ABSTRACT
Background: The use of antibiotics in dentistry is associated with the emergence and spread of antibiotic-resistant microorganisms, including commensal staphylococci.
Methods: A total of 367 oral samples were collected, from which staphylococci were isolated and identified by using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF). The antibiotic susceptibility of the isolates was determined and molecular characteristics for methicillin-resistant staphylococci was performed.
Results: A total of 103 coagulase-negative staphylococci (CoNS), among them S. warneri, S. haemolyticus, S. saprophyticus, S. pasteuri, S. epidermidis, S. hominis, S. xylosus, S. equorum, S. kloosii, S. succinus, S. cohnii, and S. simulans, were confirmed by MALDI-TOF. Resistance to most tested antibiotics was statistically higher in CoNS than in S. aureus isolates (P-value < 0.05). CoNS isolates showed high resistance to penicillin (S. saprophyticus 88.9%), erythromycin (S. haemolyticus 84.6%), fusidic acid (S. saprophyticus 77.8%), co-trimoxazole (S. epidermidis 71.4%), gentamicin (S. warneri 63.8%), and tetracycline (S. saprophyticus 55.6%). Multidrug resistance was largely observed, especially among S. haemolyticus and S. saprophyticus species. Methicillin-resistance in S. haemolyticus (38.5%), S. saprophyticus (22.2%) and S. aureus (13.5%) was associated with the presence of the mecA gene and SCCmec type IV or V.
Conclusion: Coagulase-negative staphylococci, especially S. haemolyticus and S. saprophyticus, seem to be a reservoir of methicillin resistance and multidrug resistance in the oral cavity.
Introduction
The oral cavity is a poorly understood ecological niche for staphylococci, which, under specific conditions, may become a source of local or systemic infections. Commensal bacteria, among them staphylococci, gain a growing interest in the context of oral health [Citation1,Citation2].
Staphylococci are responsible for the opportunistic community- and hospital-acquired infections. Staphylococcus aureus, one of the coagulase-positive staphylococci (CoPS), constitutes the most common cause of human infections, whereas coagulase-negative staphylococci (CoNS) are believed to be less pathogenic. However, under exposure to the so-called infection-facilitating factors, CoNS may undergo a transition from commensals to pathogens. As a result, they can be involved in a variety of infections with various locations, manifestations and outcomes. CoNS can cause severe infections, such as septicaemia, native and prosthetic valve endocarditis, shunt-associated meningitis, osteomyelitis, and urinary tract infections [Citation3,Citation4]. The infections mentioned above occur primarily in immunocompromised patients and persons with indwelling medical devices [Citation5].
Staphylococci are characterizing by alarmingly increasing rates of antibiotic resistance; this problem belongs to the most important issues related to the management of staphylococcal infections worldwide. In particular, the issue of resistance is important in the case of multidrug-resistant (MDR) and methicillin-resistant (MR) strains. Resistance to methicillin is determined by an extra penicillin-binding protein (PBP2a), encoded by the mecA gene, located on a mobile genetic element known as staphylococcal cassette chromosome (SCCmec). This cassette can be exchanged between various strains of the same species, or even between various staphylococcal species, and can carry genes encoding resistance to other antibiotics [Citation6]. Methicillin-resistant strains cannot be treated with an anti-staphylococcal penicillin (e.g. oxacillin) and show insusceptibility to all β-lactams, including antibiotics with β -lactamase inhibitors and carbapenems. Furthermore, resistance to methicillin is frequently associated with the resistance to other groups of antibiotics (macrolides, lincosamides, aminoglycosides, fluoroquinolones, and sulfonamides), and requires the use of second-line antibiotics such as vancomycin, daptomycin, or linezolid [Citation7,Citation8].
Recent reports indicate the oral cavity as a significant reservoir for antibiotic-resistant bacteria, especially methicillin-resistant S. aureus (MRSA). MRSA may colonize various ecological niches in the mouth, such as the tongue, oral mucosa, periodontal pockets, and denture surfaces [Citation1]. Coagulase-negative staphylococci are known as oral commensal bacteria but the studies reporting the prevalence of staphylococcal species in the oral cavity and their antibiotic resistance are sparse. Consequently, this study aimed to assess the prevalence, antimicrobial-resistance profiles, and molecular characteristics of different Staphylococcus species isolated from oral samples.
Materials and methods
Isolation of staphylococcal isolates
The oral Staphylococcus spp. were isolated from 367 oral microbiological samples analysed consecutively at the Laboratory of Department of Oral Microbiology of the Medical University of Gdansk during routine clinical laboratory procedures, over a period of one and a half years. The analysed staphylococci were not specifically isolated for this research, they were part of the diagnostic laboratory procedure and no humans were involved in the experiments. All samples were streaked onto Columbia blood agar (GrasoBiotech, Starogard Gd., Poland) and differential-selective media mannitol salt agar (bioMérieux, Marcy l’Etoile, France) and were incubated 18–24 h at 37°C. After incubation, colonies with typical staphylococcal morphology (size, shape, or color) were selected to identify by using MALDI-TOF MS according to the manufacturer’s recommendations. The isolates were stored at −80°C in Trypticase Soy Broth (Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with 20% glycerol.
Identification of staphylococcal isolates by MALDI-TOF MS
The staphylococcal isolates were identified using MALDI-TOF MS (Bruker Daltonics, Germany). Bacteria were prepared for identification by extraction of proteins with ethanol and formic acid, according to the manufacturer’s instructions. One loopful of a fresh culture (20–24 h growth on Brain Heart Infusion Agar at 37°C) was suspended in 150 μl of sterile deionized water, and then 450 μl of pure ethanol was added and the sample was mixed thoroughly by vortexing. After centrifugation, the bacterial pellet was resuspended in 50 μl of 70% aqueous formic acid, then 50 μl of acetonitrile was added to the precipitate, and the sample was thoroughly mixed by vortexing. After centrifugation, 1 μl of the supernatant was collected, applied to a metal plate, and allowed to dry at room temperature. Then, 1 μl of an α-cyano-4- hydroxycinnamic acid matrix solution was applied and the sample was left to dry at room temperature. Calibration was performed using a standard calibration mixture of an Escherichia coli extract (Bruker Daltonics) containing RNase and myoglobin proteins. The analysis was repeated three times for each sample. The metal plate with the samples was placed in a MALDI Biotyper chamber for analysis. Automatic measurement of the spectrum and comparative analysis with reference spectra of bacteria were performed using an Ultraflextreme mass spectrometer and MALDI-Biotyper 3.0 software (Bruker Daltonics). The reliability of identification in the MALDI Biotyper system was expressed in points. A log(score) ≥2.0 indicated identification to the species level, and a log(score) ≥1.7 and <2.0 indicated identification to the genus level.
Antimicrobial susceptibility testing
The antimicrobial susceptibility was performed by the disk diffusion method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [Citation9]. Fifteen antimicrobial agents on Mueller-Hinton agar plates (Becton Dickinson, Franklin Lakes, NJ, USA) were tested: oxacillin, cefoxitin, gentamicin, erythromycin, clindamycin, tetracycline, chloramphenicol, ciprofloxacin, trimethoprim/sulfamethoxazole, fusidic acid, linezolid, rifampicin, tigecycline and vancomycin (Bio-Rad, Marnes la Coquette, France) and penicillin G (Oxoid, Basingstoke, England). The phenotype of resistance to macrolide-lincosamide-streptogramin B was tested and interpreted according to the EUCAST. Vancomycin susceptibility was determined with E-test method (bioMerieux, Marcy-l’Etoile, France). Multidrug resistance (MDR) was defined as a resistance to three or more classes of antibiotics.
Isolation of staphylococcal DNA
Genomic DNA extraction was performed on each staphylococcal isolate using the Genomic Micro AX Staphylococcus Gravity kit (A&A Biotechnology, Gdynia, Poland) following the manufacturer’s instructions.
Methicillin-resistance and staphylococcal cassette chromosome mec (SCCmec) typing
Initially, resistance to methicillin was determined using cefoxitin (30 µg) and oxacillin (1 µg) disks, and then confirmed by the detection of PBP2a protein (OXOID ™ PBP2 ‘Latex Agglutination Test Kit, Basingstoke, England). Methicillin-resistance was verified by the detection of the mecA and mecC genes [Citation10,Citation11]. Methicillin-susceptible S. aureus ATCC25923 and methicillin-resistant S. aureus ATCC43300 were used as the reference strains. For mec positive strains, five major staphylococcal chromosomal cassette mec (I–V) was determined as described by Oliveira et al. [Citation12] and by Milheiriço et al. [Citation13]. The SCCmec type was determined on the basis of the band pattern profiles obtained.
Detection of toxin genes
Detection of toxin genes for methicillin-resistant staphylococci was performed. Genes of the enterotoxins (sea, seb, sec, sed, see), toxic shock syndrome toxin-1 (tst) were detected as described by Becker et al. [Citation14]. Detection of Panton-Valentine leukocidin genes (lukS-PV/lukF-PV) was performed as described by Lina et al. [Citation15].
Spa typing
The spa typing was performed for methicillin-resistant S. aureus strains as described previously [Citation16]. The method is based on the sequence analysis of variable region of the protein A (spa) gene, resulting in spa types, assigned by the Ridom StaphType software version 2.2.1 (http://www.ridom.de/ Ridom GmbH, Wurzburg, Germany) and the Ridom SpaServer database (http://spaserver.ridom.de/). The predicted MLST were assigned based on Ridom SpaServer.
Statistical analysis
All calculations were performed with Statistica 10 package (StatSoft, Tulsa, OK, USA) with the threshold of statistical significance set at P-value ≤ 0.05. The significance of differences in the percentages of antibiotic-resistant CoNS and S. aureus isolates was verified with Pearson chi-squared test or Fisher exact test.
Results
Distribution of staphylococcal species
One hundred ninety-two Staphylococcus spp. isolates belonging to 13 staphylococcal species were identified in this study. The most commonly detected species was S. aureus (46.4%). A total of 103 coagulase-negative staphylococci (CoNS), among them S. warneri (45.6%), S. haemolyticus (12.6%), S. saprophyticus (8.7%), S. pasteuri (7.8%), S. epidermidis (6.8%), S. hominis (4.9%), S. xylosus (4.9%), S. equorum (2.9%), S. kloosii (1.9%), S. succinus (1.9%), S. cohnii (1%), and S. simulans (1%), were confirmed by MALDI-TOF MS ().
Table 1. Antibiotic resistance of Staphylococcus species isolated from the oral cavity
Antimicrobial resistance
Overall, the 192 Staphylococcus spp. isolates showed resistance to penicillin (62.5%), gentamicin (51%), erythromycin (30.7%), tetracycline (30.2%), cefoxitin/oxacillin (13.5%), clindamycin (15.1%), trimethoprim/sulfamethoxazole (10.4%), fusidic acid (7.8%), chloramphenicol (4.7%), and ciprofloxacin (2.1%). None of the isolates were resistant to linezolid, rifampicin, tigecycline and vancomycin. Resistance to most tested antibiotics was statistically higher in CoNS than in S. aureus isolates (P < 0.05) (). Some CoNS species exhibited especially high resistance to penicillin (S. saprophyticus 88.9%), erythromycin (S. haemolyticus 84.6%), fusidic acid (S. saprophyticus 77.8%), trimethoprim/sulfamethoxazole (S. epidermidis 71.4%), gentamicin (S. warneri 63.8%), and tetracycline (S. saprophyticus 55.6%) (, ).
Figure 1. Antibiotic resistance of the six most frequent Staphylococcus species isolated from the oral cavity
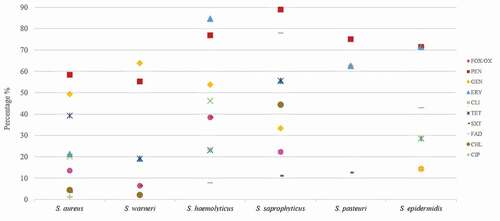
Multidrug resistance was largely observed among S. haemolyticus (61.5%), S. saprophyticus (55.6%), S. succinus (2/2), S. hominis (2/5), and S. aureus isolates (25.8%) and turned out to be higher in CoNS than in S. aureus. MDR CoNS isolates were resistant to 7, 6 and 5 groups of antibiotics ().
Table 2. Antibiotic resistance profiles of methicillin-resistant and multidrug-resistant oral coagulase-negative staphylococci (CoNS)
Resistance to macrolide-lincosamide-streptogramin was represented by cMLSB (9.7%) and iMLSB phenotypes (1.9%) in CoNS strains, including S. haemolyticus, S. epidermidis, S. hominis, S. equorum, S. succinus species. The remaining 29 CoNS isolates (28.2%) resistant to erythromycin represented an MSB resistance profile.
Methicillin-resistance and SCCmec typing
Fourteen CoNS isolates were identified as methicillin-resistant (MR), including S. haemolyticus (38.5%), S. saprophyticus (22.2%), S. epidermidis (14.3%), S. warneri, (6.4%), S. hominis (2/5) and S. succinus (1/2) species. All of MR isolates were mecA-positive, while none harboured mecC. Staphylococcal cassette chromosome mec (SCCmec) types IV (5/10) and V (5/10) were detected. No isolate represented I, II and III SCCmec types. No SCCmec type was identified in the case of the four mecA-positive CoNS ().
Twelve of the 89 S. aureus isolates were mecA-positive and mecC-negative. SCCmec types IV (66.7%) and V (33.3%) were detected, suggesting a community origin (CA-MRSA). MRSA assigned to nine spa types (t012, t091, t156, t189, t437, t888, t5644, t13670 and t18953). MRSA isolates showed iMLSB and cMLSB phenotypes of resistance to macrolide-lincosamide-streptogramin ().
Table 3. Characteristics of methicillin-resistant S. aureus (MRSA) isolated from the oral cavity
Detection of toxin genes
Genes encoding for the enterotoxin seb (2 isolates), enterotoxin sec (3 isolates), Panton-Valentine leukocidin (2 isolates), exfoliative toxin A eta (1 isolates), and toxic shock syndrome toxin-1 tst (1 isolates) were identified in MRSA isolates. Two isolates carrying lukS-PV/lukF-PV genes represented spa type t437 ().
Discussion
Staphylococci can be frequently isolated from oral cavities of both healthy and ill persons [Citation17–19]. Recent studies showed that the oral cavity should be considered a potential source of systemic bacterial spread and a reservoir of antimicrobial resistance genes [Citation20–22].
This study demonstrated a relatively high prevalence of diverse staphylococcal species in the oral cavity. Similar to previous studies, we identified S. aureus as the most frequent oral isolate [Citation17,Citation23]. Aside from S. aureus, we isolated also twelve different species of CoNS. The presence of various CoNS species in the oral cavity was also demonstrated in recent studies conducted in Poland, Japan, and Argentina [Citation23–25]. In a study of healthy persons and patients subjected to kidney transplantation, Majchrzak et al. identified thirteen staphylococcal species, with the most frequently isolated CoNS being S. epidermidis (40.2%), S. warneri (10.3%), and S. haemolyticus (9.2%) [Citation24]. Ohara-Nemoto et al. reported on nine Staphylococcus species; S. epidermidis was the most common species isolated from plaque and saliva, followed by S. hominis, S. warneri, S. intermedius, S. capitis, and S. haemolyticus (12.5–7.1%) [Citation17]. Also, Cuesta et al. [Citation25] identified S. epidermidis as the primary CoNS species in the oral cavity of dental patients. The most commonly detected non-S. aureus species in our present study were S. warneri (45.6%), S. haemolyticus (12.6%), and S. saprophyticus (8.7%). The discrepancy between our findings and the results of the studies mentioned above may result from differences in patient groups, methods of staphylococci isolation and identification, and geographic region. S. warneri was previously reported as a cause of catheter-related bacteriemia, endocarditis, multiple abscesses, and septic arthritis [Citation26,Citation27]. S. saprophyticus and S. haemolyticus are considered harmful hospital pathogens that cause severe infections with a significant level of bacteraemia [Citation28]. This evidence suggests that the CoNS colonising oral cavity may pose a substantial risk of infection, whether local or systemic one.
While antibiotic resistance of clinical staphylococcal isolates associated with various infections is a well-established fact, little is known about the antibiotic resistance of commensal CoNS present in the oral cavity. In our study, the majority of CoNS (66.1%) and S. aureus (51.7%) isolates from the oral cavity were resistant to penicillin. Resistance to most antibiotics was statistically higher in CoNS than in S. aureus isolates. Some CoNS species exhibited especially high resistance to tested antibiotics, S. saprophyticus being predominantly resistant to penicillin, tetracycline, fusidic acid and chloramphenicol, S. haemolyticus to erythromycin and clindamycin, S. warneri to gentamycin, S. epidermidis to co-trimoxazole. These findings confirm the reports by Cui et al. [Citation28], Arredondo et al. [Citation29], and Szczuka et al. [Citation30], which showed a high level of antibiotic resistance among CoNS species.
Also, the proportion of isolates resistant to multiple antibiotics varied depending on the species. The largest proportions of multidrug-resistant CoNS isolates were found in S. haemolyticus (61.5%) and S. saprophyticus (55.6%), respectively. Equally high proportions of multidrug-resistant isolates from those species were previously reported by other authors in a clinical setting [Citation3,Citation30]. Our present study adds to those findings, demonstrating that multidrug resistance may also be a common problem in commensal non-healthcare-associated CoNS from the oral cavity and does not necessarily result from antibiotic pressure.
Methicillin-resistant staphylococci constitute a major challenge in the treatment of both nosocomial and community-acquired infections. Methicillin resistance is determined by an extra penicillin-binding protein (PBP2a), encoded by the mecA gene. In the present study, the mecA gene was found in 38.5%, 22.2%, 13.5% S. haemolyticus, S. saprophyticus and S. aureus isolates, respectively. The proportions of isolates carrying this gene were nearly thrice as high as reported by Smith et al. [Citation19] but still lower than the rates documented in staphylococcal infections (40–60%) [Citation8,Citation28]. In previous studies, mecA-carriage was frequently demonstrated in CoNS belonging to S. haemolyticus, S. epidermidis, and S. hominis species, which is consistent with our findings [Citation5,Citation30,Citation31]. mecA gene carriage rate in S. saprophyticus (22.3%) was higher than in clinical isolates reported by Cui et al. [Citation28]. The presence of the mecA gene was not observed in some staphylococcal species, such as S. xylosus, S. pasteuri, S. simulans, and S. cohnii, which is consistent with the previous results [Citation32]. The MDR phenotype was widespread among methicillin-resistant CoNS strains, such as S. haemolyticus S. saprophyticus, and S. epidermidis. These results imply that CoNS may constitute a potential reservoir for methicillin resistance and play a role in the inter-species transfer of resistance genes.
SCCmec is a mobile genetic element consisting of two components, the mec gene complex and the ccr (cassette chromosome recombinase) gene complex. The combination of the genes confers various SCCmec types. SCCmec types I, II and III are predominant in hospital-acquired isolates (HA-MRSA), whereas SCCmec types IV and V are mainly associated with community-acquired isolates (HA-MRSA). SCCmec types IV and V are smaller than SCCmec types I, II and III, which facilitates their mobility and spread [Citation6]. While SCCmec types I, II and III were not identified in the present study, S. aureus and CoNS isolates were shown to harbour SCCmec type IV or V. SCCmec type V was preferentially associated with S. haemolyticus, similar to results demonstrated by Szczuka et al. [Citation30]. No SCCmec type was identified in the case of the four mecA-positive CoNS. These findings are consistent with the results of previous studies in which non-typeable ccr genes were shown to be associated with the heterogeneity of SCCmec elements in methicillin-resistant CoNS strains [Citation31,Citation33].
This study demonstrated a relatively high diversity of spa types in detected oral MRSA isolates. Particular attention should be given to two isolates of the spa type t437 having lukS-PV/lukF-PV genes. Panton-Valentine leukocidin (PVL) genes is considered as a stable genetic marker for CA-MRSA strains that can carry SCCmec type IV or V. Above half (66.6%) of S. aureus isolates in this study were assigned to SCCmec IV, whereas strains represented spa t437 harbored SCCmec V. Similar strains were isolated in Germany and in Taiwan [Citation34,Citation35], but recent report from Poland described predominance of t437 SCCmec-IV-pvl-positive strains in specimens from diabetic patients [Citation36].
In conclusion, this study showed that the oral cavity is colonised by both S. aureus and a broad spectrum of diverse CoNS species. The level of antimicrobial resistance was higher in CoNS than in S. aureus isolates. CoNS isolates, especially S. haemolyticus and S. saprophyticus, often displayed MDR, with high rates of resistance to methicillin, erythromycin, gentamicin, tetracycline, co-trimoxazole and fusidic acid. In both S. aureus and CoNS, methicillin-resistance was associated with the presence of the mecA gene and SCCmec types IV or V. The MRSA isolates identified as spa type t437 carried Panton-Valentine leucocidin genes (lukS-PV/lukF-PV). Although S. aureus strains appear to be more prevalent in the oral cavity, these are coagulase-negative staphylococci, especially S. haemolyticus and S. saprophyticus, which seem to be a reservoir of methicillin resistance and multidrug resistance. Our findings warrant monitoring for both colonisation and resistance rates of staphylococci in the oral cavity.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kearney A, Kinnevey P, Shore A, et al. The oral cavity revealed as a significant reservoir of Staphylococcus aureus in an acute hospital by extensive patient, healthcare worker and environmental sampling. J Hosp Infect. 2020;6:S0195–6701(20)30103–1.
- Radaic A, Kapila YL. The oralome and its dysbiosis: new insights into oral microbiome-host interactions. Comput Struct Biotechnol J. 2021;19:1335–8.
- Michels R, Last K, Becker SL, et al. Update on coagulase-negative staphylococci-what the clinician should know. Microorganisms. 2021;9(4):830.
- Lisowska-Łysiak K, Lauterbach R, Międzobrodzki J, et al. Epidemiology and pathogenesis of staphylococcus bloodstream infections in humans: a review. Pol J Microbiol. 2021;70(1):13–23.
- Becker K, Both A, Weißelberg S, et al. Emergence of coagulase-negative staphylococci. Expert Rev Anti Infect Ther. 2020;18(4):349–366.
- Saber H, Jasni AS, Jamaluddin TZ, et al. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) species. Malays J Med Sci. 2017;24(5):7–18.
- Kosecka-Strojek M, Sadowy E, Gawryszewska I, et al. Emergence of linezolid-resistant Staphylococcus epidermidis in the tertiary children’s hospital in Cracow, Poland. Eur J Clin Microbiol Infect Dis. 2020;39(9):1717–1725.
- Khoshnood S, Shahi F, Jomehzadeh N, et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among methicillin-resistant Staphylococcus aureus strains isolated from burn patients. Acta Microbiol Immunol Hung. 2019;66(3):387–398.
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) http://www.eucast.org/ (2021).
- Khairalla A, Wash R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep. 2017;7(1):7390.
- Stegger M, Andersen PS, Kearns A, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or new mecA homologue mecALGA251. Clin Microbiol Infect. 2012;18(4):395–400.
- Oliveira DC, de Lencastre H. Multiplex PCR Strategy for Rapid Identification of Structural Types and Variants of the mec Element in Methicillin-Resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155–2161.
- Milheirico C, Oliveira DC, De Lancastre H. Update to the Multiplex PCR Strategy for Assignment of mec Element Types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–3377.
- Becker K, Roth R, Peters G. Rapid and Specific Detection of Toxigenic Staphylococcus aureus: use of Two Multiplex PCR Enzyme Immunoassays for Amplification and Hybridization of Staphylococcal Enterotoxin Genes, Exfoliative Toxin Genes, and Toxic Shock Syndrome Toxin 1 Gene. J Clin Microbiol. 1998;36(9):2548–2553.
- Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine Leukocidin--Producing Staphylococcus aureus in Primary Skin Infections and Pneumonia. Clin Infect Dis. 1999;29(5):1128–1132.
- Aires-de-sousa M, Boye K, de Lencastre H, et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006;44(2):619–621.
- Ohara-Nemoto Y, Haraga H, Kimura S, et al. Occurrence of staphylococci in the oral cavities of healthy adults and nasal–oral trafficking of the bacteria. J Med Microbiol. 2008;57(1):95–99.
- Blomqvist S, Å Å, Arirachakaran P, et al. Phenotype, genotype, and antibiotic susceptibility of Swedish and Thai oral isolates of Staphylococcus aureus. J Oral Microbiol. 2015;7(1):26250.
- Smith AJ, Robertson D, Tang MK, et al. Staphylococcus aureus in the oral cavity: a three-year retrospective analysis of clinical laboratory data. Br Dent J. 2003;195(12):701–703.
- Kwapisz E, Garbacz K, Kosecka-Strojek M, et al. Presence of egc-positive major clones ST 45, 30 and 22 among methicillin-resistant and methicillin-susceptible oral Staphylococcus aureus strains. Sci Rep. 2020;10(1):18889.
- Garbacz K, Jarzembowski T, Kwapisz E, et al. Do the oral Staphylococcus aureus strains from denture wearers have a greater pathogenicity potential? J Oral Microbiol. 2018;11(1):1536193.
- Kwapisz E, Garbacz K, Wierzbowska M, et al. Prevalence of methicillin-resistant Staphylococcus aureus among adult patients with symptoms of oral infection. Med Dośw Mikrobiol. 2017;69:209–214.
- Hirose M, Aung MS, Fukuda A, et al. Prevalence and genetic characteristics of methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci isolated from oral cavity of healthy children in Japan. Microb Drug Resist. 2019;25(3):400–407.
- Majchrzak K, Mierzwinska-Nastalska E, Chmura A, et al. Comparison of staphylococcal flora in denture plaque and the surface of the pharyngeal mucous membrane in kidney transplant recipients. Transplant Proc. 2016;48(5):1590–1597.
- Cuesta AI, Jewtuchowicz V, Brusca MI, et al. Prevalence of Staphylococcus spp and Candida spp in the oral cavity and periodontal pockets of periodontal disease patients. Acta Odontol Latinoam. 2010;23(1):20–26.
- Yamamoto J, Endo A, Sugawara H, et al. Native valve endocarditis due to staphylococcus warneri developing in a patient with type 1 diabetes. Intern Med. 2020;59(18):2269–2274.
- Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926.
- Cui J, Liang Z, Mo Z, et al. The species distribution, antimicrobial resistance and risk factors for poor outcome of coagulase-negative staphylococci bacteraemia in China. Antimicrob Resist Infect Control. 2019;8(1):65.
- Arredondo A, Blanc V, Mor C, et al. Tetracycline and multidrug resistance in the oral microbiota: differences between healthy subjects and patients with periodontitis in Spain. J Oral Microbiol. 2020;13(1):1847431.
- Szczuka E, Jabłońska L, Kaznowski A. Coagulase-negative staphylococci: pathogenesis, occurrence of antibiotic resistance genes and in vitro effects of antimicrobial agents on biofilm-growing bacteria. J Med Microbiol. 2016;65(12):1405–1413.
- Abbasi Montazeri E, Seyed-Mohammadi S, Asarehzadegan Dezfuli A, et al. Investigation of SCCmec types I-IV in clinical isolates of methicillin-resistant coagulase-negative staphylococci in Ahvaz, Southwest Iran. Biosci Rep. 2020;40(5):BSR20200847.
- Xu Z, Shah HN, Misra R, et al. The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Antimicrob Resist Infect Control. 2018 13; 7(1):73.
- Lee SI, Kim SD, Park JH, et al. Species distribution, antimicrobial resistance, and enterotoxigenicity of non-aureus staphylococci in retail chicken meat. Antibiotics (Basel). 2020;9(11):809.
- Klein S, Menz MD, Zanger P, et al. Increase in the prevalence of Panton–Valentine leukocidin and clonal shift in community-onset methicillin-resistant Staphylococcus aureus causing skin and soft-tissue infections in the Rhine-Neckar Region, Germany, 2012–2016. Int J Antimicrob Agents. 2019;53(3):261–267.
- Chen FJ. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012;50(5):1679–1683.
- Stańkowska M, Garbacz K, Piechowicz L, et al. Dissemination of t437-SCCmecIV and coagulase-negative t037- SCCmecIII types among borderline oxacillin-resistant staphylococcus aureus isolated from skin infections and diabetic foot ulcers. Infect Drug Resist. 2019;12:3197–3203.
