ABSTRACT
Introduction: Cystic fibrosis (CF) is an autosomal genetic disease, associated with the production of excessively thick mucosa and with life-threatening chronic lung infections. The microbiota of the oral cavity can act as a reservoir or as a barrier for infectious microorganisms that can colonize the lungs. However, the specific composition of the oral microbiome in CF is poorly understood.Methods: In collaboration with CF associations in Spain, we collected oral rinse samples from 31 CF persons (age range 7-47) and matched controls, and then performed 16S rRNA metabarcoding and high-throughput sequencing, combined with culture and proteomics-based identification of fungi to survey the bacterial and fungal oral microbiome.Results: We found that CF is associated with less diverse oral microbiomes, which were characterized by higher prevalence of Candida albicans and differential abundances of a number of bacterial taxa that have implications in both the connection to lung infections in CF, as well as potential oral health concerns, particularly periodontitis and dental caries.Conclusion: Overall, our study provides a first global snapshot of the oral microbiome in CF. Future studies are required to establish the relationships between the composition of the oral and lung microbiomes in CF.
Introduction
Cystic fibrosis (CF) is a severe autosomal recessive genetic disease caused by mutations in the CF transmembrane conductance regulator (CFTR) gene [Citation1]. CF is one of the most common rare genetic disorders, particularly in the Caucasian population, affecting one in 2000–3000 newborns in the European Union [Citation2]. The CFTR protein acts as a chloride channel that transports ions across the apical membrane of epithelial cells throughout the body [Citation3]. This channel is involved in the production of several secretions including sweat, digestive fluids, and mucus. If the channel is impaired, these secretions increase their thickness, mostly affecting the function of the lungs, but also other organs such as the pancreas, liver, kidneys, and intestine. Lung infections are common and often develop into chronic and severe, life-threatening forms due to a deficient mucociliary clearance of the thick mucus [Citation4]. Several bacterial and fungal species are commonly associated with chronic respiratory infection of the lower airways in CF, including Aspergillus fumigatus, Candida albicans, Haemophilus influenzae, Staphylococcus aureus, Burkholderia cepacia complex, Stenotrophomonas maltophilia, Achromobacter xylosoxidans and Pseudomonas aeruginosa, with P. aeruginosa playing a major role in the morbidity and mortality of patients [Citation5,Citation6]. Hence, P. aeruginosa is a key pathogen in CF lung disease and has been found to be involved in the progressive obstructive pulmonary disease and bronchiectasis resulting from chronic endobronchial infection [Citation7]. These infections start early in life and progressively increase with patient age, with P. aeruginosa present in the lungs of up to 80% of patients over the age of 18 years [Citation8].
Despite the fact that P. aeruginosa is one of the most widespread and destructive opportunistic pathogens, it does not colonize the airways alone. Microbes commonly present in the oral cavity are also present in sputum from CF patients [Citation9]. In fact, the oral cavity has been proposed as a reservoir of bacteria, both commensal and pathogenic, that can colonize the lower airways due to micro-aspirations [Citation10,Citation11]. A model has been proposed comparing the biodiversity of the respiratory tract to island biogeography [Citation12], in which the mouth and throat, much like the mainland, are sources of relatively high species diversity, whereas the diversity of the airways decreases with the distance from the mouth, just as distant islands display more specific subsets of the mainland’s diversity. An investigation of this notion showed that samples taken from distal lung sites were more distinct from the upper respiratory tract than proximal sites [Citation13]. Thus, there might be a strong connection between the oral microbiome and many of the pulmonary pathogens acting in CF.
Beyond the mouth’s microbiota, the physiology of the oral cavity is also a factor in CF. Both salivary pH and the pH of the airways in CF are typically lower than those of non-CF individuals [Citation14,Citation15], due in part to malfunction of the CFTR protein, leading to defective secretion of the buffer molecule bicarbonate [Citation16]. The airway microbiota in CF have also shown a decrease in alpha diversity, a measure of the number of organisms present in a sample, and a value that decreases more with diminishing lung function [Citation17–21]. Taken together, these factors have important implications on the microbiome and oral health in general, though there is some debate in the literature. The acidic environment may leave CF individuals more susceptible to both dental caries and periodontitis [Citation22,Citation23]. The combination of low pH and low alpha diversity may predispose CF individuals to dental caries in particular, as according to the ‘ecological plaque hypothesis,’ acidogenic bacteria foster an acidic environment and the diversity drops as many species intolerant to the change are unable to grow [Citation24]. Periodontitis, on the other hand, has been shown to present with greater alpha diversity in the oral cavity [Citation25–28]. While some studies suggest a greater risk for dental caries in CF [Citation14,Citation29], a systematic literature review found that CF generally had lower incidence of caries, or no difference [Citation30]. The CFTR protein’s role of pH regulation is vital in odontoblasts, cells which secrete dentin during tooth development, and ameloblasts, cells which deposit enamel, and in fact, abnormal enamel mineralization has been seen in CF as a result of the defective CFTR protein [Citation31,Citation32]. Conversely, it has been suggested that the CFTR protein may actually promote periodontitis, and indeed a study of gingival biopsies showed greater and more widespread CFTR expression in patients with periodontitis compared to healthy controls [Citation33], suggesting that a mutation in the CFTR gene could predispose CF individuals to have better periodontal health. The review of oral health in CF also suggested that treatments for CF, such as antibiotics and inhaled anti-inflammatory medications, may protect CF patients from the colonization of early caries and periodontitis pathogens [Citation30]. One study found that airway pH increased in CF patients after antibiotic treatment [Citation15], and there have been instances of higher pH in CF as compared to controls, which have been attributed to treatment with supplementary pancreatic enzymes [Citation34]. The evidence suggests that the combination of factors typical to CF may inhibit cariogenic and periodontal pathogens in the oral cavity, but still produces an environment with a low pH and low alpha diversity that promotes damage to the outer layers of the teeth.
Here we present the first metabarcoding study, to our knowledge, of the oral microbiome in cystic fibrosis with a comparison to matched control samples. We compare the overall composition of oral bacteria between the groups and explore the differential abundances of bacterial taxa, as well as the presence or absence of yeast and mold species. We also calculate co-occurrence networks to study the underlying ecologies present in the CF and non-CF groups and further analyze the results alongside metadata collected through a citizen science approach.
Materials and methods
Sample collection
We contacted CF individuals and their relatives, through local associations of CF families integrated in the Spanish Federation for CF (https://fibrosisquistica.org/). Additionally, we analyzed samples from matched groups of individuals (see below) from a larger study targeting the general population (www.sacalalengua.org). All participants signed an informed consent form allowing the use of their saliva samples for microbiological research. For participants under the age of 18, the consent form was also signed by one of the parents or a legal guardian. This project was approved by the ethics committee of the Barcelona Biomedical Research Park (PRBB). Samples were collected from January to November 2017. Participants were asked not to ingest any food or beverage (except water) for 1 h before collecting the sample. All donors received clear indications about the sample collection procedure in person, and the collection of the samples was carried out with the assistance of a researcher involved in the project, following a demonstration. All participants responded to a uniform questionnaire (see below), which was adapted for CF in collaboration with CF partner associations. Before collection of the oral rinse, the pH of the saliva was measured using pH test strips (MColorpHast, Merck, range 5.0–10.0; 0.5 accuracy units), the accuracy of which have been previously validated [Citation35]. Saliva samples were collected using a mouthwash as described earlier [Citation35]. In brief, the protocol is as follows: participants rinsed their mouth with 15 mL of sterile phosphate-buffered saline (PBS) solution, for 1 min. Then, they returned the liquid into a 50 mL tube. The samples were then centrifuged at 4,500 g for 12 min at room temperature (r.t.) in an Eppendorf 5430 centrifuge equipped with an Eppendorf F-35-6-30 rotor. The supernatant was discarded and the pellets were resuspended with the remaining PBS, transferred to 1.5 ml tubes and centrifuged at 4,500 g for an additional 5 min at r.t. using an Eppendorf FA-45-24-11-HS rotor. Supernatants were discarded, and pellets were frozen and stored at − 80°C until further analysis.
A total of 31 oral rinse samples were collected from individuals with CF (ages 7–47) during the second edition of ‘Stick out your tongue’, a citizen science project in Spain (SLL2 – from ‘Saca La Lengua’ in Spanish, see http://www.sacalalengua.org [Citation35]), and in collaboration with CF family associations in Spain. Citizen science aims to involve the public in large-scale investigations to obtain a wide range of data as well as to increase public understanding of relatively esoteric scientific undertakings [Citation36,Citation37]. Sample collection was coupled with science communication activities with CF individuals and their relatives, aiming to raise awareness about the microbiome, its role on health and disease, and its potential particularities in CF. The SLL2 project questionnaire about health and lifestyle was adapted to CF with the help of CF associations.
In order to determine the effects of familial relations in the context of this study, we also collected samples from some relatives. Some of the 31 CF individuals were related to others also with CF, including 8 siblings and 2 individuals that were partners. There were 36 other relatives without CF that also participated in the study, which primarily consisted of parents, but also included 2 other partners, 4 siblings, and 1 grandmother. The ages of the siblings with CF ranged from 7 to 41 years old, while the siblings without CF ranged from 22 to 32 years old. The parents ranged from 41 to 71 years old.
DNA extraction and sequencing
The DNA extraction and amplification and sequencing of the V3-V4 region of the 16S ribosomal RNA gene were performed as previously described [Citation38]. Briefly, for sample DNA extraction we used the ZR-96 Fungal/Bacterial DNA kit (Zymo research Ref D6006), following manufacturer’s instructions. Two DNA samples derived from bacterial mock communities from the BEI Resources of the Human Microbiome Project were included as controls: ‘HM-782D’ and ‘HM-783D’. V3-V4 16S primers, PCR conditions, and library preparation for further Illumina MiSeq sequencing in multiplex with 2 × 300 bp reads using v3 chemistry were done following our previous protocols already described [Citation38]. We also included negative controls for the DNA extraction and PCR amplification steps, which provided no visible band or quantifiable amount of DNA by gel visualization or Bioanalyzer, whereas all samples resulted in clearly visible bands after 20 cycles. Twelve such controls were subjected to library preparation and sequenced. Expectedly, these sequenced non-template controls systematically yielded very few reads (a range of 155–1005 reads per sample), in contrast to an average of ~64,000 reads/sample in sample-derived libraries.
Fungal composition analysis
To assess the fungal composition in our samples, we used traditional culture-based methods to enrich for possible fungal species instead of fungal metagenomics technologies (such as ITS amplification), which produced unsatisfactory results in this type of samples in previous studies [Citation38]. In this way, we could overcome the limitations of the low presence of fungi in oral rinse samples and the difficulty to break the fungal cell wall to access the DNA in comparison to bacteria. We used previously optimized protocols in our group [Citation38], which mainly consists of plating 6% of the original sample onto a YPD sterile plate with antibiotics (100 μg/ml of chloramphenicol and 100 μg/ml of ampicillin). After 7 days of incubation at 30°C, we assessed the number of colonies, their phenotypes, and the presence of bacteria. A maximum of 10 colonies were randomly selected per sample and re-grown onto a fresh plate for 24 h in the same conditions. We used MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization-Time Of Flight) mass spectrometry analysis for fungal identification of each colony and, if the results were inconclusive after two attempts, we complemented this analysis with colony PCR to amplify the Internal Transcribed Spacer (ITS) hypervariable region of the 5.8S ribosomal gene (fungal marker) and further Sanger sequencing.
MALDI-TOF analysis was performed with a MALDI Biotyper (Bruker Daltonik MALDI Biotyper) in the Centre for Omics Sciences (COS) in EureCat (Centre Tecnològic de Catalunya, Reus, Spain), following previously described protocols [Citation38]: total proteins from the fungal colonies were extracted in our lab following standard protocols and then samples were sent to COS for the analysis. Samples were deposited in duplicate on a Polished Steel Target Plate (Bruker), coated with the matrix α-Cyano-4-hydroxycinnamic (HCCA) and analyzed with MALDI-TOF/TOF (MALDI ultrafleXtreme, Bruker, Germany). Spectrum identification was performed using the real-time classification software MALDI Biotyper (Bruker, Germany). Thus, for each colony sample, we obtained two alternative scores, which were interpreted as follows: i) 2.3–3.0, highly probable species identification; ii) 2.0–2.299, secure genus identification, probable species identification; iii) 1.7–1.999, probable genus identification; iv) <1.7, not reliable information. Samples identified with scores lower than 2.0 were manually re-analyzed, and its spectrum was classified using the Biotyper Offline Classification software. We considered that the results were consistent with species identification when the best match had a score >2.0 and the second best match was at least >1.7 with at least the same genus as the first one. The remaining samples were re-grown, and the whole process was repeated. If after two more experiments with MALDI-TOF analysis the identification of the fungal species was still inconclusive, we performed ITS amplification and sequencing.
For ITS-Sequencing, we performed a colony PCR from fresh colonies, directly using biomass material from a recently replated colony (maximum of 12–24 hours old). To do this colony PCR we used DongSheng Biotech (DSBio) Taq mix (#2012) and 20 pmol of each primer (ITS1: 5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ; NL4: 5ʹ-GGTCCGTGTTTCAAGACGG-3ʹ) in a total volume of 40 μl, and the following conditions: 5 min at 94°C, then 30 cycles of 30 s at 94°C, 30 s at 55°C, 1.5 min at 72°C, and a final extension of 5 min at 72°C. PCR products were purified with QIAquick PCR Purification Kit (Qiagen), following the manufacturer’s instructions. Samples were sent for Sanger Sequencing to Eurofins Genomics sequencing service, following SUPREMERUN recommendations. The resulting sequences were evaluated with the webtool Blast [Citation39]. Only samples with a high score (>95%) of identity were considered as correctly identified.
In summary, yeast colonies analyzed from a total of 75 samples could be properly identified by MALDI-TOF. For the rest of the samples (17) further analysis with ITS amplification and Sanger sequencing was required: 11 of them could be identified with certainty (percentage of identity >95% in Blast). Additionally, for some samples, we were able to identify the predominant species responsible for bacterial growth on the YPD plates with antibiotics.
Pre-processing of 16S rRNA sequence reads and taxonomy assignment
Sequence reads from fastq files were filtered using the ‘dada2’ R package (version 1.10.1) [Citation40] to produce counts of amplicon sequence variants (ASVs). Low quality reads were first removed by applying the filterAndTrim function with the following parameters: forward and reverse reads were trimmed to lengths of 275 and 230 nucleotides, respectively (truncLen = c(275,230)); the leading 10 nucleotides were trimmed in both reads (trimLeft = c(10,10)); reads with maximum expected errors greater than 5 in both reads were discarded (maxEE = c(5,5)); all other parameters used the default values. The remainder of the pipeline followed the suggestions in the tutorial from the authors of the tool ([Citation41], https://benjjneb.github.io/dada2/tutorial.html). Taxonomy was assigned using the dada2-formatted database of SILVA version 132 [Citation42]. A phylogenetic tree for use in UniFrac distance calculations was generated by following a protocol [Citation43] that uses the ‘DECIPHER’ (version 2.10.2) [Citation44] and ‘phangorn’ (version 2.5.5) [Citation45] R packages. After processing reads with the dada2 pipeline, only those samples with at least 1,000 reads were retained. At the end of this process, there were a total of 1,648 samples, though only a portion of those were used for the analyses of this study, as described below in the section ‘Statistical analyses’.
For analyses regarding the abundances of taxa, a centered log ratio transformation was applied to the ASV counts. Zeros were first replaced with the ‘count zero multiplicative’ method in the cmultRepl function from the ‘zCompositions’ R package (version 1.3.4) [Citation46]. Then centered log ratios were calculated using the codaSeq.clr function from the ‘CoDaSeq’ R package [Citation47,Citation48].
Diversity measures
Alpha diversity measures were calculated using the estimate_richness function from the ‘phyloseq’ R package (version 1.30.0) [Citation49]. For beta diversity measures, both the weighted and unweighted UniFrac distances, which weight dissimilarity between samples by phylogenetic distances between taxa, were calculated using the UniFrac function from the ‘phyloseq’ package. The weighted UniFrac distances give additional weight based on taxa abundances. Bray-Curtis and Jaccard distances were calculated using the vegdist function from the ‘vegan’ R package (version 2.5–6) [Citation50]. As the Jaccard distance is based on the presence or absence of taxa, the decostand function, also from the ‘vegan’ package, was applied to the ASV counts table, using the method ‘pa’ for presence/absence, before the vegdist function was applied. The Aitchison distance was calculated using the aDist function from the ‘robCompositions’ R package (version 2.2.1) [Citation51].
Statistical analyses
When running statistical tests, we first randomly selected representative matched non-CF samples as controls 100 times to ensure consistency in the results. These same 100 sub-samples were used for each of the relevant tests and were matched for geographical location, age, and gender by the following process. Of the 1,617 non-CF samples in the SLL2 dataset, we removed those samples with any other chronic disorder, leaving 1,335 samples. The CF samples came from seven autonomous communities in Spain (Andalucia, Aragón, Cantabria, Catalunya, Madrid, Galicia, and the Basque Country), so from the healthy controls, we first randomly selected two times the proportion of CF samples from each of those locations (i.e. 2x 4/31, 1/31, 11/31, 6/31, 6/31, 2/31, and 1/31, respectively). To ensure a comparable age range, we determined rough age brackets of youth (under 20), adult (20–60), and senior (60 and over), and randomly selected from the geographically matched samples the same proportions of each age group from CF samples (i.e. 10/31 youths, 21/31 adults, 0/31 seniors). Among the CF samples, there were 14 females and 17 males. Thus, a given sub-sampling was finally rejected and reselected if the proportions of males and females were not similar to that of CF samples (i.e. (14±2)/31 females and (17±2)/31 males). From all of the 100 sub-samplings, a total of 352 samples were used as matched controls.
For each of these sub-samples, a number of statistical tests were run with the CF and matched controls together. First, we performed a permutational multivariate analysis of variance (permanova) based on each of the five distance metrics mentioned above using the adonis function from the ‘vegan’ package. The model included the following fixed effects: CF/non-CF, reported use of antibiotics, gender, age, and population of the city/town from which the sample came (as a generalized proxy of both location and lifestyle). There were 21 CF samples that reported antibiotic use and 10 that did not.
Then, to determine differential abundances of taxa and variation in other variables like alpha diversity and pH, we performed a linear model using the function lm from the base R package ‘stats’ (version 3.6.3) [Citation52], again using the same fixed effects as for the permanova test. The abundance values used for these tests were the centered log ratios of the ASV counts, as described above. The Anova function from the ‘car’ R package (version 3.0–7) [Citation53] was used to calculate type-II anova tables, from which p-values were taken for each fixed effect in the model. These p-values were corrected for multiple testing with the p.adjust function from the ‘stats’ package, using the ‘fdr’ method.
Inferred co-occurrence networks
To produce co-occurrence networks, we first filtered out very rare taxa to avoid spurious associations in taxa that do not appear regularly, by using the filterTaxonMatrix function from the ‘seqtime’ R package (version 0.1.1) [Citation54]. We retained those taxa that had at least 15 counts in at least 20 of the 383 samples that included the CF and matched control samples. Then, we calculated the networks for the CF samples and each of the 100 matched control sub-samplings using the spice.easi function from the ‘SpiecEasi’ package (version 1.0.7) [Citation55]. To produce the network figures, we used a tutorial from the authors of the ‘SpiecEasi’ package as a guide [Citation56]. To calculate the Hamming distances between networks, we used the netdist function from the ‘nettools’ package (version 1.1.0) [Citation57].
Results
Increased abundances of airway pathogens and decreased abundances of some periodontal pathogens in CF
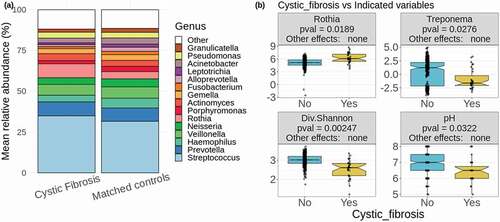
Our analyses identified 26 bacterial genera that consistently differed significantly in abundance between CF and controls among the 100 sub-samples (, see Methods section for explanation of this process) and many of these organisms have implications in the pathogenesis of CF and a number of oral health conditions, which will be examined in the discussion section. The genera found at higher abundance in CF as compared to matched non-CF sets included Chryseobacterium, Microbacterium, Brevundimonas, Blvii28 wastewater-sludge group, Stenotrophomonas, Streptococcus, Rothia, Staphylococcus, Delftia, Comamonas, Scardovia, Desulfobulbus, genera of the family Clostridiales_vadinBB60_group, Mobiluncus, Sphingobacterium, and Mogibacterium (, ), Supplementary Figure 1). On the other hand, the genera Peptostreptococcus, genera of the Clostridiales family Family_XIII, Alloprevotella, Treponema, Aggregatibacter, Parvimonas, Bergeyella, genera of the order Saccharimonadales, and Fusobacterium were found at lower abundance in CF, as well as, to a lesser extent, Haemophilus (, ), Supplementary Figure 1). At the phylum level, Firmicutes and Actinobacteria, and unclassified sequences, were found at higher abundances in CF, while Spirochaetes, Fusobacteria and Patescibacteria presented lower abundances in CF (Supplementary Figure 2). There were no taxa from the genus to the phylum level that differed significantly between the non-CF relatives of CF individuals and the same 100 sub-samplings of matched controls. All these differences were not affected by the fixed effects controlled for in the calculations (antibiotic use, gender, age, and population).
Table 1. Significance of differentially abundant taxa and other variables between CF and matched controls. Columns indicate, in this order, the taxonomic level or the type of variable considered, the organism name or the variable name, the tendency of the difference in CF (↗: higher in CF, ↘: lower in CF, permanova results are not directional), the mean adjusted p-value of the statistical comparison between CF and matched controls, and the numbers of matched controls sub-samples for which the test is significant. Rows are ordered first by the tendency in CF samples, with organisms/variables that were greater in CF first, and then by mean adjusted p-value within each variable group
Figure 1. Cystic fibrosis differs in factors affecting both oral and lung health. (a) Mean relative abundances of 15 most abundant genera in CF samples and matched controls. The remaining genera are grouped together and colored in white. (b) Two of the significantly differentially abundant genera are shown (centered log ratio values of Rothia and Treponema), as well as alpha diversity as calculated by the Shannon diversity index and the salivary pH. In the header of each boxplot, ‘Other effects’ refers to the significance of the fixed effects included in the calculations (antibiotic use, gender, age, and population). The label ‘none’ indicates that none of these had a significant effect on average for the given taxon or variable
Candida albicans more prevalent in CF
We used a culture-based approach paired with proteomics to determine the presence of yeast and mold species in our oral rinse samples (, see Methods section). Candida albicans was consistently significantly more prevalent CF samples than matched controls (, mean adjusted p among 100 sub-samples = 0.0310, significant in 82 of 100 sub-sample tests). C. albicans was detected in 17 of the 31 CF samples, while it was found in 50 of the 352 matched controls that were used in all of the sub-sampling tests (p = 0.0000875 for multinomial log-linear model including all CF and matched controls). We also detected yeast, regardless of species, more frequently in CF samples than matched controls, though less consistently than C. albicans in particular. Yeasts were found in 18 of 31 CF samples and in 74 of 352 matched controls (, mean adjusted p among 100 sub-samples = 0.0765, significant in 61 sub-sample tests, p = 0.000636 for multinomial log-linear model including all CF and matched control samples). These differences were not significantly affected by the fixed effects included in the calculations (antibiotic use, gender, age, and population).
Table 2. Analysis of colonies grown onto YPD + antibiotics plates. The table summarises the number (n) and frequency (%) of samples which formed colonies for a) yeasts, – with b) indicating the mean and the range of the number of colonies for the yeast positive samples – c) mold, d) bacteria, and e-p) identified fungal species as determined by MALDI-TOF per group: subjects with CF (Cystic Fibrosis) and matched control individuals
Lower alpha diversity and salivary pH in CF
CF samples consistently had lower alpha diversity than the matched controls, as determined by the Shannon diversity index, Faith’s phylogenetic diversity, which incorporates weights based on phylogenetic distances between taxa, and species richness, which is a count of the unique taxa identified in each sample. The CF samples also consistently had lower pH than matched controls ()). There was no difference in these alpha diversity values or in pH between the non-CF relatives of CF individuals and the same 100 sub-samplings of matched controls. The fixed effects included in the calculations (antibiotic use, gender, age, and population) did not have significant effects in any of these differences. There was also a significant difference in the overall composition of CF samples compared to matched controls based on a permanova test of the five distance metrics described in the methods, each of which measures distances in a different way ().
Networks of co-occurring taxa differ significantly between CF and controls
We inferred taxon co-occurrence networks among the 31 CF samples as well as each of the 100 sub-samples to explore underlying differences in the ecology of the oral microbiome in these conditions (). Using a Hamming distance calculation, which measures the degree to which the connections within two networks differ from each other, we found that the networks of the 100 matched control sub-samples were significantly more similar to each other than they were to the CF network (Kruskal–Wallis p-value = 2.2e-16, )). The mean Hamming distance between matched control networks was 0.018 ± 0.005, while the mean distance between the CF network and those of the matched controls was 0.025 ± 0.004.
Figure 2. Networks of co-occurring taxa differ significantly between cystic fibrosis and matched control samples. Co-occurrence networks of taxa in (a) the 31 CF samples and (b) all 352 matched control samples. Only those vertices representing taxa mentioned in the text are labeled, as these were the relevant connections that differed between CF and controls. Vertices are colored by the phylum to which they belong. Vertex sizes are proportional to the abundances of those taxa. Edges are colored according to the trend of the association between indicated taxa, where blue edges are positive and red edges are negative. Edge widths are proportional to the strength of the associations. For the matched control network in (b), this comes from a network calculated for all 352 controls together, merely for the sake of visualization. All statistics included in the text are based on the networks from the 100 matched control sub-samples. (c) Distributions of Hamming distances between the CF network and the 100 matched control networks (CF vs mC – yellow) and between each of the matched control networks (mC vs mC – blue). The p-value indicates the significance of the difference between these distributions
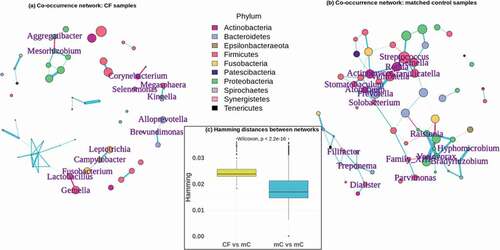
There were a number of specific correlations between taxa that differed between the CF network and those of the matched controls, which have potential implications for the oral cavity as a reservoir of microorganisms for the lower airways, as well as for oral health conditions. Some connections that occurred exclusively in the CF network included negative associations between Alloprevotella and Brevundimonas, Lactobacillus and both Fusobacterium and Gemella, and Aggregatibacter and Mesorhizobium, as well as positive associations between Campylobacter and Leptotrichia, Megasphaera and Kingella, and Corynebacterium and Selenomonas. These connections are summarized in , in the ‘CF networks’ section. There were also some connections that did not occur in the CF network, but did occur consistently in the matched control networks. These are also highlighted in , in the ‘Matched control networks’ section, which also shows the number of times a given association was significant among the 100 control networks.
Table 3. Associations of co-occurring taxa that differ between CF and matched control networks. The column ‘Association’ lists the co-occurring taxa. The column ‘Direction of correlation’ indicates the direction of that association. The final column shows the number of matched control networks (of the 100 sub-samplings that were performed) in which the correlation was significant
Discussion
Our results provide further evidence for the relationship between the microbiota of the oral cavity and the lower respiratory tract and suggest a potential reservoir function of the oral microbiome [Citation10–13]. We found a number of organisms that have been associated with lung infections in CF at higher abundance and prevalence in the oral rinse samples of CF individuals as compared to matched controls (), including the bacterial genera Chryseobacterium [Citation58–60], Microbacterium [Citation61,Citation62], Brevundimonas [Citation60,Citation63,Citation64], Stenotrophomonas [Citation19,Citation21,Citation65–67], Streptococcus [Citation21,Citation68–71], Rothia [Citation68,Citation71,Citation72], Staphylococcus [Citation21,Citation65,Citation68,Citation73], Delftia [Citation60,Citation64,Citation74–76], Comamonas [Citation19,Citation59,Citation64,Citation76], Scardovia [Citation64,Citation76,Citation77], Mobiluncus [Citation76,Citation78], Sphingobacterium [Citation60,Citation79], Mogibacterium [Citation76,Citation80], and the fungal species Candida albicans [Citation68,Citation81–84]. In addition, we found significant differences in the overall composition of the oral microbiome based on CF using a permanova test on five different distance metrics, each of which focuses on different aspects of the composition. Taken together, this highlights the strong differences across the entirety of the oral microbiomes of CF and non-CF individuals. Although we did not find a significant difference in the oral cavity between CF and non-CF in the abundance of Pseudomonas, the genus of the primary infective agent in CF, P. aeruginosa, the differences in these other taxa highlight the dramatic shift in the microbial equilibrium in CF. There is, of course, the possibility that the increased abundances of these organisms results from colonization of bacteria originating in the lower airways, but we feel that it is more likely that the mouth acts as a source of bacteria for the lungs, which become more susceptible to infection under the conditions of CF. For one, all of the organisms mentioned here are detected in both CF and non-CF oral cavities, which would preclude their appearance as a result of CF lung infections. Moreover, the absence of some of the primary infectors in CF in both sample groups, such as Burkholderia or Achromobacter, and the equivalent abundances of Pseudomonas would support the directionality we have suggested.
In fact, P. aeruginosa may be one of the few pathogens prominent in CF lung infections for which the abundance in the oral cavity is not an indicative biomarker, though it interacts with a number of oral species in the lung. Along with some of the other most significant CF pathogens like Staphylococcus and Stenotrophomonas, Pseudomonas is actually a late colonizer of the lungs. These taxa dominate the lung microbiome in CF after the age of 6, while the lungs of CF patients under the age of 2 are primarily composed of oral commensal species from the genera Streptococcus, Prevotella and Veillonella [Citation21]. When Streptococcus colonizes before Pseudomonas, the oral species S. salivarius is able to inhibit the growth of gram-negative bacteria, such as P. aeruginosa, by the production of lactic acid which disrupts their cell membranes [Citation85,Citation86]. This effect may inhibit the initial spread of some CF pathogens to the lower airways, but it primarily acts within the mouth and throat [Citation87], and so other mechanisms allow growth in the lungs as the patient ages. Even as some Streptococcal species inhibit Pseudomonas in the upper airways, others can promote its growth in the lower airways. One model that has been proposed for the physiology of CF lung infections suggests that 2,3-butanediol produced by oral and airway Streptococcus is abundant due to increased fermentation under more anaerobic conditions. The 2,3-butanediol can act as a buffer for the decreased pH, but then becomes a carbon source for both P. aeruginosa and Rothia mucilaginosa, and adds to the virulence of P. aeruginosa by allowing it to produce more reactive oxygen species and to provide additional electron acceptors to other anaerobic CF pathogens [Citation71].
Rothia mucilaginosa is an oral commensal species and perhaps the most closely tied to P. aeruginosa in CF lung infections as it forms similar biofilms [Citation88], may do so alongside P. aeruginosa [Citation72], and can act as a source of metabolites necessary for the production of glutamate by P. aeruginosa, which is used as a component of its cell wall and may increase its virulence [Citation89]. R. mucilaginosa has been associated with a decline in lung function in CF [Citation68] and has been suggested to adapt very efficiently to the CF lung environment, to the point that individual CF patients may have unique strains of the species, as seen with P. aeruginosa and Staphylococcus aureus [Citation72]. It is able to do so by the use of extracellular lactate, which is present at higher concentrations in CF individuals with poor lung function [Citation90], in order to undergo fermentation under anaerobic conditions, as well as adapting to resist antibiotics, protect against foreign nucleic acids, and to take advantage of the Fe2+ produced by the reduction of Fe3+ by both P. aeruginosa and S. maltophilia in the low-pH CF lung [Citation72].
Desulfobulbus is one of the only taxa found at higher abundance in the oral cavity of CF individuals in this study that, to our knowledge, has not been reported as a CF lung pathogen, though the evidence suggests that it may be well suited to a CF environment. Desulfobulbus is a sulfate-reducer [Citation91], which is relevant because P. aeruginosa is able to use sulfate from mucins in the airways [Citation92], and the resulting volatile sulfur compounds may be utilized by the fungus Aspergillus fumigatus, a common CF pathogen [Citation93]. In the same vein, it has been shown that there is increased sulfation of mucus glycoproteins in CF [Citation94,Citation95].
The frequent treatment of CF with antibiotics may result in the development of antibiotic resistance in a number of pathogens, with evidence that rates of resistance are increasing in some species [Citation66], including Candida albicans [Citation96], also found at greater prevalence in CF samples in our study. C. albicans has been implicated in CF lung infections [Citation81,Citation82,Citation84], and in particular, it has been associated with a substantial decline in lung function [Citation68,Citation83], as well as pancreatic insufficiency, exacerbation of lung infection, lower BMI and lower percentage of predicted forced expiratory volume in 1 second (FEV1) [Citation83].
Although in this study we did not have specific information regarding the oral health status of the donors, as they were taken from a larger exploratory study that was not focused explicitly on oral diseases (SLL2 – from ‘Saca La Lengua’ in Spanish, see http://www.sacalalengua.org [Citation35]), the results presented here broadly support the notion seen throughout the literature that, in CF patients, there is a lower incidence of periodontitis, though they develop an acidic oral environment prone to harm the enamel and dentin of the teeth. Factors that may indicate lower incidence of periodontitis in CF include low alpha diversity [Citation25–28], as seen in our data, malfunction of the CFTR protein [Citation33], and generally higher use of anti-inflammatory and antibiotic medications [Citation30]. Indeed, nearly all of the genera found at lower abundance in CF samples here have been associated with periodontal disease. Many of these are members of the various bacterial complexes which form biofilms at different stages of the disease [Citation97–103]. Some of the organisms found at higher abundance in CF samples in this study have been implicated in dental caries and carious lesions on the teeth [Citation104–116] and, conversely, others found at lower abundance in CF samples have been linked to the absence of caries and tooth damage [Citation109,Citation117–121].
Alloprevotella presents an interesting case that may merit further study on its own regarding CF. Species in this genus reduce nitrate in the saliva which contributes to the anti-inflammatory response to periodontitis [Citation122–124], so its lower abundance in CF here could be linked in part to lower incidence of periodontitis. However, the nitrate reduction in CF patients seems to be a more complicated process. When it is metabolized to nitric oxide (NO), it has an anti-inflammatory effect in the airways of CF individuals. Some studies have shown that precursors to NO, like nitrate and nitrite, are higher in the saliva and exhaled breath condensate (EBC) of CF patients than in those of controls, but nonetheless, the amount of exhaled NO was lower in CF [Citation125,Citation126]. One proposed explanation is that NO may be produced normally in CF, but its diffusion is inhibited in the thick mucus produced in CF airways [Citation127], though these first two studies suggest that there is an impairment in the formation of NO in CF patients. Another study found that increasing the intake of dietary nitrate led to an increase in exhaled NO as compared to placebo treatments [Citation128]. From this information, we cannot extrapolate to determine the exact mechanism in the impairment of the NO cycle in CF, but the low abundance of the nitrate-reducing genus Alloprevotella forms a link to this process that may warrant a deeper investigation.
Significant differences in many co-occurrences of taxa among the CF samples and the 100 matched control groups suggest underlying ecological differences in these two conditions. The Hamming distance calculations showed that the 100 matched control networks were more similar to each other than to the CF network ()), and we also highlighted a number of particular co-occurrences that were specific to either the CF or control networks, and these may have implications for the connections between the mouth and lung in CF, as well as for oral health conditions. In the CF network (), ), the negative association between the genera Alloprevotella and Brevundimonas follows with the results from our study and the literature, wherein Alloprevotella was lower and Brevundimonas was higher in CF [Citation60,Citation63,Citation64]. It is unclear whether this connection is related to the NO cycle in CF, but at least some strains of Brevundimonas species are nitrate reducers [Citation129], so it may be that Brevundimonas is able to outcompete Alloprevotella in the CF environment. There may be a more concrete explanation for the negative associations between Lactobacillus and both Fusobacterium and Gemella, which are periodontal pathogens [Citation97,Citation130,Citation131]. Many studies explore the use of Lactobacillus species as probiotic treatments and have shown that they can inhibit periodontal pathogens [Citation132] by co-aggregating with them, promoting immune responses and interrupting biofilm formations [Citation133]. Campylobacter and Leptotrichia also have a meaningful connection in the CF oral microbiome, as they can be found in association in dental caries [Citation134], where they can metabolize sugars to lactic acid, aiding in the process of cariogenesis [Citation135,Citation136], and we have already mentioned that extracellular lactate levels are higher in CF sputum [Citation90]. Megasphaera is sometimes associated with CF [Citation137,Citation138] and has been considered a member of the core microbiome of the lower lung, regardless of CF status [Citation139]. This genus can work to neutralize acidic conditions that damage teeth [Citation140] and Kingella is acidogenic [Citation141], so their co-occurrence in the CF network may indicate that they complement each other in the acidic CF oral cavity. The association between Corynebacterium and Selenomonas in CF is reasonable, as both have been implicated in CF [Citation142–144].
Among the matched control samples, the co-occurrence networks primarily support our speculations on the underlying mechanisms that might lead to lower incidence of periodontitis and greater damage to enamel in CF patients as compared to non-CF controls. Prevotella and Veillonella are among the four most abundant genera in our dataset and were significantly associated in 92 of the matched control networks. Prevotella, Veillonella, and Solobacterium are all periodontal pathogens [Citation98,Citation145,Citation146], and Prevotella melaninogenica and Solobacterium moorei, whose genera were significantly associated in 61 matched control networks, utilize cysteine to produce hydrogen sulfide (H2S), resulting in halitosis [Citation147,Citation148]. Cysteine is a precursor to glutathione [Citation149], a peptide that helps to protect the lung from oxidants, and is found at lower levels in the lungs of CF patients [Citation150], and acetylcysteine, a prodrug to cysteine, has been used to improve lung function in CF patients [Citation151]. So perhaps there is a connection between greater overall availability of cysteine in non-CF individuals and the production of H2S by organisms like P. melaninogenica and S. moorei. Veillonella species also produce volatile sulfur compounds that cause halitosis [Citation146]. Streptococcus and Gemella are among the most abundant taxa present in the oral microbiome ()) so it is not strange that they would have a significant association in 92 of the 100 control networks, though it is curious that they did not in the CF network. As mentioned above, Streptococcus was significantly more abundant in our CF samples and has been associated with CF lung infections [Citation21,Citation68–71], and Gemella may play a role in exacerbations of CF infections [Citation152]. Nonetheless, they both have some involvement in periodontitis as well [Citation130,Citation131,Citation153,Citation154], and species of Streptococcus in particular make up the ‘yellow-complex’ of periodontal pathogens, which are early colonizers in that disease [Citation145,Citation155]. Somewhat unexpected associations also occurred exclusively in the matched control networks between Gemella and Granulicatella (occurs in 72 of the 100 matched control networks), and between Rothia and Granulicatella (55 of the 100 control networks), as Granulicatella has also been associated with CF [Citation73,Citation156,Citation157] and caries due to its acidogenic nature [Citation158]. The significant associations seen exclusively in our matched control networks involving Ralstonia and Variovorax [Citation159–162], Atopobium [Citation163,Citation164], Bradyrhizobium [Citation165], Hyphomicrobium [Citation166], Actinomyces [Citation167], Stomatobaculum [Citation168], Rothia [Citation102,Citation163,Citation164], Clostridiales Family_XIII [Citation101], Parvimonas [Citation97,Citation101,Citation169], Treponema [Citation97,Citation101], Filifactor [Citation101,Citation170,Citation171], and Dialister [Citation172,Citation173], all relate to organisms implicated in periodontitis.
Conclusion
In this study, we found significant differences in the oral microbiomes of CF and non-CF individuals from all around Spain, which have implications for the potential of the oral cavity to act as a reservoir of microorganisms for the lower airways, as well as for oral health in CF. Differential abundances of bacteria and fungi follow trends in the literature regarding CF lung infections, presenting similar microbial activity in the oral cavity. We highlighted underlying physiological differences that are apparent in the co-occurrences of taxa among CF samples and matched control samples, which add greater evidence to those trends discussed here. These results provide a snapshot of the unique composition of the CF oral microbiome and how it relates to oral health and the lower airways.
Author Contributions
Conceptualization, T.G. and J.R.W.; formal analysis, J.R.W.; validation, J.R.W.; visualization J.R.W., T.G.; methodology, E.S., S.I.G., E.C., E.K., L.C. L.A.B., N.A.S., M.A.T., A.P.S., A.B., C.C., J.P., T.G., M.A.; data curation, L.C., J.P.; writing original draft, J.R.W., S.I.G., E.S., E.C., T.G.; supervision, T.G.; project administration, T.G.; funding acquisition, T.G.; resources T.G., J.P., J.H. All authors have read and agreed to the published version of the manuscript.
Disclosure of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Supplemental Material
Download MS Word (423.9 KB)Acknowledgments
We are thankful to all citizens that participated in the second edition of the “Saca La Lengua’’ project by contributing samples and sharing ideas. In particular, for the work described here we are extremely thankful to the Spanish Federation of Cystic Fibrosis (www.fibrosisquistica.org) CF and their affiliated local associations around Spain that actively participated in the project. Only with their effort are studies like this possible. The authors acknowledge the CRG Genomics Core Facility, CRG Bioinformatics Core Facility, CRG biomolecular screening and protein technologies unit, CRG communication and public relationships department, and UCT ICTS High Performance Computing unit for providing access to the computing facilities. CRG authors acknowledge the Spanish Ministry for Economy, Industry and Competitiveness (MEIC) for the EMBL partnership, and Centro de Excelencia Severo Ochoa.
Data availability statement
The fastq files for the samples used for the analyses in this study were uploaded to the Sequence Read Archive (SRA) with the BioProject accession number PRJNA667,146 and can be found here: http://www.ncbi.nlm.nih.gov/bioproject/667146.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Zielenski J, Rozmahel R, Bozon D, et al. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics. 1991;10:214–17.
- Bassett DE Jr, Boguski MS, Hieter P. Yeast genes and human disease. Nature. 1996;379:589–590.
- Saint-Criq V, Gray MA. Role of CFTR in epithelial physiology. Cell Mol Life Sci. 2017;74:93–115.
- Tilley AE, Walters MS, Shaykhiev R, et al. Cilia dysfunction in lung disease. Annu Rev Physiol. 2015;77:379–406.
- Taylor CJ, McGaw J, Howden R, et al. Bacterial reservoirs in cystic fibrosis. Arch Dis Child. 1990;65:175–177.
- Marshall BC, Elbert A, Petren K, et al. Cystic fibrosis foundation patient registry 2014 annual data report. Bethesda, Maryland: Cystic Fibrosis Foundation; 2015.
- Rosenfeld M, Emerson J, McNamara S, et al. Risk factors for age at initial Pseudomonas acquisition in the cystic fibrosis epic observational cohort. J Cyst Fibros. 2012;11:446–453.
- Saiman L, Siegel J. Cystic fibrosis foundation consensus conference on infection control participants. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Am J Infect Control. 2003;31:S1–62. Available from: https://www.ncbi.nlm.nih.gov/pubmed/12762292
- Rivas Caldas R, Le Gall F, Revert K, et al. Pseudomonas aeruginosa and periodontal pathogens in the oral cavity and lungs of cystic fibrosis patients: a case-control study. J Clin Microbiol. 2015;53:1898–1907.
- Boutin S, Graeber SY, Weitnauer M, et al. Comparison of microbiomes from different niches of upper and lower airways in children and adolescents with cystic fibrosis. PLoS One. 2015;10:e0116029.
- Gomes-Filho IS, Passos JS, Seixas Da Cruz S. Respiratory disease and the role of oral bacteria. J Oral Microbiol. 2010;2:5811.
- Whiteson KL, Bailey B, Bergkessel M, et al. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med. 2014;189:1309–1315.
- Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12:821–830.
- Pawlaczyk-Kamieńska T, Borysewicz-Lewicka M, Batura-Gabryel H. Salivary biomarkers and oral microbial load in relation to the dental status of adults with cystic fibrosis. Microorganisms. 2019;7:7.
- Tate S, MacGregor G, Davis M, et al. Airways in cystic fibrosis are acidified: detection by exhaled breath condensate. Thorax. 2002;57:926–929.
- Kunzelmann K, Schreiber R, Hadorn HB. Bicarbonate in cystic fibrosis. J Cyst Fibros. 2017;16:653–662.
- Cuthbertson L, Walker AW, Oliver AE, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome. 2020;8:45.
- Blainey PC, Milla CE, Cornfield DN, et al. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med. 2012;4:153ra130.
- Coburn B, Wang PW, Diaz Caballero J, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5:10241.
- Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044.
- Zemanick ET, Wagner BD, Robertson CE, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. 2017;50:50.
- Baliga S, Muglikar S, Kale R. Salivary pH: a diagnostic biomarker. J Indian Soc Periodontol. 2013;17:461–465.
- Prasad M, Toshi SR, Mallik M, et al. Periodontal disease and salivary pH: case control study. Int Arch Integr Med. 2019;6:1–6. Available from: https://imsear.searo.who.int/jspui/handle/123456789/187183
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271.
- LaMonte MJ, Genco RJ, Zheng W, et al. Substantial differences in the subgingival microbiome measured by 16S metagenomics according to periodontitis status in older women. Dent J. 2018;6. DOI:https://doi.org/10.3390/dj6040058.
- Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7:1016–1025.
- Genco RJ, LaMonte MJ, McSkimming DI, et al. The subgingival microbiome relationship to periodontal disease in older women. J Dent Res. 2019;98:975–984.
- Takeshita T, Kageyama S, Furuta M, et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama study. Sci Rep. 2016;6:22164.
- Catalán MA, Scott-Anne K, Klein MI, et al. Elevated incidence of dental caries in a mouse model of cystic fibrosis. PLoS One. 2011;6:e16549.
- Pawlaczyk-Kamieńska T, Borysewicz-Lewicka M, Śniatała R, et al. Dental and periodontal manifestations in patients with cystic fibrosis - A systematic review. J Cyst Fibros. 2019;18:762–771.
- Arquitt CK, Boyd C, Wright JT. Cystic fibrosis transmembrane regulator gene (CFTR) is associated with abnormal enamel formation. J Dent Res. 2002;81:492–496.
- Bronckers A, Kalogeraki L, Jorna HJN, et al. The cystic fibrosis transmembrane conductance regulator (CFTR) is expressed in maturation stage ameloblasts, odontoblasts and bone cells. Bone. 2010;46:1188–1196.
- Ajonuma L, Lu Q, Cheung BPK, et al. Expression and localization of cystic fibrosis transmembrane conductance regulator in human gingiva. Cell Biol Int. 2010;34:147–152.
- Herman K, Kowalczyk-Zając M, Pytrus T. Oral cavity health among cystic fibrosis patients: literature overview. Adv Clin Exp Med. 2017;26:1147–1153.
- Willis JR, González-Torres P, Pittis AA, et al. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome. 2018;6:218.
- National Academies of Sciences, Engineering, and Medicine, Division of Behavioral and Social Sciences and Education, Board on Science Education, Committee on Designing Citizen Science to Support Science Learning. Learning through citizen science: enhancing opportunities by design. Dibner KA, Pandya R, editors. Washington (DC): National Academies Press (US); 2019. doi:https://doi.org/10.17226/25183.
- Gura T. Citizen science: amateur experts. Nature. 2013;496:259–261.
- Willis JR, Saus E, Iraola-Guzmán S, et al. Oral microbiome in down syndrome and its implications on oral health. Manuscript submitted for publication. J Oral Microbiol. 2021;13(1):1865690.
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410.
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583.
- Callahan BJ. DADA2 Pipeline tutorial. 1(16). In: DADA2 [Internet]. [cited 2020 Jul 9]. Available from: https://benjjneb.github.io/dada2/tutorial.html
- Callahan B. Silva taxonomic training data formatted for DADA2 (Silva version 132). Zenodo; 2018. DOI:https://doi.org/10.5281/ZENODO.1172783.
- Callahan BJ, Sankaran K, Fukuyama JA, et al. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 2016;5:1492.
- Wright ES. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R J. 2016;8:352. Available from: https://journal.r-project.org/archive/2016/RJ-2016-025/index.html
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593.
- Palarea-Albaladejo J, Martín-Fernández JA. zCompositions — r package for multivariate imputation of left-censored data under a compositional approach. Chemometrics Intellig Lab Syst. 2015;143:85–96.
- Gloor GB, Wu JR, Pawlowsky-Glahn V, et al. It’s all relative: analyzing microbiome data as compositions. Ann Epidemiol. 2016;26:322–329.
- Gloor GB, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol. 2016;62:692–703.
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217.
- Oksanen J, Blanchet FG, Friendly M, et al. Vegan: community ecology package. 2019. Available from: https://CRAN.R-project.org/package=vegan
- Templ M, Hron K, Filzmoser P. robCompositions: an R-package for robust statistical analysis of compositional data. In: Templ M, Hron K, Filzmoser P, editors. Compositional data analysis: theory and applications. London: John Wiley and Sons; 2011. p. 341–355.
- R Core Team. R: a language and environment for statistical computing. R foundation for statistical computing; 2020. Available from: https://www.R-project.org/
- Fox J, Weisberg S. An R companion to applied regression. Third ed. Thousand Oaks, CA: Sage; 2019. Available from: https://socialsciences.mcmaster.ca/jfox/Books/Companion/
- Faust K, Bauchinger F, De Buyl S, et al. seqtime: time series analysis of sequencing data. Github; 2020. Available from: https://github.com/hallucigenia-sparsa/seqtime
- Kurtz ZD, Müller CL, Miraldi ER, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226.
- Faust K Microbial association network construction tutorial. 2017. Available from: http://psbweb05.psb.ugent.be/conet/microbialnetworks/spieceasi.php
- Filosi M, Visintainer R, Riccadonna S. nettools: a network comparison framework. Cran; 2017.
- Lambiase A, Del Pezzo M, Raia V, et al. Chryseobacterium respiratory tract infections in patients with cystic fibrosis. J Infect. 2007;55:518–523.
- Coenye T, Goris J, Spilker T, et al. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of inquilinus limosus gen. nov., sp. nov. J Clin Microbiol. 2002;40:2062–2069.
- Fernández-Olmos A, García-Castillo M, Morosini M-I, et al. MALDI-TOF MS improves routine identification of non-fermenting gram negative isolates from cystic fibrosis patients. J Cyst Fibros. 2012;11:59–62.
- Sharma P, Diene SM, Gimenez G, et al. Genome sequence of microbacterium yannicii, a bacterium isolated from a cystic fibrosis patient. J Bacteriol. 2012;194:4785.
- Sharma P, Diene SM, Thibeaut S, et al. Phenotypic and genotypic properties of Microbacterium yannicii, a recently described multidrug resistant bacterium isolated from a lung transplanted patient with cystic fibrosis in France. BMC Microbiol. 2013;13:97.
- Menuet M, Bittar F, Stremler N, et al. First isolation of two colistin-resistant emerging pathogens, brevundimonas diminuta and ochrobactrum anthropi, in a woman with cystic fibrosis: a case report. J Med Case Rep. 2008;2:373.
- Carter C. Bioinformatics analysis of homologies between pathogen antigens, autoantigens and the CFTR cystic fibrosis protein: a role for immunoadsorption therapy? Nat Precedings. 2010. DOI:https://doi.org/10.1038/npre.2010.5352.1
- Razvi S, Quittell L, Sewall A, et al. Respiratory microbiology of patients with cystic fibrosis in the USA, 1995 to 2005. Chest. 2009;136:1554–1560.
- Hauser AR, Jain M, Bar-Meir M, et al. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev. 2011;24:29–70.
- Talmaciu I, Varlotta L, Mortensen J, et al. Risk factors for emergence of Stenotrophomonas maltophilia in cystic fibrosis. Pediatr Pulmonol. 2000;30:10–15.
- Paganin P, Fiscarelli EV, Tuccio V, et al. Changes in cystic fibrosis airway microbial community associated with a severe decline in lung function. PLoS One. 2015;10:e0124348.
- Maeda Y, Elborn JS, Parkins MD, et al. Population structure and characterization of viridans group streptococci (VGS) including streptococcus pneumoniae isolated from adult patients with cystic fibrosis (CF). J Cyst Fibros. 2011;10:133–139.
- García-Castillo M, Morosini MI, Valverde A, et al. Differences in biofilm development and antibiotic susceptibility among streptococcus pneumoniae isolates from cystic fibrosis samples and blood cultures. J Antimicrob Chemother. 2007;59:301–304.
- Whiteson KL, Meinardi S, Lim YW, et al. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. Isme J. 2014;8:1247–1258.
- Lim YW, Schmieder R, Haynes M, et al. Mechanistic model of Rothia mucilaginosa adaptation toward persistence in the CF lung, based on a genome reconstructed from metagenomic data. PLoS One. 2013;8:e64285.
- Sánchez-Bautista A, Rodríguez-Díaz JC, Garcia-Heredia I, et al. Airway microbiota in patients with paediatric cystic fibrosis: relationship with clinical status. Enferm Infecc Microbiol Clin. 2019;37:167–171.
- De Dios Caballero J, Del Campo R, Royuela A, et al. Bronchopulmonary infection-colonization patterns in Spanish cystic fibrosis patients: results from a national multicenter study. J Cyst Fibros. 2016;15:357–365.
- Tabatabaei M, Dastbarsar M, Moslehi MA. Isolation and identification of Pandoraea spp. From bronchoalveolar lavage of cystic fibrosis patients in Iran. Ital J Pediatr. 2019;45:118.
- Filkins LM, Hampton TH, Gifford AH, et al. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol. 2012;194:4709–4717.
- Soret P, Vandenborght L-E, Francis F, et al. Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci Rep. 2020;10:3589.
- Worlitzsch D, Rintelen C, Böhm K, et al. Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clin Microbiol Infect. 2009;15:454–460.
- Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A. 2012;109:5809–5814.
- Coffey MJ, Nielsen S, Wemheuer B, et al. Gut microbiota in children with cystic fibrosis: a taxonomic and functional dysbiosis. Sci Rep. 2019;9:18593.
- Bakare N, Rickerts V, Bargon J, et al. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses. 2003;46:19–23.
- Valenza G, Tappe D, Turnwald D, et al. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 2008;7:123–127.
- Chotirmall SH, O’Donoghue E, Bennett K, et al. Sputum Candida albicans presages FEV₁ decline and hospital-treated exacerbations in cystic fibrosis. Chest. 2010;138:1186–1195.
- Lepesqueur LSS, Tanaka MH, Lima De MGG, et al. Oral prevalence and antifungal susceptibility of Candida species in cystic fibrosis patients. Arch Oral Biol. 2020;116:104772.
- Scoffield JA, Wu H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun. 2015;83:101–107.
- Whiley RA, Fleming EV, Makhija R, et al. Environment and colonisation sequence are key parameters driving cooperation and competition between Pseudomonas aeruginosa cystic fibrosis strains and oral commensal streptococci. PLoS One. 2015;10:e0115513.
- Boutin S, Dalpke AH. Acquisition and adaptation of the airway microbiota in the early life of cystic fibrosis patients. Mol Cell Pediatr. 2017;4:1.
- Yuan Z, Panchal D, Syed MA, et al. Induction of cyclooxygenase-2 signaling by Stomatococcus mucilaginosus highlights the pathogenic potential of an oral commensal. J Immunol. 2013;191:3810–3817.
- Gao B, Gallagher T, Zhang Y, et al. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere. 2018;3. DOI:https://doi.org/10.1128/mSphere.00151-18.
- Bensel T, Stotz M, Borneff-Lipp M, et al. Lactate in cystic fibrosis sputum. J Cyst Fibros. 2011;10:37–44.
- Widdel F, Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch Microbiol. 1982;131:360–365.
- Robinson CV, Elkins MR, Bialkowski KM, et al. Desulfurization of mucin by Pseudomonas aeruginosa: influence of sulfate in the lungs of cystic fibrosis patients. J Med Microbiol. 2012;61:1644–1653.
- Scott J, Sueiro-Olivares M, Ahmed W, et al. Pseudomonas aeruginosa-derived volatile sulfur compounds promote distal aspergillus fumigatus growth and a synergistic pathogen-pathogen interaction that increases pathogenicity in co-infection. Front Microbiol. 2019;10:2311.
- Mohapatra NK, Cheng PW, Parker JC, et al. Alteration of sulfation of glycoconjugates, but not sulfate transport and intracellular inorganic sulfate content in cystic fibrosis airway epithelial cells. Pediatr Res. 1995;38:42–48.
- Zhang Y, Doranz B, Yankaskas JR, et al. Genotypic analysis of respiratory mucous sulfation defects in cystic fibrosis. J Clin Invest. 1995;96:2997–3004.
- Foweraker J. Recent advances in the microbiology of respiratory tract infection in cystic fibrosis. Br Med Bull. 2009;89:93–110.
- Mohanty R, Asopa SJ, Joseph MD, et al. Red complex: polymicrobial conglomerate in oral flora: a review. J Family Med Prim Care. 2019;8:3480–3486.
- Colombo APV, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432.
- Koyanagi T, Sakamoto M, Takeuchi Y, et al. Comprehensive microbiological findings in peri-implantitis and periodontitis. J Clin Periodontol. 2013;40:218–226.
- Riggio MP, Lennon A. Development of a PCR assay specific for peptostreptococcus anaerobius. J Med Microbiol. 2002;51:1097–1101.
- Bizzarro S, Laine ML, Buijs MJ, et al. Microbial profiles at baseline and not the use of antibiotics determine the clinical outcome of the treatment of chronic periodontitis. Sci Rep. 2016;6:20205.
- Ramanan P, Barreto JN, Osmon DR, et al. Rothia bacteremia: a 10-year experience at Mayo Clinic, Rochester, Minnesota. J Clin Microbiol. 2014;52:3184–3189.
- Ruhl S, Eidt A, Melzl H, et al. Probing of microbial biofilm communities for coadhesion partners. Appl Environ Microbiol. 2014;80:6583–6590.
- Zhou J, Jiang N, Wang S, et al. Exploration of human salivary microbiomes–insights into the novel characteristics of microbial community structure in caries and caries-free subjects. PLoS One. 2016;11:e0147039.
- Chen H, Ling Z, Wen J, et al. Oral microbiota in chinese she children with dental caries. IADR/AADR/CADR General Session and Exhibition. J Dent Res;2013. [cited 2020 Aug 14].
- Jiang Q, Liu J, Chen L, et al. The oral microbiome in the elderly with dental caries and health. Front Cell Infect Microbiol. 2018;8:442.
- Mantzourani M, Gilbert SC, Sulong HNH, et al. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 2009;43:308–313.
- Tanner ACR, Kent RL Jr, Holgerson PL, et al. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 2011;90:1298–1305.
- Jiang W, Ling Z, Lin X, et al. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb Ecol. 2014;67:962–969.
- Henne K, Rheinberg A, Melzer-Krick B, et al. Aciduric microbial taxa including scardovia wiggsiae and bifidobacterium spp. in caries and caries free subjects. Anaerobe. 2015;35:60–65.
- Richards VP, Alvarez AJ, Luce AR, et al. Microbiomes of site-specific dental plaques from children with different caries status. Infect Immun. 2017;85:85.
- Wang Y, Zhang J, Chen X, et al. Profiling of oral microbiota in early childhood caries using single-molecule real-time sequencing. Front Microbiol. 2017;8:2244.
- Naidu BV, Reginald BA. Quantification and correlation of oral Candida with caries index among different age groups of school children: a case-control study. Ann Med Health Sci Res. 2016;6:80–84.
- Lozano Moraga CP, Rodríguez Martínez GA, Lefimil Puente CA, et al. Prevalence of Candida albicans and carriage of Candida non-albicans in the saliva of preschool children, according to their caries status. Acta Odontol Scand. 2017;75:30–35.
- Xiao J, Huang X, Alkhers N, et al. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 2018;52:102–112.
- Eidt G, De Andrade CG, De Negrini TC, et al. Role of Candida albicans on enamel demineralization and on acidogenic potential of streptococcus mutans in vitro biofilms. J Appl Oral Sci. 2019;27:e20180593.
- Xu L, Chen X, Wang Y, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study. Front Physiol. 2018;9:342.
- Xu H, Hao W, Zhou Q, et al. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. PLoS One. 2014;9:e89269.
- Lif Holgerson P, Öhman C, Rönnlund A, et al. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10:e0128534.
- Schoilew K, Ueffing H, Dalpke A, et al. Bacterial biofilm composition in healthy subjects with and without caries experience. J Oral Microbiol. 2019;11:1633194.
- Hernández M, Planells P, Martínez E, et al. Microbiology of molar-incisor hypomineralization lesions. A pilot study. J Oral Microbiol. 2020;12:1766166.
- Henskens YM, Van Den Keijbus PA, Veerman EC, et al. Protein composition of whole and parotid saliva in healthy and periodontitis subjects. Determination of cystatins, albumin, amylase and IgA. J Periodontal Res. 1996;31:57–65.
- Qu XM, Wu ZF, Pang BX, et al. From nitrate to nitric oxide: the role of salivary glands and oral bacteria. J Dent Res. 2016;95:1452–1456.
- Espinoza JL, Harkins DM, Torralba M, et al. Supragingival plaque microbiome ecology and functional potential in the context of health and disease. MBio. 2018;9:9.
- Grasemann H, Ioannidis I, Tomkiewicz RP, et al. Nitric oxide metabolites in cystic fibrosis lung disease. Arch Dis Child. 1998;78:49–53.
- Zetterquist W, Marteus H, Kalm-Stephens P, et al. Oral bacteria–the missing link to ambiguous findings of exhaled nitrogen oxides in cystic fibrosis. Respir Med. 2009;103:187–193.
- De Winter-de Groot KM, Van Der Ent CK. Nitric oxide in cystic fibrosis. J Cyst Fibros. 2005;4(Suppl 2):25–29.
- Kerley CP, Kilbride E, Greally P, et al. Dietary nitrate acutely and markedly increased exhaled nitric oxide in a cystic fibrosis case. Clin Med Res. 2016;14:151–155.
- Nawaz A, Hasan F, Shah AA. Degradation of poly(ɛ-caprolactone) (PCL) by a newly isolated Brevundimonas sp. strain MRL-AN1 from soil. FEMS Microbiol Lett. 2015;362:1–7.
- Al-Jebouri M. The relationship between periodontal disease and predisposing factors. Tikrit J Dental Sci. 2016;4:68–80.
- Colombo APV, Bennet S, Cotton SL, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–1287.
- Kõll‐Klais P, Mändar R, Leibur E. Oral lactobacilli in chronic periodontitis and periodontal health: species composition and antimicrobial activity. Oral Microbiol Immunol. 2005;20:354–361.
- Chatterjee A, Bhattacharya H Probiotics in periodontal health and disease. society of periodontology. 2011. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/pmc3134041/
- Eribe ERK, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14:131–137.
- Peterson SN, Snesrud E, Liu J, et al. The dental plaque microbiome in health and disease. PLoS One. 2013;8:e58487.
- Thompson J, Pikis A. Metabolism of sugars by genetically diverse species of oral Leptotrichia. Mol Oral Microbiol. 2012;27:34–44.
- Losada PM, Chouvarine P, Dorda M, et al. The cystic fibrosis lower airways microbial metagenome. ERJ Open Res. 2016;2. DOI:https://doi.org/10.1183/23120541.00096-2015.
- Nielsen S, Needham B, Leach ST, et al. Disrupted progression of the intestinal microbiota with age in children with cystic fibrosis. Sci Rep. 2016;6:24857.
- Zakharkina T, Heinzel E, Koczulla RA, et al. Analysis of the airway microbiota of healthy individuals and patients with chronic obstructive pulmonary disease by T-RFLP and clone sequencing. PLoS One. 2013;8:e68302.
- Nallabelli N, Patil PP, Pal VK, et al. Biochemical and genome sequence analyses of Megasphaera sp. strain DISK18 from dental plaque of a healthy individual reveals commensal lifestyle. Sci Rep. 2016;6:33665.
- Dewhirst FE, Chen C-KC, Paster BJ, et al. Phylogeny of species in the family neisseriaceae isolated from human dental plaque and description of kingella orale sp. nov. Int J Syst Bacteriol. 1993;43:490–499.
- Bittar F, Cassagne C, Bosdure E. Outbreak of corynebacterium pseudodiphtheriticum infection in cystic fibrosis patients, France. Emerg Infect Dis. 2010;16:1231–1236. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/pmc3298292/
- Pivot D, Fanton A, Badell-Ocando E, et al. Carriage of a single strain of nontoxigenic corynebacterium diphtheriae bv. Belfanti (corynebacterium belfantii) in four patients with cystic fibrosis. J Clin Microbiol. 2019;57. DOI:https://doi.org/10.1128/JCM.00042-19.
- Layeghifard M, Li H, Wang PW, et al. Microbiome networks and change-point analysis reveal key community changes associated with cystic fibrosis pulmonary exacerbations. NPJ Biofilms Microbiomes. 2019;5:1–12.
- Carrouel F, Viennot S, Santamaria J, et al. Quantitative molecular detection of 19 major pathogens in the interdental biofilm of periodontally healthy young adults. Front Microbiol. 2016;7:840.
- Mashima I, Fujita M, Nakatsuka Y, et al. The distribution and frequency of oral veillonella spp. associated with chronic periodontitis. Int J Curr Microbiol App Sci. 2015 . [cited 2020 Oct 15]. Available from: https://www.semanticscholar.org/paper/e8d55ce20a438733917336d7671f9228506628ae
- Haraszthy VI, Zambon JJ, Sreenivasan PK, et al. Identification of oral bacterial species associated with halitosis. J Am Dent Assoc. 2007;138:1113–1120.
- Stephen AS, Naughton DP, Pizzey RL, et al. In vitro growth characteristics and volatile sulfur compound production of Solobacterium moorei. Anaerobe. 2014;26:53–57.
- Stipanuk MH, Dominy JE Jr, Lee J-I, et al. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr. 2006;136:1652S–1659S.
- McKone EF, Shao J, Frangolias DD, et al. Variants in the glutamate-cysteine-ligase gene are associated with cystic fibrosis lung disease. Am J Respir Crit Care Med. 2006;174:415–419.
- Conrad C, Lymp J, Thompson V, et al. Long-term treatment with oral N-acetylcysteine: affects lung function but not sputum inflammation in cystic fibrosis subjects. A phase II randomized placebo-controlled trial. J Cyst Fibros. 2015;14:219–227.
- Carmody LA, Zhao J, Schloss PD, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann ATS. 2013;10:179–187.
- Dani S, Prabhu A, Chaitra KR, et al. Assessment of streptococcus mutans in healthy versus gingivitis and chronic periodontitis: a clinico-microbiological study. Contemp Clin Dent. 2016;7:529–534.
- Contardo MS, Díaz N, Lobos O, et al. Oral colonization by streptococcus mutans and its association with the severity of periodontal disease in adults. Revista Clínica de Periodoncia, Implantología y Rehabilitación Oral. 2011;4:9–12.
- Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontol 2000. 2002;28:12–55.
- Vandeplassche E, Tavernier S, Coenye T, et al. Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens. Eur Respir Rev. 2019;28:28.
- Bevivino A, Bacci G, Drevinek P, et al. Deciphering the ecology of cystic fibrosis bacterial communities: towards systems-level integration. Trends Mol Med. 2019;25:1110–1122.
- Jagathrakshakan SN, Sethumadhava RJ, Mehta DT, et al. 16S rRNA gene-based metagenomic analysis identifies a novel bacterial co-prevalence pattern in dental caries. Eur J Dent. 2015;9:127.
- Basavaraju M, Sisnity VS, Palaparthy R, et al. Quorum quenching: signal jamming in dental plaque biofilms. J Dent Sci. 2016;11:349–352.
- Utari PD, Vogel J, Quax WJ. Deciphering physiological functions of AHL quorum quenching acylases. Front Microbiol. 2017;8:1123.
- Muras A, Otero-Casal P, Blanc V, et al. Acyl homoserine lactone-mediated quorum sensing in the oral cavity: a paradigm revisited. Sci Rep. 2020;10:9800.
- Muras A, Mayer C, Otero-Casal P, et al. Short-Chain N-acylhomoserine lactone quorum-sensing molecules promote periodontal pathogens in in vitro oral biofilms. Appl Environ Microbiol. 2020;86:86.
- Camelo-Castillo AJ, Mira A, Pico A, et al. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol. 2015;6:119.
- Ai D, Huang R, Wen J, et al. Integrated metagenomic data analysis demonstrates that a loss of diversity in oral microbiota is associated with periodontitis. BMC Genomics. 2017;18:1041.
- Shin M, Han D, Yi H, et al. Characterization of subgingival microorganisms according to periodontitis and cardiovascular status. IADR/AADR/CADR General Session. J Dent Res; 2013.
- Anesti V, McDonald IR, Ramaswamy M, et al. Isolation and molecular detection of methylotrophic bacteria occurring in the human mouth. Environ Microbiol. 2005;7:1227–1238.
- Vielkind P, Jentsch H, Eschrich K, et al. Prevalence of actinomyces spp. in patients with chronic periodontitis. Int J Med Microbiol. 2015;305:682–688.
- Chen W-P, Chang S-H, Tang C-Y, et al. Composition analysis and feature selection of the oral microbiota associated with periodontal disease. Biomed Res Int. 2018;2018:3130607.
- Tindall BJ, Euzéby JP. Proposal of parvimonas gen. nov. and quatrionicoccus gen. nov. as replacements for the illegitimate, prokaryotic, generic names micromonas Murdoch and Shah 2000 and Quadricoccus Maszenan et al. 2002, respectively. Int J Syst Evol Microbiol. 2006;56:2711–2713.
- Deng Z-L, Szafrański SP, Jarek M, et al. Dysbiosis in chronic periodontitis: key microbial players and interactions with the human host. Sci Rep. 2017;7:3703.
- Naginyte M, Do T, Meade J, et al. Enrichment of periodontal pathogens from the biofilms of healthy adults. Sci Rep. 2019;9:5491.
- Ghayoumi N, Chen C, Slots J. Dialister pneumosintes, a new putative periodontal pathogen. J Periodontal Res. 2002;37:75–78.
- Oswal P, Katti S, Joshi V, et al. Identification of dialister pneumosintes in healthy and chronic periodontitis patients with type 2 diabetes mellitus and its correlation with the red complex bacteria. J Interdiscip Dent. 2020;10:17.
