival plaque of adolescents with Down’s syndrome. Oral Microbiol Immunol. 1992;7:244–248.
- Meskin LH, Farsht EM, Anderson DL. Prevalence of Bacteroides melaninogenicus in the gingival crevice area of institutionalized trisomy 21 and cerebral palsy patients and normal children. J Periodontol. 1968;39:326–328.
- Sakellari D, Arapostathis KN, Konstantinidis A. Periodontal conditions and subgingival microflora in Down syndrome patients. A case-control study. J Clin Periodontol. 2005;32:684–690.
- Amano A, Kishima T, Akiyama S, et al. Relationship of periodontopathic bacteria with early-onset periodontitis in Down’s syndrome. J Periodontol. 2001;72:368–373.
- Khocht A. Down syndrome and periodontal disease. Genetics and etiology of Down syndrome. Subrata Dey, InTechOpen; 2011. DOI: https://doi.org/10.5772/17371. Available from: https://www.intechopen.com/books/genetics-and-etiology-of-down-syndrome/down-syndrome-and-periodontal-disease.
- Agholme MB, Dahllöf G, Modéer T. Changes of periodontal status in patients with Down syndrome during a 7-year period. Eur J Oral Sci. 1999;107:82–88.
- Reuland-Bosma W, Dijk J. Periodontal disease in down’s syndrome: a review. J Clin Periodontol. 1986;13:64–73.
- Martinez-Martinez RE, Loyola-Rodriguez JP, Bonilla-Garro SE, et al. Characterization of periodontal biofilm in down syndrome patients: a comparative study. J Clin Pediatr Dent. 2013;37(3):289–296.
- Amano A, Murakami J, Akiyama S, et al. Etiologic factors of early-onset periodontal disease in Down syndrome. Japan Dent Sci Rev. 2008;44(2):118–127.
- Deps TD, Angelo GL, Martins CC, et al. Association between dental caries and down syndrome: a systematic review and meta-analysis. PLoS One. 2015;10(6):e0127484.
- Moreira MJS, Schwertner C, Jardim JJ, et al. Dental caries in individuals with down syndrome: a systematic review. Int J Paediatr Dent. 2016;26:3–12.
- Cheng RHW, Yiu CKY, Leung WK. Oral health in individuals with Down syndrome. In: Prenatal diagnosis and screening for down syndrome. Rijeka, Croatia: Subrata Dey, InTechOpen; 2011. p. 59–76.
- Vigild M. Dental caries experience among children with down’s syndrome. J Ment Defic Res. 1986;30(Pt 3):271–276.
- Willis JR, González-Torres P, Pittis AA, et al. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome. 2018;6(1):218.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410.
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583.
- Callahan BJ DADA2 pipeline tutorial (1.16) Available online: https://benjjneb.github.io/dada2/tutorial.html (accessed on 2020 Jul 9
- Callahan B Silva taxonomic training data formatted for DADA2 (Silva version 132); 2018;.
- Callahan BJ, Sankaran K, Fukuyama JA, et al. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 2016;5:1492.
- Wright ES. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R J. 2016;8:352.
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593.
- Palarea-Albaladejo J, Martín-Fernández JA. zCompositions — R package for multivariate imputation of left-censored data under a compositional approach. Chemometrics Intellig Lab Syst. 2015;143:85–96.
- Gloor GB, Wu JR, Pawlowsky-Glahn V, et al. It’s all relative: analyzing microbiome data as compositions. Ann Epidemiol. 2016;26:322–329.
- Gloor GB, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol. 2016;62:692–703.
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217.
- Oksanen J, Blanchet FG, Friendly M, et al. Vegan. Community Ecol Package. 2019; 10(631-637); 719.
- Templ M, Hron K, Filzmoser P. robCompositions: an R-package for robust statistical analysis of compositional data. Compositional data analysis: theory and applications. John Wiley and Sons; 2011. 341–355. ISBN 9780470711354.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020. Vienna, Austria.
- Fox J, Weisberg S. An R Companion to Applied Regression; Third. Sage; 2019. Thousand Oaks, CA.
- Areias C, Sampaio-Maia B, Pereira MDL, et al. Reduced salivary flow and colonization by mutans streptococci in children with Down syndrome. Clinics. 2012;67(9):1007–1011.
- Chaushu S, Becker A, Chaushu G, et al. Stimulated parotid salivary flow rate in patients with Down syndrome. Spec Care Dentist. 2002;22:41–44.
- Domingues NB, Mariusso MR, Tanaka MH, et al. Reduced salivary flow rate and high levels of oxidative stress in whole saliva of children with Down syndrome. Spec Care Dentist. 2017;37:269–276.
- Down’s heart group dental care for children and adults with heart problems and down’s syndrome - Down’s Heart Group Available online: https://dhg.org.uk/information/dental-care/(accessed on 2020 Jul 7).
- Dental PE. Care for the patient with down syndrome. Downs Syndr Res Pract. 1998;5:111–116.
- Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417.
- Lif Holgerson P, Öhman C, Rönnlund A, et al. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10(5):e0128534.
- Torlakovic L, Klepac-Ceraj V, Ogaard B, et al. Microbial community succession on developing lesions on human enamel. J Oral Microbiol. 2012;4:16125.
- Johansson I, Witkowska E, Kaveh B, et al. The microbiome in populations with a low and high prevalence of caries. J Dent Res. 2016;95:80–86.
- Xu L, Chen X, Wang Y, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study. Front Physiol. 2018;9:342.
- Espinoza JL, Harkins DM, Torralba M, et al. Supragingival plaque microbiome ecology and functional potential in the context of health and disease. MBio. 2018;9(6). DOI:https://doi.org/10.1128/mBio.01631-18
- Li Y, Zou C-G, Fu Y, et al. Oral microbial community typing of caries and pigment in primary dentition. BMC Genomics. 2016;17(1):558.
- Chambers ST, Murdoch D, Morris A, et al. HACEK infective endocarditis: characteristics and outcomes from a large, multi-national cohort. PLoS One. 2013;8(5):e63181.
- Dhotre SV, Davane MS, Nagoba BS. Periodontitis, bacteremia and infective endocarditis: a review study. Arch Pediatr Infect Dis. 2017;5. DOI:https://doi.org/10.5812/pedinfect.41067.
- Carinci F, Martinelli M, Contaldo M, et al. Focus on periodontal disease and development of endocarditis. J Biol Regul Homeost Agents. 2018;32:143–147.
- Ninomiya M, Hashimoto M, Yamanouchi K, et al. Relationship of oral conditions to the incidence of infective endocarditis in periodontitis patients with valvular heart disease: a cross-sectional study. Clin Oral Investig. 2020;24:833–840.
- Down’s heart group infective endocarditis in children and adults with heart problems and down’s syndrome - Down’s Heart Group Available online: https://dhg.org.uk/information/infective-endocarditis/(accessed on 2020 Jul 7
- Chen C. Distribution of a newly described species, Kingella oralis, in the human oral cavity. Oral Microbiol Immunol. 1996;11:425–427.
- Ruhl S, Eidt A, Melzl H, et al. Probing of microbial biofilm communities for coadhesion partners. Appl Environ Microbiol. 2014;80:6583–6590.
- Coelho ED, Arrais JP, Matos S, et al. Computational prediction of the human-microbial oral interactome. BMC Syst Biol. 2014;8:24.
- Dewhirst FE, Chen C-KC, Paster BJ, et al. Phylogeny of species in the family neisseriaceae isolated from human dental plaque and description of kingella orale sp. nov. Int J Syst Bacteriol. 1993;43:490–499.
- Lourenço TGB, Heller D, Silva-Boghossian CM, et al. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 2014;41:1027–1036.
- Han XY, Falsen E. Characterization of oral strains of Cardiobacterium valvarum and emended description of the organism. J Clin Microbiol. 2005;43:2370–2374.
- Nørskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56:2135–2146.
- Fine DH, Patil AG, Velusamy SK. Aggregatibacter actinomycetemcomitans (Aa) under the radar: myths and misunderstandings of Aa and its role in aggressive periodontitis. Front Immunol. 2019;10:728.
- Vieira Colombo AP, Magalhães CB, Hartenbach FARR, et al. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb Pathog. 2016;94:27–34.
- Fritschi BZ, Albert-Kiszely A, Persson GR. Staphylococcus aureus and other bacteria in untreated periodontitis. J Dent Res. 2008;87:589–593.
- Colombo AV, Barbosa GM, Higashi D, et al. Quantitative detection of Staphylococcus aureus, Enterococcus faecalis and Pseudomonas aeruginosa in human oral epithelial cells from subjects with periodontitis and periodontal health. J Med Microbiol. 2013;62:1592–1600.
- Loberto JCS, Martins, C.A. de P, Santos SSFD, et al. Staphylococcus spp. in the oral cavity and periodontal pockets of chronic periodontitis patients. Braz J Microbiol. 2004;35:64–68.
- Dos Santos BRM, Demeda CF, da Silva EENF, et al. Prevalence of subgingival Staphylococcus at periodontally healthy and diseased sites. Braz Dent J. 2014;25:271–276.
- Kim G-Y, Lee CH. Antimicrobial susceptibility and pathogenic genes of Staphylococcus aureus isolated from the oral cavity of patients with periodontitis. J Periodontal Implant Sci. 2015;45:223–228.
- Rajani R, Klein JL. Infective endocarditis: A contemporary update. Clin Med. 2020;20:31–35.
- Kinane DF, Riggio MP, Walker KF, et al. Bacteraemia following periodontal procedures. J Clin Periodontol. 2005;32:708–713.
- Lockhart PB, Brennan MT, Sasser HC, et al. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117:3118–3125.
- Lockhart PB, Brennan MT, Thornhill M, et al. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J Am Dent Assoc. 2009;140:1238–1244.
- Kumar PS, Griffen AL, Moeschberger ML, et al. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955.
- Kistler JO, Booth V, Bradshaw DJ, et al. Bacterial community development in experimental gingivitis. PLoS One. 2013;8:e71227.
- Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. Isme J. 2012;6:1176–1185.
- Al-Jebouri M. The Relationship between Periodontal Disease and Predisposing Factors. Tikrit J Dental Sci. 2016;4:68–80.
- Ramanan P, Barreto JN, Osmon DR, et al. Rothia bacteremia: a 10-year experience at Mayo clinic, Rochester, Minnesota. J Clin Microbiol. 2014;52:3184–3189.
- Al Soub H, El-Shafie SS, Al-Khal A-LM, et al. Gemella morbillorum endocarditis. Saudi Med J. 2003;24:1135–1137.
- Akiyama K, Taniyasu N, Hirota J, et al. Recurrent aortic valve endocarditis caused by Gemella morbillorum–report of a case and review of the literature. Jpn Circ J. 2001;65:997–1000.
- Ricaurte JC, Klein O, LaBombardi V, et al. Rothia dentocariosa endocarditis complicated by multiple intracranial hemorrhages. South Med J. 2001;94:438–440.
- Fridman D, Chaudhry A, Makaryus J, et al. Rothia dentocariosa endocarditis: an especially rare case in a previously healthy man. Tex Heart Inst J. 2016;43:255–257.
- Colombo APV, Bennet S, Cotton SL, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287.
- Kataoka H, Taniguchi M, Fukamachi H, et al. Rothia dentocariosa induces TNF-alpha production in a TLR2-dependent manner. Pathog Dis. 2014;71:65–68.
- Baker JL, Morton JT, Dinis M, et al. Deep metagenomics examines the oral microbiome during dental caries, revealing novel taxa and co-occurrences with host molecules. Genome Res. 2020 Nov 25. doi: https://doi.org/10.1101/gr.265645.120.
- Uranga CC, Arroyo P Jr, Duggan BM, et al. Commensal oral rothia mucilaginosa produces enterobactin, a metal-chelating siderophore. mSystems. 2020;5. DOI:https://doi.org/10.1128/mSystems.00161-20.
- Shen S, Samaranayake LP, Yip H-K. Coaggregation profiles of the microflora from root surface caries lesions. Arch Oral Biol. 2005;50:23–32.
- Chen C, Hemme C, Beleno J, et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. Isme J. 2018;12:1210–1224.
- Camelo-Castillo AJ, Mira A, Pico A, et al. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. 2015;6:119.
- Wolff B, Boutin S, Lorenz H-M, et al. FRI0698 Prevotella and alloprevotella species characterize the oral microbiome of early rheumatoid arthritis. Ann Rheum Dis. 2017;76:754.
- Li Y, Tan X, Zhao X, et al. Composition and function of oral microbiota between gingival squamous cell carcinoma and periodontitis. Oral Oncol. 2020;107:104710.
- Qu XM, Wu ZF, Pang BX, et al. From nitrate to nitric oxide: the role of salivary glands and oral bacteria. J Dent Res. 2016;95:1452–1456.
- Henskens YM, van den Keijbus PA, Veerman EC, et al. Protein composition of whole and parotid saliva in healthy and periodontitis subjects. Determination of cystatins, albumin, amylase and IgA. J Periodontal Res. 1996;31:57–65.
- Kerley CP, Kilbride E, Greally P, et al. Dietary nitrate acutely and markedly increased exhaled nitric oxide in a cystic fibrosis case. Clin Med Res. 2016;14:151–155.
- Uchida-Fukuhara Y, Ekuni D, Islam MM, et al. Caries increment and salivary microbiome during university life: a prospective cohort study. Int J Environ Res Public Health. 2020;17:3713.
- Kianoush N, Adler CJ, Nguyen K-AT, et al. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One. 2014;9:e92940.
- Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437.
- Schoilew K, Ueffing H, Dalpke A, et al. Bacterial biofilm composition in healthy subjects with and without caries experience. J Oral Microbiol. 2019;11:1633194.
- Li Y, Ge Y, Saxena D, et al. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–87.
- Hurley E, Barrett MPJ, Kinirons M, et al. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 2019;19:13.
- Gross EL, Leys EJ, Gasparovich SR, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128.
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271.
- LaMonte MJ, Genco RJ, Zheng W, et al. Substantial differences in the subgingival microbiome measured by 16S metagenomics according to periodontitis status in older women. Dent J. 2018;6:58.
- Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7:1016–1025.
- Genco RJ, LaMonte MJ, McSkimming DI, et al. The subgingival microbiome relationship to periodontal disease in older women. J Dent Res. 2019;98:975–984.
- Takeshita T, Kageyama S, Furuta M, et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama study. Sci Rep. 2016;6:22164.
- Baliga S, Muglikar S, Kale R. Salivary pH: A diagnostic biomarker. J Indian Soc Periodontol. 2013;17:461–465.
- Prasad M, Toshi, Sehgal R, et al. Periodontal disease and salivary pH: case control study. Prasad Mayuri, Toshi, Sehgal Rajat, Mallik Manisha, Nabi Aaysha Tabinda, Singh Sneha. Periodontal disease and salivary pH: case control study. Int Arch Integr Med. 2019;6:1–6.
- Davidovich E, Aframian DJ, Shapira J, et al. A comparison of the sialochemistry, oral pH, and oral health status of Down syndrome children to healthy children. Int J Paediatr Dent. 2010;20:235–241.
- Siqueira WLJ, Nicolau J. Stimulated whole saliva components in children with Down syndrome. Spec Care Dentist. 2002;22:226–230.
- Maranhão FCDA, Mendonça NM, Teixeira TC, et al. Molecular identification of candida species in the oral microbiota of individuals with down syndrome: a case-control study. Mycopathologia. 2020;185:537–543.
- Brusca MI, Rosa A, Albaina O, et al. The impact of oral contraceptives on women’s periodontal health and the subgingival occurrence of aggressive periodontopathogens and Candida species. J Periodontol. 2010;81:1010–1018.
- Urzúa B, Hermosilla G, Gamonal J, et al. Yeast diversity in the oral microbiota of subjects with periodontitis: candida albicans and Candida dubliniensis colonize the periodontal pockets. Med Mycol. 2008;46:783–793.
- Jewtuchowicz VM, Mujica MT, Brusca MI, et al. Phenotypic and genotypic identification of Candida dubliniensis from subgingival sites in immunocompetent subjects in Argentina. Oral Microbiol Immunol. 2008;23:505–509.
- McManus BA, Maguire R, Cashin PJ, et al. Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J Clin Microbiol. 2012;50:3335–3344.
- Naidu BV, Reginald BA. Quantification and correlation of oral candida with caries index among different age groups of school children: a case-control study. Ann Med Health Sci Res. 2016;6:80–84.
- Lozano Moraga CP, Rodríguez Martínez GA, Lefimil Puente CA, et al. Prevalence of Candida albicans and carriage of Candida non-albicans in the saliva of preschool children, according to their caries status. Acta Odontol Scand. 2017;75:30–35.
- Al-Ahmad A, Auschill TM, Dakhel R, et al. Prevalence of Candida albicans and Candida dubliniensis in caries-free and caries-active children in relation to the oral microbiota-a clinical study. Clin Oral Investig. 1963–1971;2016(20). DOI:https://doi.org/10.1007/s00784-015-1696-9
- De-La-Torre J, Quindós G, Marcos-Arias C, et al. Oral Candida colonization in patients with chronic periodontitis. Is there any relationship? Rev Iberoam Micol. 2018;35:134–139.
- Zhang X-B, Yu S-J, Yu J-X, et al. Retrospective analysis of epidemiology and prognostic factors for candidemia at a hospital in China, 2000-2009. Jpn J Infect Dis. 2012;65:510–515.
- Krom BP, Kidwai S, Ten Cate JM. Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res. 2014;93:445–451.
- Nadig SD, Ashwathappa DT, Manjunath M, et al. A relationship between salivary flow rates and Candida counts in patients with xerostomia. J Oral Maxillofac Pathol. 2017;21:316.
- Torres SR, Peixoto CB, Caldas DM, et al. Relationship between salivary flow rates and Candida counts in subjects with xerostomia. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;93:149–154.
- Wu T, Samaranayake LP. The expression of secreted aspartyl proteinases of Candida species in human whole saliva. J Med Microbiol. 1999;48:711–720.
- Caroline de Abreu Brandi T, Portela MB, Lima PM, et al. Demineralizing potential of dental biofilm added with Candida albicans and Candida parapsilosis isolated from preschool children with and without caries. Microb Pathog. 2016;100:51–55.
