ABSTRACT
Background The physical appearance of tongue coatings is vital for traditional Chinese medicine (TCM) to diagnose health and disease status. The microbiota of different tongue coatings could also influence coating formation and be further associated with specific diseases. Previous studies have focused on bacteria from different tongue coatings in the context of specific diseases, but the normal variations in healthy individuals remain unknown.Aim: We examined the tongue microbiota by metagenomics in 94 healthy individuals classified into eight different tongue types.Results: The overall composition of the tongue coating microbiome is not drastically different among different coating types, similar to the findings of previous studies in healthy populations. Further analysis revealed microbiota characteristics of each coating type, and many of the key bacteria are reported to be implicated in diseases. Moreover, further inclusion of diabetic patients revealed disease-specific enrichment of Capnocytophaga, even though the same tongue coatings were studied.Conclusions: This work revealed the characteristic compositions of distinctive tongue coatings in a healthy population, which serves as a basis for understanding the tongue coating formation mechanism and provides a valuable reference to further investigate disease-specific tongue coating bacterial markers.
Introduction
While the gut microbiota has drawn tremendous attention, studies of the oral microbiota have only emerged relatively recently [Citation1,Citation2]. The tongue coating microbiota, an essential component of the tongue coating, represents a large microbial fraction of the body that mainly contains Bacteroides, Fusobacteria, Actinobacteria and Firmicutes. The microbiota composition is shaped by the uptake of nutrients from the host epithelium, saliva and their interactions [Citation3]. The corresponding various colors and thicknesses of tongue coatings have long been used in traditional Chinese medicine (TCM) as indicators of the health status of an individual and to assist in the prediction or diagnosis [Citation4] of diseases such as gastritis [Citation5], esophagitis [Citation6], pancreatic organ dysfunction [Citation7] or even early-stage breast cancer [Citation8]. Usually, according to its color, thickness and moisture appearance, tongue coating is classified into white thin (W-thin), white thin greasy (W-thin greasy), white thick greasy (W-thick greasy), white thick dry (W-thick dry), yellow thin (Y-thin), yellow thin greasy (Y-thin greasy), yellow thick greasy (Y-thick greasy), and yellow thick dry (Y-thick dry) by TCM.
There have been a number of studies investigating the microbiota characteristics with regard to specific disease types and with a strong focus on a particular coating type, taking W-thin coating in a healthy population as a control [Citation9–12]. However, the identified marker bacteria may be disease specific and affected by different stages of disease progression, yet only a few studies have taken this into consideration [Citation13]. The marker bacteria among distinctive tongue coating types thus could vary in different diseases. Microbiota related to halitosis were found to belong to Alloprevotella tannerae and Solobacterium moorei, tonsillitis was associated with Corynebacterium argentoratense, and periodontal diseases were associated with Prevotella nigrescens and Capnocytophaga sputigena. These bacteria were found to be aggregated in the yellow tongue coating (YTC) of gastritis patients [Citation9]. Another comparison of 13 chronic erosive gastritis patients with typical YTC with 10 healthy volunteers with white tongue coating (WTC) demonstrated Bacillus as a potential YTC diagnostic marker [Citation10]. A further study of 28 chronic hepatitis B (CHB) YTC patients and 25 CHB WTC patients with 22 healthy controls showed that Proteobacteria was enriched in YTC and correlated with higher HBV-DNA levels [Citation11]. In 115 gastric cancer patients, Proteobacteria abundance declined from the W-thin group to the Y-thin group, with the lowest abundance in the Y-thick group [Citation12]. Therefore, the representative microbes for certain tongue coatings vary among different diseases.
However, different tongue coating types also exist in the healthy population. Studying the microbial basis of tongue coating types in this population is vital in demonstrating the important roles of microbes in reflecting health status, excluding the impact of diagnosed disease. In our study, we investigated the natural distribution and microbial characteristics of different tongue coating types in a relatively large healthy population devoid of disease influences. Specifically, we divided the tongue coatings of a healthy population into eight types according to color, thickness and moisture appearance and investigated the microbiota compositions by metagenomic sequencing. The characteristics of bacteria for distinctive tongue coatings were identified, and the co-occurrence relationship between microbes was constructed. This provides a valuable reference for deciphering the tongue coating formation mechanism and facilitates future diagnosis. We further compared marker bacteria in healthy individuals and diabetic patients with the same tongue coating types, demonstrating the disease-specific effect on tongue coating microbes and highlighting the necessity of identifying marker bacteria for tongue coating types within a healthy population.
Materials and methods
Study participants
All participants provided written informed consent, and protocols were approved by the ethical commission of the Institute of Biophysics, Chinese Academy of Sciences. The target population was healthy residents in the Xinjiekou district, Beijing, China, with occupations including government officials, nursing home staff, hotel service staff and department store staff. Participants who reported no severe diseases relating to the heart, lung, liver, kidney, brain or other organs, with no chronic disease and no acute cardiovascular events or myocardial infarction in the past six months were included. Participants with medical histories of using glucocorticoids and antibiotics within the last three months and a long history of smoking, alcohol abuse or drug abuse were excluded. Pregnant or breast-feeding women were also excluded. All enrolled individuals had healthy oral tissues and were free of nonrestored carious lesions [Citation14]. For Y-thick greasy tongue coating samples from diabetic patients, participants diagnosed with diabetes (criteria: typical diabetes symptoms including thirst, hunger, hyperphagia, body weight loss with a random blood glucose test result higher than 11.1 mmol/L, fasting blood sugar higher than 7 mmol/L, or 2 h oral glucose tolerance test higher than 11.1 mmol/L) with no complicated diseases were enrolled from endocrine outpatients at the Guang’anmen Hospital, China Academy of Chinese Medical Sciences. The rest of the filter criteria were as above. Tongue coating samples from four diabetic males residing in Beijing, China and aged 33, 48, 53, and 54 years were selected and subjected to further sequencing and analysis. The 20 individuals who were part of the salivary flow rate cohort were recruited from volunteers residing in Beijing at Guang’anmen Hospital, China Academy of Chinese Medical Sciences.
Tongue imaging
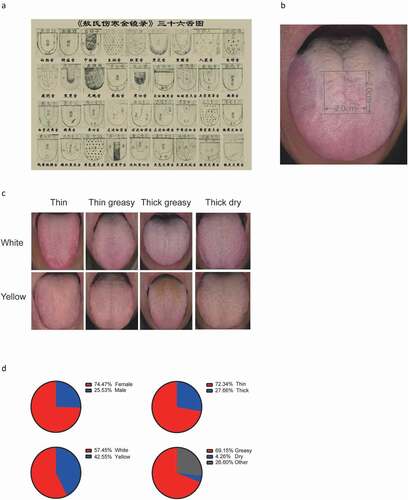
A DS01-G tongue manifestation acquisition instrument (DaoSheng Medical Technology Co. Ltd., China) was used to photograph the tongue coatings of the participants. Tongue coating types were prediagnosed at the site. The tongue coating images were then segregated into W-thin, W-thin greasy, W-thick greasy, W-thick dry, Y-thin, Y-thin greasy, Y-thick greasy and Y-thick dry by three certified and experienced TCM doctors according to the middle site of the tongue (). The images with consistent diagnostic results from the three TCM doctors and the doctor at the site were selected, and their corresponding samples were collected.
Figure 1. Example of traditional Chinese medicine tongue type classification and tongue-coating classification of our participants. (A) Thirty-six tongue types presented in the ancient medical book of Ao Shi Shang Han Jin Jing Lu (1341 A.D.). (B) Diagnosis and sampling site of the tongue dorsum. (C) Examples of eight tongue coating types (upper from left to right: white thin, white thin greasy, white thick greasy, white thick dry; lower from left to right: yellow thin, yellow thin greasy, yellow thick greasy, yellow thick dry). (D) Proportion of sex and specific tongue coating characteristics in the recruited healthy population
Sample collection
Tongue coating samples were collected after a mouth wash and using a protocol that had been tested during a pilot phase of this study. The sampling protocol is described as follows: volunteers who met the filter criteria were required to rinse their mouths three times with mineral water at 9:00–10:00 in the morning. After resting for 30 min, tongue images were collected with a Daosheng Tongue DS01-G Image Instrument. A sterile swab was used to collect the microbiome at the middle site of the tongue. The microbiome was collected with the sterile swab by scraping back and forth 15 times, followed by a 180-degree rotation to scrape for another 15 times. Each sterile swab was stored in a TinyGene microbiome preserved Eppendorf tube containing buffer and proteinase K (TinyGen Bio-Tech (Shanghai) Co. Ltd., China) and was immediately placed on dry ice for transportation to a − 80°C freezer within two hours.
For unstimulated saliva sample collection, the participants were asked to rinse their mouths with water, followed by relaxation for 5 min, during which time a questionnaire was completed and tongue images were collected. Then, the participants were asked to swallow to void the mouth of saliva, collect the unstimulated saliva for 5 min and spit into a preweighed 15 ml Falcon tube. The salivary flow rate (g/min) was calculated as (saliva-tube weight subtracted with tube weight) g/5 min.
DNA extraction
Total DNA was extracted from samples using protease K cleavage combined with the phenol chloroform extraction method according to the manufacturer’s protocols. DNA completeness and purity were checked by running the samples on 1.2% agarose gels. DNA concentration was checked with a Qubit fluorometer.
Metagenomic library preparation and sequencing
Extracted DNA was sheared on a Covaris M220 machine (Covaris, USA) programmed to generate 300-bp fragments. The sequencing libraries were constructed with a NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (NEB, USA). The products were purified using AgarosAgencourt AMPure XP (Beckman, USA) and quantified using the GenNextTM NGS Library Quantification Kit (Toyobo, Japan). The libraries were sequenced using the Illumina NovaSeq 6000 platform and 150-bp paired-end technology at TinyGen Bio-Tech (Shanghai) Co., Ltd.
Bioinformatics analysis
The raw FastQ files were demultiplexed based on the index. The raw, paired-end reads were trimmed and quality controlled using Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) and cutadapt software. The host sequences were removed by using BWA (http://bio-bwa.sourceforge.net/). The clean data were assembled and predicted by the Megahit (https://github.com/voutcn/megahit) and METAProdigal (http://prodigal.ornl.gov/) software.
The predicted genes were clustered, and a nonredundant gene catalog was constructed with the CD-HIT package (http://www.bioinformatics.org/cd-hit/) (parameters: identity = 95%, coverage = 90%). The abundances of genes were estimated quickly by genomeCoverageBed in BEDTools. The representative sequences were identified taxonomically against the NR database with an e-value of 1e-5 by the DIAMOND software. The taxonomic information included domain, kingdom, phylum, class, order, family, genus, and species.
Co-occurrence networks
Spearman’s correlation analysis and p value calculation performed with cor.test in R language were applied to the abundance values of the bacterial species with a linear discriminant analysis (LDA) value higher than 2 in each group. Bacteria with a Spearman’s correlation ≥ 0.6 and p value ≤ 0.05 were further selected for co-occurrence network construction with igraph (version 1.2.5) in R.
Statistical analyses
Principal component analysis (PCA) was performed using prcomp in R with the taxonomy abundance information, and plotting was performed by ggplot2 in R (version 3.4.1). The significance of separation between each pair of groups was calculated by Analysis of Similarities (ANOSIM). The microbiota results were analyzed using the nonparametric factorial Kruskal-Wallis (KW) sum-rank test, and the LEfSe method was applied with LEfSe v1.0. p values < 0.05 were considered significant.
Results
Tongue coating types are unevenly distributed in the healthy population
The study population was recruited from healthy volunteers in the Xinjiekou community in Beijing. A total of 94 out of 180 individuals passed the quality filters (see Materials and methods) and were included in the tongue coating microbiome analysis. The average residence time in Beijing was approximately 20–30 years. This maximally reduced the living environmental impact on the oral microbiota, such as the composition of tap water [Citation15]. To identify characteristic bacteria in the distinctive tongue coatings, we first classified the tongue coatings into eight types according to color, thickness and moisture appearance () [Citation16]. This resulted in W-thin, W-thin greasy, W-thick greasy, W-thick dry, Y-thin, Y-thin greasy, Y-thick greasy and Y-thick dry tongue coatings, which are the most prevalent tongue coating types. Because microbe biomass is most abundant at the middle site of the tongue dorsum, the middle site of the tongue coating () was used in tongue coating typing classification () and for biofilm sampling. In terms of general characteristics, the cohort contained 74.47% females and 25.53% males (, upper left). In total, 72.34% of the tongue coatings could be defined as thin, whereas 27.66% could be defined as a thick tongue coating (, upper right); the percentage of white tongue coating was highest at 57.45%, followed by that of yellow tongue coating at 42.55% (, lower left), while in terms of moisture, greasy tongue coating accounted for 69.15% of the population (, lower right).
In the recruited population, the most prevalent tongue coatings were W-thin, W-thin greasy and Y-thin greasy, numbering 20 in each, while the numbers for W-thick greasy, Y-thin and Y-thick greasy were approximately 10 cases in each. In contrast, there were a few cases of W-thick dry and Y-thick dry, with one and three cases each (). The age distribution of each group generally ranged from 45 to 55 years, with no significant differences between groups (). Of note, the one W-thick dry volunteer 36 years old and three Y-thick dry volunteers, all approximately 54 years old, were females, suggesting that rare tongue coatings are potentially age- and/or sex-related. Due to the unpaired age and few recruited numbers of W-thick dry and Y-thick dry tongue coating groups, we excluded them from the microbiota analysis. The percentage of females was highest in the W-thin group at 94.47% and lowest in the W-thick greasy group at 41.67%, suggesting different compositions of females and males in each tongue coating type (). The BMI value was highest in the W-thin greasy group at an average of 25 kg/m2, with no significant differences among the groups.
Table 1. General information on the healthy population used in our study
Characteristics of the tongue coating microbiome
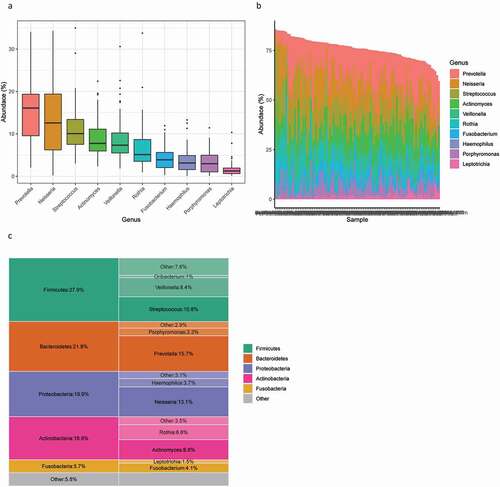
Microbiome analysis revealed that the most common phylum found on the tongue was Firmicutes, reaching the highest abundance of 27.9%, followed by Bacteroidetes, Proteobacteria, Actinobacteria and Fusobacteria. At a finer taxonomic level, the most enriched genus of the study population was Prevotella, with an average abundance of 15.7%, followed by Neisseria at 13.1%, Streptococcus at 10.8%, Actinomyces at 8.8%, Veillonella at 8.4%, Rothia at 6.6%, Fusobacterium at 4.1%, Haemophilus at 3.7%, Porphyromonas at 3.3% and Leptotrichia at 1.5% (). The most common genus found in our study is consistent with that in other reports [Citation11,Citation17].
Figure 2. Overall microbial composition related to tongue coating. (A) Box plot of the ten most abundant microbes at the genus level. (B) Stacked bars of the ten most abundant genera in each sample. (C) The five most abundant phyla and their most abundant genera
Since we did not find a significant difference in microbial beta diversity with respect to different tongue coatings, as revealed by PCA (Supplementary Figure 1), we applied LDA by comparison between each group and the other groups to further identify the specific marker microbes of a specific group (Supplementary Figure 2). The analysis results showed very few bacteria with LDA scores higher than 3 in the W-thin tongue coating, except for the Bacillales at order level. For W-thin greasy, three functionally unknown species were found to be abundant (). For W-thick greasy, the abundance of Megasphaera micronuciformis and members of the Veillonellaceae family, as well as the Firmicutes phylum, plus Streptococcus infantis under the Firmicutes phylum were all found to be enriched. Moreover, two bacteria related to oral dental caries, Actinomyces sp. ICM58 and Actinomyces sp. oral taxon 172 belonging to Actinobacteria, were enriched in the W-thick greasy group [Citation18].
Figure 3. Top differentially enriched taxa in different tongue coating types. Enriched taxa in the designated tongue coating type with LDA scores higher than 3 from LEfSe analysis are presented
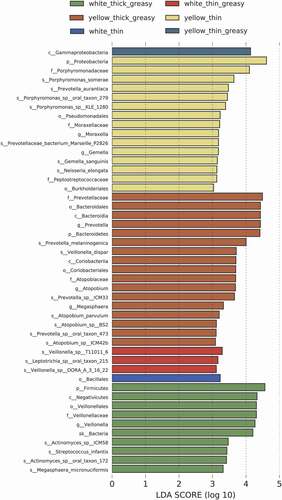
Additionally, we found that the phylum Proteobacteria was enriched in the Y-thin tongue coating group. Within Proteobacteria, the Moraxella genus and Porphyromonassomerae, as well as Porphyromonas sp. KLE 1280, Porphyromonas sp. oral taxon 278, and Porphyromonas oral taxon 279, all belonging to the family Porphyromonadaceae, were highly enriched. Similarly, within the Bacteroidetes phylum, Prevotella genus, the human oral cavity resident species Prevotella aurantiaca were more abundant in the Y-thin group, while in the phylum Firmicutes, the species Gemella sanguinis, together with upper class Gemella at the genus level and members of the family Peptostreptococcaceae, were enriched. For the Y-thick greasy tongue coating group, Bacteroidetes was the most abundant phylum, and at the species level, Prevotella melaninogenica was highly enriched. In Firmicutes, the species Veillonella dispar, which mostly resides on the tongue dorsum, and Atopobium parvulum and its closely related species Atopobium sp. BS2 and Atopobium sp. ICM42b, which belong to the Actinobacteria phylum, were highly abundant in the Y-thick greasy group ().
Disease-related characteristic bacteria in specific tongue coating types
We then carried out an in-depth literature search and confirmed that among the enriched bacteria in specific tongue coating types, many are indicated in certain diseases (these are summarized in ). In the W-thick greasy type, the associated microbial biomarkers Vellonella, M. micronuciformis and Streptococcus infantis are associated with gastritis. Specifically, the genus Veillonella is causative for gastritis of a specific subtype [Citation9]. M. micronuciformis is enriched in the tongue coating of gastritis cancer patients [Citation12], while S. infantis indicates the precancerous cascade [Citation19]. Moreover, the white coating tongue appearance has been demonstrated to be enriched in gastritis patients in a cohort of 4,742 individuals by logistic regression analysis, making this appearance indicative of a higher risk of gastritis [Citation6].
Table 2. Disease related characteristic bacteria in specific tongue coating types
Within the biomarker of the Y-thin tongue coating type, the genus Moraxella and species Neisseria elongate are common bacterial microbiota of the oropharynx. Moraxella sometimes gives rise to opportunistic infections [Citation20], and numerous reports indicate endocarditis [Citation21,Citation22], osteomyelitis [Citation23] and pancreatic cancer [Citation13] induced by Neisseria elongata, typically arising after dental procedures or dental abscesses. In Firmicutes, the species G. sanguinis was again an opportunistic pathogen leading to endocarditis [Citation24], and the family Peptostreptococcaceae was found to be overrepresented in the guts of colorectal cancer patients [Citation25]. Finally, the enriched Bacteroidetes species P. somerae is related to soft tissue infection [Citation26].
The esophageal squamous cell carcinoma-inducing oral bacteria P. melaninogenica and V. dispar [Citation27] were coenriched in the Y-thick greasy group. Multifunctional P. melaninogenica in saliva is also associated with oral squamous cell carcinoma [Citation28] and is causative of periodontitis [Citation29], Alzheimer’s disease mortality [Citation30], hyperglycemia [Citation31], [and MHCII, CD80 and IFN-γ upregulation in salivary gland epithelial cells in Sjögren’s syndrome [Citation32]. The species A. parvulum may be associated with sepsis [Citation33]. Moreover, the yellow tongue coating was revealed to be a tongue coating appearance that can predict esophagitis [Citation6].
Bacterial correlational network within the tongue coating microbiome
To identify the potential correlations and thus the interactions of marker bacteria within each group, correlation analysis was carried out (). We identified interesting correlational networks that might indicate important bacterial cofunctions within microbiomes of different tongue coating types. Our analysis showed that the highly enriched gastritis-related S. infantis in the W-thick greasy group was positively associated with Streptococcus peroris with undetermined pathogenic potency. These two bacteria have been found to coexist on the tooth surface and in the pharynx of humans [Citation34]. Within the Y-thin tongue coating bacteria, the representative Y-thin bacterium N. elongata was positively correlated with C. sputigena, a pathogenic species associated with horioamnionitis, abortion, periodontitis, pleuropneumonitis [Citation35,Citation36,Citation37,Citation38] and abscess [Citation39,Citation40], and with the oral cavity resident Lautropia mirabilis [Citation41–44]. In the Y-thick greasy group, the abundant species P. melaninogenica, V. dispar and Atopobium parvulum were significantly positively correlated with each other, demonstrating the coexistence and potential interactions of these bacteria. P. melaninogenica and V. dispar were both positively correlated with Prevotella fusca, while P. melaninogenica and A. parvulum were both positively associated with Prevotella scopos. P. fusca and P. scopos coexist in human oral cavity and P. fusca is implicated in periodontitis [Citation45,Citation46] and can serve as additional marker bacteria for the Y-thick greasy tongue type.
Table 3. Co-occurrence of enriched bacteria in different tongue coating types
To further investigate the underlying reason why different tongue coatings were characterized by different bacteria, the unstimulated salivary flow rate, which impacts the oral microbiota abundance and variation [Citation47,Citation48], was examined in additional individuals with the different tongue coating types (Supplementary Table 1). The results showed that individuals with thick tongue coatings tended to have slower salivary flow rates than those with thin tongue coatings (Supplementary Figure 3), and more than half of the participants with thick tongue coatings demonstrated hyposalivation (< 0.1 g/min). Specifically, the flow rate of the Y-thick tongue coating group decreased significantly compared with that of the white thin and yellow thin tongue coating types (Supplementary Figure 3). This indicates a potential role of the salivary flow rate in influencing the microbiome in thick tongue coating groups, especially in the Y-thick tongue coating group.
Type 2 diabetes bacterial markers in the tongue coating microbiome are distinct from those in the healthy population
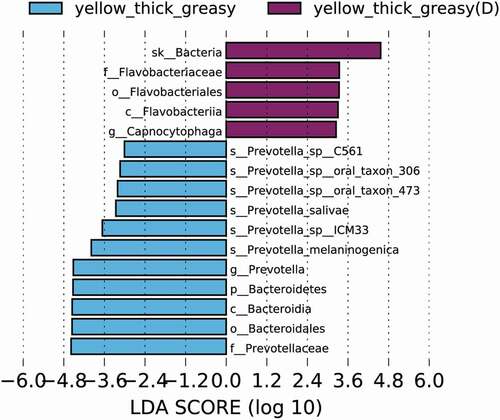
Because most existing studies focusing on tongue coating marker bacteria were carried out in the context of specific diseases, it is difficult to distinguish specific tongue coating microbiomes from the interferences of diseases. To investigate the potential differences in the same tongue coating between healthy individuals and individuals with disease, we carried out a primary comparative study of the tongue coating microbiome in the same Y-thick greasy group in the healthy group and in an additional diabetic group. To our surprise, the microbiota in the two groups showed clear distinctions (Supplementary Figure 4, p = 0.098). In the Y-thick greasy group of diabetic patients, the Capnocytophaga genus was highly enriched (). This potentially pathogenic genus is reported to be related to periodontitis and type 1 diabetes mellitus [Citation49], and it was recently found in a diabetic patient with peritonitis [Citation50]. The species P. melaninogenica was enriched in the healthy population. Such differences in microbe composition were still present between sex-matched groups, which included four males from the healthy Y-thick greasy group and four males from the diabetic Y-thick greasy group (Supplementary Figure 5A). Specifically, Capnocytophaga again was indicative of diabetic patients, together with N. elongata and Neisseria mucosa, which may be specific to male diabetic patients (Supplementary Figure 5B). Therefore, many of the previous disease-based studies might be misleading and cannot offer reliable baseline information on the tongue coating microbiome, thereby potentially causing confusion in many related interpretations.
Discussion
In our current study, we profiled the tongue coating microbiome in healthy populations and demonstrated their differences with respect to physical appearances, a judging criterion used in TCM throughout history. The consistency of marker microbes in our study compared with previous reports demonstrates the stable residence of these microbes in the tongue coating. The most common phyla identified in our study, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and Fusobacteria (), steadily reappeared in other reports [Citation11,Citation15,Citation19]. In addition, we did not find major differences in composition among respective tongue coating types by overall comparison (Supplementary Figure 1), which is in agreement with previous reports that low beta-diversity differences (between samples) were found in oral samples of healthy populations [Citation15,Citation51,Citation52].
Despite finding significant bacterial markers only in three tongue coating groups, these marker species are distinct from those in previous studies. Among the four potential biomarkers of the W-thick coating in gastric cancer patients [Citation12], only M. micronuciformis reappears in our study. As M. micronuciformis and Veillonella were repeatedly found to reside in W-thick coatings in gastric inflammation patients [Citation9], gastritis cancer patients [Citation12] and the healthy population (in our study), they may serve as significant distinguishable microbes for W-thick tongue coatings. The W-thick coating tongue appearance indeed can predict stomach dysfunction [Citation5,Citation6], and M. micronuciformis and Veillonella may thus serve as pre-gastritis markers in the healthy population. Moreover, whether and how these species contribute to the formation of W-thick tongue coatings is worthy of further investigation. [Proteobacteria was higher in abundance in the Y-thin group than in the Y-thick group in gastric cancer patients [Citation12] and was enriched in the Y-tongue coating of chronic hepatitis B patients [Citation11]. In our study, the Y-thin group similarly had the highest level of Proteobacteria (). Within this phylum, we further pinpointed the enrichment of the genus Moraxella and the species N. elongata in Y-thin coatings, which was not identified previously. Bacteroidetes was upregulated in the Y-thick group in our study but decreased in chronic hepatitis B patients, indicating that HBV may drive the declination of Bacteroidetes [Citation11]. Moreover, for the Y-thick greasy tongue coating, the enrichment of cancer and sepsis associated P. melaninogenica, V. dispar and A. parvulum suggested the potential occurrence of these serious diseases, which requires close follow-up. Coincidentally, the yellow tongue coating population is at a high risk for esophageal cancer [Citation6]. Finally, the significant enrichment of the diabetic marker bacteria Capnocytophaga in the Y-thick greasy group of diabetic patients emphasizes the importance of our analysis and the need to distinguish disease-specific and tongue coating-specific bacteria in future studies (). Collectively, the identified bacterial markers together with the tongue coating appearance in the healthy population may indicate potential dysfunction of specific organs and the occurrence of specific diseases, which requires further health monitoring follow-up study of the healthy population.
The reasons behind different tongue coating types enriched by distinctive marker microbes may partly be attributed to different salivary flow rates [Citation53]. It has been reported that more frequent self-reported symptoms of low salivary flow rate was associated with higher abundance of tongue coating P. melaninogenica [Citation54]. Consistently, we demonstrated that the Y-thick greasy group had significantly less salivary secretions (Supplementary Figure 3) and that P. melaninogenica was distinguishable as the characteristic marker microbe (). However, although the salivary flow rate seems to decline in the W-thick greasy group (Supplementary Figure 3), the characterized marker microbe, M. micronuciformis (), is not reported to be associated with impaired salivary secretion [Citation55]. Therefore, how the salivary flow rate affects each group and, in particular, different marker species requires further studies and interpretations.
More investigations and in-depth studies need to be performed to determine the mechanism leading to different tongue coating appearances. It is unknown whether the characteristic bacteria and their associated partners shown by our comparative analysis would be sufficient to produce factors such as pigment or metabolic molecules to form a specific color or thickness on the tongue. For example, Capnocytophaga or P. melaninogenica alone may be dispensable for the Y-thick greasy appearance (), as the Y-thick greasy appearance could occur with either of these strains. On the other hand, nutrient acquisition from host epithelial material and from the oral cavity through saliva, as well as interaction with neighboring microbes, are essential for the patch-like growth of bacteria [Citation3]. Additionally, apoptotic epithelial cells may contribute to different tongue coatings [Citation56].
Taken together, the results of this study revealed the characteristic microbiome of distinctive tongue coatings in a healthy population, which sheds light on the tongue coating formation mechanism. This study also provides a valuable reference for investigating disease-specific tongue coating bacterial markers for future studies.
Author contributions
Yixin Zeng and Xiaolin Tong conceived the study. Hairong Chen, Min Li, Qingwei Li and Jun Wang designed the study. Hairong Chen and Qingwei Li performed experiments and analyzed data. Min Li and Sheng Liu recruited the diabetic population. Min Li and Chensi Yao recruited volunteers participating in the salivary flow rate experiments. Zixiong Wang, Ping Liu, Zhuoya Zhao and Fan Yang helped with the sample collection and data analysis. Jun Wang supervised the bioinformatics analysis. Hairong Chen and Qingwei Li drafted the manuscript. Hairong Chen, Jun Wang, Min Li, Xinjian Li, Xiaolin Tong and Yixin Zeng revised the MS. The authors report no conflict of interest.
Suuplementary material
Supplemental data for this article can be accessed here
Supplemental Material
Download Zip (773.1 KB)Acknowledgments
This work was supported by the Key Research Program of the Chinese Academy of Sciences under Grant KJZD-SW-L05 to Yixin Zeng and KJZD-SW-L05-04 to Hairong Chen. We thank Dr. Bo Yu (Institute of Microbiology, Chinese Academy of Sciences) for technical supports at the initiation phase of the study. We thank chief doctor Haiyan Zhao (Xinjiekou Community Health Service Center, Xicheng District, Beijing, China) for assisting in recruitment of research participants.
Supplemental data
Supplemental data for this article can be accessed here.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Gao L, Xu T, Huang G, et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–12.
- Jia G, Zhi A, Lai PFH, et al. The oral microbiota - a mechanistic role for systemic diseases. Br Dent J. 2018;224:447–455.
- Wilbert SA, Mark Welch JL, Borisy GG. Spatial Ecology of the Human Tongue Dorsum Microbiome. Cell Rep. 2020;30:4003–4015 e3.
- Solos I, Liang Y. A historical evaluation of Chinese tongue diagnosis in the treatment of septicemic plague in the pre-antibiotic era, and as a new direction for revolutionary clinical research applications. J Integr Med. 2018;16:141–146.
- Shi D, Tang C, Blackley SV, et al. An annotated dataset of tongue images supporting geriatric disease diagnosis. Data Brief. 2020;32:106153.
- Deng B, Chen Z, Wang X, et al. Tongue image information for prediction esophageal precancerous lesion: a multi-centric logistic regression analysis of esophageal cancer high risk population in China. J Clin Oncol. 2017;35:e15579–e15579.
- Saritha B, Kannan B. Disease Analysis Using Tongue Image. Int J Eng Res Technol. 2013;2:671-676. https://www.ijert.org/research/disease-analysis-using-tongue-image-IJERTV2IS4317.pdf.
- Lo L-C, Cheng T-L, Chen Y-J, et al. TCM tongue diagnosis index of early-stage breast cancer. Complement Ther Med. 2015;23(5):705–713.
- Jiang B, Liang X, Chen Y, et al. Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Sci Rep. 2012;2:936.
- Ye J, Cai X, Yang J, et al. Bacillus as a potential diagnostic marker for yellow tongue coating. Sci Rep. 2016;6:32496.
- Zhao Y, Mao YF, Tang YS, et al. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol. 2018;24:3448–3461.
- Xu J, Xiang C, Zhang C, et al. Microbial biomarkers of common tongue coatings in patients with gastric cancer. Microb Pathog. 2019;127:97–105.
- Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–588.
- Lu H, Ren Z, Li A, et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016;6:33142.
- Willis JR, Gonzalez-Torres P, Pittis AA, et al. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome. 2018;6:218.
- Ioannis Solos, Yuan Liang. A historical evaluation of Chinese tongue diagnosis in the treatment of septicemic plague in the pre-antibiotic era, and as a new direction for revolutionary clinical research applications. J Integr Med. 2008;16,141-146
- Rabe A, Gesell Salazar M, Michalik S, et al. Metaproteomics analysis of microbial diversity of human saliva and tongue dorsum in young healthy individuals. J Oral Microbiol. 2019;11:1654786.
- Al-Hebshi NN, Baraniya D, Chen T, et al. Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol. 2019;11:1557986.
- Cui J, Cui H, Yang M, et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell. 2019;10:496–509.
- Oldfield NJ, Ala’Aldeen DAA. Neisseria and moraxella. Medical Microbiology. 2012;256–264. DOI:https://doi.org/10.1016/b978-0-7020-4089-4.00038-x
- Yoo YP, Kang KW, Yoon HS, et al. Infective endocarditis caused by Neisseria elongata on a native tricuspid valve and confirmed by DNA sequencing. Tex Heart Inst J. 2014;41:227–230.
- Foissac LB M, Mourguet M, Prudhomme L, et al. Native mitral valve endocarditis due to Neisseria elongata following an untreated dental abscess. Germs. 2020;10:123-125. https://pubmed.ncbi.nlm.nih.gov/32656111/
- Spielman AF, Ghumman A, Panthaki Z, et al. Neisseria elongata osteomyelitis: literature review and case report in a 63-year-old male presenting with progressive right-handed redness, swelling and pain. Int J Surg Case Rep. 2020;73:228–230.
- Yang CH, Tsai KT. Gemella sanguinis endocarditis: first case report in Taiwan and review of the literature. J Formos Med Assoc. 2014;113:562–565.
- Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–1911.
- Paula H Summanen, Bengül Durmaz, Marja-Liisa Väisänen, et al. Porphyromonas somerae sp. nov., a pathogen isolated from humans and distinct from porphyromonas levii. J Clin Microbiol. 2005;43(9):4455-9.
- Yang W, Chen C-H, Jia M, et al. Tumor-Associated Microbiota in Esophageal Squamous Cell Carcinoma. Front Cell Dev Biol. 2021;9:641270. https://pubmed.ncbi.nlm.nih.gov/33681225/
- Rai AK, Panda M, Das AK, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. 2020. DOI:https://doi.org/10.1007/s00203-020-02011-w
- Vuotto C, Barbanti F, Mastrantonio P, et al. Lactobacillus brevis CD2 inhibits Prevotella melaninogenica biofilm. Oral Dis. 2014;20:668–674.
- Beydoun MA, Beydoun HA, Hossain S, et al. Clinical and Bacterial Markers of Periodontitis and Their Association with Incident All-Cause and Alzheimer’s Disease Dementia in a Large National Survey. J Alzheimers Dis. 2020;75:157–172.
- Wei YSHY, Su GW, Chang YR, et al. Identification of hyperglycemia-associated microbiota alterations in saliva and gingival sulcus. Arch Biochem Biophys. 2020;682:108278. https://pubmed.ncbi.nlm.nih.gov/31981541/
- Appel S, Alam J, Lee A, et al. Dysbiotic oral microbiota and infected salivary glands in Sjögren’s syndrome. Plos One. 2020;15. DOI:https://doi.org/10.1371/journal.pone.0244423
- Oyaert M, Cools P, Breyne J, et al. Sepsis with an Atopobium-like species in a patient with Fournier’s gangrene. J Clin Microbiol. 2014;52:364–366.
- Kawamura Y, Hou XG, Todome Y, et al. Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int J Syst Bacteriol. 1998;48(Pt 3):921–927.
- Serge Douvier CN, Filipuzzi L, Kisterman J-P. Chorioamnionitis with intact membranes caused by Capnocytophaga sputigena. Eur J Obstet Gynecol Reprod Biol. 1999;83(1):109-12. https://pubmed.ncbi.nlm.nih.gov/10221619/.
- Anna Alanen EL. Second-trimester abortion caused by Capnocytophaga sputigena: case report. Am J Perinatol. 1999;16(4):181-183. https://pubmed.ncbi.nlm.nih.gov/10458530/.
- Idate U, Bhat K, Kotrashetti V, et al. Molecular identification of Capnocytophaga species from the oral cavity of patients with chronic periodontitis and healthy individuals. J Oral Maxillofacial Pathol. 2020;24:397–398.
- Atmani S, Wanin S, Bellon G, et al. [Capnocytophaga sputigena pleuropneumonitis: a case report]. Archives De Pédiatrie Organe Officiel De La Sociéte Franaise De Pédiatrie. 2008;15: 1535–1537.
- Chan JFW, Wong SSY, Leung SSM, et al. Capnocytophaga sputigena primary iliopsoas abscess. J Med Microbiol. 2010;59:1368.
- Migiyama Y, Anai M, Kashiwabara K, et al. Lung abscess following bronchoscopy due to multidrug-resistant Capnocytophaga sputigena adjacent to lung cancer with high PD-L1 expression. J Infect Chemother. 2018;24:852–855.
- Rossmann SN, Wilson PH, Hicks J, et al. Isolation of Lautropia mirabilis from oral cavities of human immunodeficiency virus-infected children. J Clin Microbiol. 1998;36:1756–1760.
- Shaddox LM, Huang H, Lin T, et al. Microbiological characterization in children with aggressive periodontitis. J Dent Res. 2012;91:927–933.
- Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432.
- Perera M, Al-Hebshi NN, Perera I, et al. Inflammatory Bacteriome and Oral Squamous Cell Carcinoma. J Dent Res. 2018;97:725–732.
- Downes J, Wade WG. Prevotella fusca sp. nov. and Prevotella scopos sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2011;61:854–858.
- Ibrahim M, Subramanian A, Anishetty S. Comparative pan genome analysis of oral Prevotella species implicated in periodontitis. Funct Integr Genomics. 2017;17:513–536.
- Almstahl A, Wikstrom M. Oral Microflora in Subjects with Reduced Salivary Secretion. J Dent Res. 1999;78:1410–1416.
- Proctor DM, Fukuyama JA, Loomer PM, et al. A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat Commun. 2018;9:681.
- Sakalauskiene J, Kubilius R, Gleiznys A, et al. Relationship of clinical and microbiological variables in patients with type 1 diabetes mellitus and periodontitis. Med Sci Monit. 2014;20:1871–1877.
- Tanabe K, Okamoto S, Hiramatsu Asano S, et al. Capnocytophaga canimorsus peritonitis diagnosed by mass spectrometry in a diabetic patient undergoing peritoneal dialysis: a case report. BMC Nephrol. 2019;20:219.
- Moon JH, Lee JH. Probing the diversity of healthy oral microbiome with bioinformatics approaches. BMB Rep. 2016;49:662–670.
- Human Microbiome PC. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214.
- Marsh PD, Do T, Beighton D, et al. 2016. Influence of saliva on the oral microbiota. Periodontology 2000 70:80–92. Doi:https://doi.org/10.1111/prd.12098
- Vanhatalo A, Blackwell JR, L’Heureux JE, et al. Nitrate-responsive oral microbiome modulates nitric oxide homeostasis and blood pressure in humans. Free Radic Biol Med. 2018;124:21–30.
- Rusthen S, Kristoffersen AK, Young A, et al. Dysbiotic salivary microbiota in dry mouth and primary Sjogren’s syndrome patients. PLoS One. 2019;14:e0218319.
- Yanhua Huang, Ziyi Fu, Wei Dong, Zhenming Zhang, Jinquan Mu and Junfeng Zhang. Serum starvation-induces down-regulation of Bcl-2/Bax confers apoptosis in tongue coating-related cells in vitro. Mol Med Rep. 2018 Apr; 17(4): 5057–5064. Additional information: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5865968/