ABSTRACT
Oral streptococci are gram-positive facultative anaerobic bacteria that are normal inhabitants of the human oral cavity and play an important role in maintaining oral microecological balance and pathogenesis. Transposon mutagenesis is an effective genetic manipulation strategy for studying the function of genomic features. In order to study cariogenic related genes and crucial biological element genes of oral Streptococcus, transposon mutagenesis was widely used to identify functional genes. With the advent of next-generation sequencing (NGS) technology and the development of transposon random mutation library construction methods, transposon insertion sequencing (TIS) came into being. Benefiting from high-throughput advances in NGS, TIS was able to evaluate the fitness contribution and essentiality of genetic features in the bacterial genome. The application of transposon mutagenesis, including TIS, to oral streptococci provided a massive amount of valuable detailed linkage data between genetic fitness and genetic backgrounds, further clarify the processes of colonization, virulence, and persistence and provides a more reliable basis for investigating relationships with host ecology and disease status. This review focuses on transposon mutagenesis, including TIS, and its applicability in oral streptococci.
Introduction
Oral streptococci are the microorganisms that colonize the oral surface, comprising the main bacteria in the human oral cavity, where they play important roles in maintaining microecological balance and causing diseases [Citation1,Citation2]. Over 100 oral bacteria have been identified as Streptococcus [Citation3]. Currently, oral streptococci have been divided into six groups: anginosus, bovis, mitis, mutans, salivarius, and pyogenic, based on biochemical testing and 16S rRNA gene sequencing analysis [Citation4,Citation5]. Streptococcus mutans is cariogenic and can easily adhere to the surface of teeth, form biofilms, release acidic compounds after carbohydrate metabolism, and enamel demineralization [Citation6]. Conclusive epidemiological evidence has shown that S. mutans plays a crucial role in the onset and development of dental caries [Citation7]. The mitis and sanguinis groups, such as S. mitis, S. gordonii, and S. sanguinis, are common commensals that can compete with pathogenic bacteria by producing bactericidal hydrogen peroxide for colonisation of the oral cavity [Citation8]. These bacteria are also associated with the formation of biofilms in the oral cavity, which are abundant in both supragingival and subgingival plaques [Citation9,Citation10]. Streptococcus gordonii is an initial colonising bacterium on the surface of teeth that can proliferate along with other oral microorganisms, leading to periodontal disease and caries [Citation11]. It can also enter the bloodstream through oral bleeding and increases the risk of invasive infections and systemic diseases, including infective endocarditis [Citation12,Citation13]. The S. anginosus group is an important component of the oropharyngeal flora that is commonly associated with various suppurative infections and abscesses in the brain, heart, meninges, liver, spleen, and lung via periapical odontogenic lesions and bacteraemia [Citation14]. Streptococcus constellatus and S. intermedius in dental plaques are associated with the occurrence and development of periodontal disease [Citation15]. In contrast, the S. salivarius group, which predominates the oral mucosal surface and saliva, is associated with oral health rather than disease [Citation16].
Transposon mutagenesis is an effective forward genetic strategy for studying gene function by observing the phenotypic changes in mutated genes. Random mutants in a variety of prokaryotes have been created by using different transposon genes such as Tn3 derivatives, IS (insertion sequence) elements, Tn7, Tn5, and mariner. Since the advent of genome sequencing, techniques such as genetic footprinting, signature-tagged Mutagenesis (STM), transposon site hybridization (TraSH), and scanning Linker mutagenesis (SLM) have been developed [Citation17]. And with the advent of next-generation sequencing (NGS), transposon insertion sequencing (TIS) combines it with large-scale transposon insertion mutations to evaluate the essentiality of genetic features and fitness contribution in the bacterial genome in the saturated random mutant libraries. The four TIS techniques published in 2009 include insertion sequencing (INSeq) in Bacteroides thetaiotaomicron [Citation18], high-throughput insertion tracking by deep sequencing (HITS) in Haemophilus influenzae [Citation19], transposon sequencing (Tn-Seq) in S. pneumoniae [Citation20], and transposon-directed insertion site sequencing (TraDIS) in S. Typhi [Citation21]. Those techniques have been widely used in various bacteria to study fitness and virulence, including Enterococcus faecalis [Citation22], Vibrio parahaemolyticus [Citation23], Salmonella enteritidis [Citation24], Edwardsiella piscicida [Citation25], Ralstonia solanacearum [Citation26] and Pantoea [Citation27]. Ultimately, TIS is a key tool for interpreting the rapidly increasing amount of genome sequencing data and is expected to shed light on the function of individual genome features. With the development of transposon technology, TIS has been reviewed from the perspectives of design and analysis [Citation28,Citation29]. Cain et al. discussed recent applications of TIS in answering general biological questions [Citation30]. The present review focuses on oral microorganisms and highlights the application of transposon mutagenesis, including TIS, to oral streptococci, as well as research progress, aiming to better understand the relationship between oral streptococcal phenotype and genotype, which can help clarify the processes of colonization, virulence, and persistence and provides a more reliable basis for investigating relationships with host ecology and disease status. and show some articles and conclusions regarding transposon mutagenesis applied to oral streptococci.
Figure 1. Functional genes identified by transposon mutant library screening in oral streptococcus. Red genes are associated with bacterial virulence, green genes with bacteria–bacteria interactions, and blue genes with drug resistance.
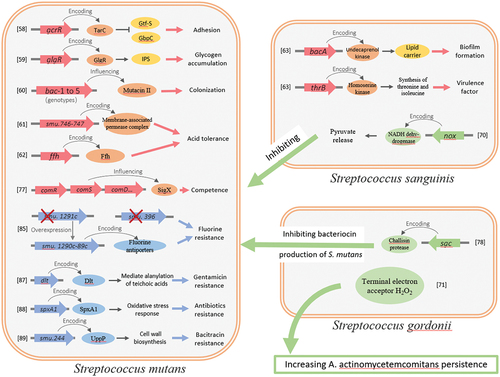
Table 1. Transposon mutagenesis in oral Streptococci.
Transposon mutagenesis and NGS
Transposons are mobile genetic factors that can move within genomes through ‘cut and paste’ or copy mechanisms. A transposase encoded by a transposon can recognise specific inverted repeat sequences at both ends of the transposon, separate the transposon from adjacent sequences, and insert it into a DNA target site [Citation31]. The most common application of transposons is insertional mutagenesis, which can be used to create libraries of mutant strains. The success of transposon mutant library screening depends on the number of mutants screened and diversity of the library.
Various transposon subsystems have been described [Citation17]. Examples of transposon subsystems include TN916, TN917, and ISS1, which have been used to study oral Streptococcus [Citation32–34]. However, certain features of TN916 and TN917 prevent the creation of unbiased libraries of randomly inserted transposons. For example, Tn 916 preferentially uses A: T-rich targets but has an insertion hotspot in some bacteria [Citation35]. TN917 inserts nonrandomly in chromosomes and is far more prevalent in specific DNA regions [Citation36], and ISS1 mediates transpositions through a replication mechanism, whereby the entire plasmid or sequence of plasmids is integrated into the bacterial genome. Moreover, some bacteria containing endogenous ISS1 copies can become targets for recombination events [Citation37].
Due to the small scale of those transposon subsystems creating mutation libraries, mariner or Tn5 transposon without insertion site bias have been utilized to generate a saturated random mutant library. And TIS combines NGS with large-scale mariner or Tn5 transposon insertion mutations, with which the essentiality of genetic features and fitness contribution in the bacterial genome can be evaluated. shows the basic workflow of the TIS. Briefly, it starts with the construction of a saturated library of random transposon insertions, where the genome of each mutant strain contains a transposon insertion [Citation38,Citation39]. After libraries from various environments are selected, the frequency variation of each inserted mutant is counted by sequencing the overall transposon-flanking region, and these variations are used to estimate the fitness of each mutant. By sequencing before and after selection for a specific condition, changes in the population insertion frequency during selection can define the importance of these genetic elements under that condition. A feature with a reduced insertion frequency is considered important for fitness under these conditions and vice versa [Citation40]. Transposon insertion sequencing is a high-throughput approach that reveals phenotypic and genotypic relationships and is applicable to a series of species.
Figure 2. Schema of transposon insertion sequencing. (a) Transposon containing inverted repeats at both ends and an antibiotic resistance selection marker is inserted into bacterial genomic DNA to disrupt Gene B. (b) Transposon insertion points of each mutant are determined and mapped through breaking, adding adaptors, PCR amplification, and sequencing. (c) Bacterial mutant libraries are grown in vitro or in vivo, and the analysis of the relative abundance of insertion mutants under each growth condition can define the fitness of genetic elements.
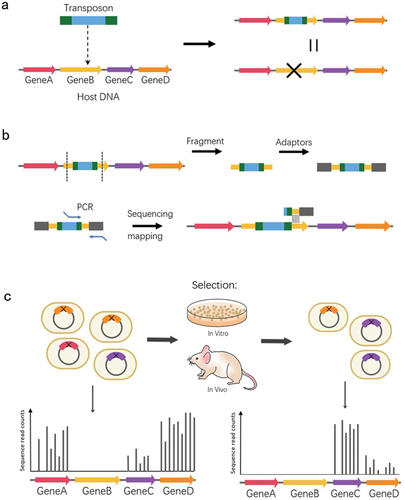
The four major TISs are Tn-Seq, TraDIS, INSeq, and HITS, which vary by transposition, library amplification, sequencing, and bioinformatics methods [Citation41,Citation42]. HITS and TraDIS are applicable to any transposon, but commercial Tn5 transposon is commonly used. In order to locate the transposon insertion, a series of steps are necessary before sequencing, including shearing DNA, ligating adaptors, amplifying by PCR, affinity purification, and removing extraneous DNA [Citation19,Citation21]. However, the DNA shearing produces a range of fragments sizes, may resulting in PCR bias. INseq and Tn-Seq use the mariner transposon exclusively which contains MmeI recognition sites in the terminal inverted repeats. When DNA from mutation library is digested with MmeI, 16 bp of flanking genomic DNA are produced, amplified by PCR after ligating adaptors, and then isolate the 120 bp product by agarose gel or PAGE gel for NGS [Citation18,Citation20]. The mariner transposon, which recognises and inserts into TA sites, is applicable to transposition to organisms with low GC in the genome, including oral Streptococci [Citation43]. The mariner transposon mutagenesis system can be transposed in streptococci both in vitro and in vivo [Citation20,Citation44]. In addition, Tn-seq sample preparation protocol is simple, and is easily to isolate the final product of precise length by agarose gel purification, making it an ideal TISs for studying oral streptococcus.
The anatomical and physiological characteristics, as well as temperature, humidity, pH, and rich nutrition of the oral cavity, provide a suitable habitat for microorganisms. The resident microorganisms in the oral cavity are numerous and complex. To date, the origin, colonisation, distribution, species, number, succession, and the relationship and dynamic balance between microorganisms and host tissues and cells are not fully understood. Transposon mutagenesis, including TIS, including powerful Tn-Seq, has been widely applied to Streptococcus and provided a massive amount of valuable detailed linkage data between genetic fitness and genetic backgrounds. This has significantly contributed to the study of oral Streptococcus physiological characteristics, interactions between microbes, and their interrelationships with hosts.
Uncovering general functions of essential genes and antibacterial drugs development
Transposon mutagenesis combined with NGS has played a significant role in determining the essential genomes of microorganisms [Citation45–47]. Several approaches can be applied to determine the necessary genes using TIS, such as annotation-dependent and independent methods [Citation29]. Essential genes that cannot be mutated determine the basic life processes of bacteria and may be targets of new antimicrobial therapies. Identifying essential genes of pathogenic microorganisms can reveal key genes and pathways to control pathogenic bacteria and the minimum genome of organisms, and these genes might serve as targets for the development of antibacterial drugs.
Van Opijnen et al. first proposed Tn-Seq to determine the fitness of each gene in S. pneumoniae and accurately quantified the genetic interactions across the genome [Citation20]. The genomes of S. sanguinis and S. mutans have also been analysed for gene essentiality. In fact, 9% of the S. sanguinis SK36 genome is essential for translation, transcription, glycan biosynthesis, protein folding, sorting, and degradation [Citation48]. In S. mutans UA159, 11% of the genome is essential and genes encode products that are closely associated with replication, translation, cell wall synthesis, and lipid metabolism. According to S. mutans core genome identified by Cornejo et al, 87% of the essential genes are part of the core genome, and the remaining 13% belong to an accessory genome [Citation49,Citation50]. Predictions indicate that most of the essential genes are part of the core genome; they encode proteins that are needed for basic biological functions and metabolism, and are conserved among strains. shows the major biological pathways of the essential genes in oral streptococci. Some essential genes in the accessory genome, which are also important to the gene-gene network, may be related to coping with unique environmental conditions, such as medium and culture conditions, as well as endogenous metabolic end products. Some genes are condition-specific; that is, they might be necessary for an organism to grow in one environment but not in others. Conditionally essential genes are discrepancies when the mutant libraries are cultured under different conditions, such as in rich or defined medium, acidic conditions or oxidative stress, and rodent models in vivo [Citation51,Citation52].
Figure 3. Major biological pathways of essential genes in oral streptococcus (based on [Citation48] and [Citation49]).
![Figure 3. Major biological pathways of essential genes in oral streptococcus (based on [Citation48] and [Citation49]).](/cms/asset/214119bb-a918-4dbe-b1e5-83269f3adf5b/zjom_a_2104951_f0003_oc.jpg)
The essential genes conserved between strains and species can be effective targets for antimicrobial agents to control various streptococcal infections. For example, 202 of 218 most essential genes for S. sanguini have homologous genes in most other streptococcal genomes [Citation48]. These genes are associated with basic biological processes, including replication, transcription, translation, peptidoglycan synthesis, acetyl coenzyme A biochemical pathways, and lipid synthesis and are highly conserved in most species. Drugs that target specific essential genes found only in one strain, such as those found only in S. mutans that encode arginine repressors, superoxide dismutase, l-lactate dehydrogenase, and the shikimate pathway, can control infection without interfering with other beneficial oral bacteria [Citation49,Citation53,Citation54].
Investigating virulence genes and host adaptation
Streptococcus contains a series of virulence factors, including adhesion and surface invasion proteins, as well as proteins for the delivery of toxins to the cell surface and extracellular environment. These factors are related to Streptococcus colonisation at different sites, biofilm formation, host tissue destruction, and host immune inflammation [Citation55]. Streptococcus mutans is closely related to the occurrence of human dental caries for its physiological properties of adhesive, being acidogenic and aciduric, capable of producing exopolysaccharides [Citation56]. Traditional transposon insertion mutations have been used to explore the genes associated with adhesion [Citation57], glycogen accumulation [Citation58], bacteriocin-like substances synthesis [Citation59], biofilm formation [Citation60], and acid tolerance [Citation61] in S. mutans ().
In niche screening, especially in vivo screening, is extremely valuable for exploring the physiological metabolism of pathogenic microorganisms and discovering the mechanisms of virulence. Streptococcus sanguinis is a common resident of the human oral cavity and one of the major pathogenic bacteria in infective endocarditis. Paik, Sehmi et al. identified six genes associated with virulence 800 mutants in the rat and the rabbit endocarditis model using a modified transposon mutagenesis system (signature-tagged mutagenesis) and dot blot analysis [Citation62]. Next-generation sequencing made it easier and faster to screen virulence factors of bacterial pathogens on a large scale. Through screening the fitness of S. sanguinis mutants in human serum by ORF‐seq (similar to Tn‐seq), 178 mutants with significant abundance changes have been observed. Analysis of the functions of these fitness genes suggests that the virulence factors of S. sanguinis are closely associated with its ability to survive under anaerobic conditions and synthesize cell walls, nucleic acids, and amino acids [Citation63]. In S. mutans, fitness determinants required for establishment or persistence under acidic and oxidative stress conditions and in rodent models have also been identified by Tn-Seq. Surprisingly, >75% of the genes in S. mutans UA159 were required for colonization of the mouse oral cavity, possibly because of considerable selection pressure to compete with commensal bacteria for survival during the initial colonization [Citation49,Citation51]. The fitness of group A streptococcus (GAS) in human saliva has been studied using TraDIS, and 92 GAS genes were found to be associated with wild-type fitness. Most of the identified genes are related to the transport and metabolism of carbohydrates, inorganic ions, and amino acids [Citation64]. And in S. pyogenes, gacH has been identified as being sensitive to phospholipase A2 secreted by the bactericidal enzyme human IIA group and zinc resistant by Tn-seq. The gacH gene in the group A carbohydrate (GAC) biosynthesis cluster encodes a new class of glycerophosphoric (GroP) transferases linked to the C6 hydroxyl group of 30% of the GAC N-acetylglucosamine side chain. GroP transferases have also been found in serotype c carbohydrate of S. mutans, depending on the presence of their respective gacH homologues [Citation65]. This structural change affects the interactions between the host and pathogen, and the development of antimicrobials.
In vivo screening is extremely valuable for exploring the physiological metabolism of pathogenic microorganisms and discovering the mechanisms of virulence. However, a major problem with the use of TIS in vivo is the effect of bottlenecks. Hampered by the removal or killing of large numbers of bacteria during the establishment of animal models, it is difficult to identify whether these missing mutants are accidental or if their fitness is low [Citation66]. Although this can be partially compensated by optimizing the analytical methods, the main bottleneck effects might irreversibly bias the experiments. Despite its limitations, the application of TIS in animal models has potential value for investigating virulence genes, antimicrobial drugs, and vaccines.
Understanding bacteria–bacteria interactions
Oral microorganisms do not exist in isolation but constantly interact and form communities with other microorganisms, and these interactions are considered important factors in the formation of disease states. Interactions among colonised oral microbes can continuously accelerate or inhibit biofilm development, and transposon mutagenesis offers the potential to identify such interactions.
Streptococcus sanguinis and S. gordonii generate H2O2, which inhibits the growth of S. mutans through pyruvate oxidase encoded by spxB [Citation67,Citation68]. The catabolite control protein A (CcpA) represses spxB expression and H2O2 release [Citation68]. While the ccpA deletion mutants of S. gordonii and S. sanguinis could directly detoxify H2O2 via pyruvate release and confer protection in trans to other bacteria. Targeted and transposon mutagenesis suggests that nox, which is presumed to encode H2O-forming NADH dehydrogenase, is essential for oxidative protection and pyruvate release, with other genes such as dps and sodA having secondary effects [Citation69]. This study revealed a novel aspect of the competitive interaction between pathogens and oral commensals and offers a direction for further study of the mechanisms underlying the varying degrees of inhibition potential between strains of commensal oral streptococci that produce H2O2. A new mechanism that leads to the combined growth of oral microbes Aggregatibacter actinomycetemcomitans and S. gordonii has also been revealed by Tn-Seq. The latter co-infects A. actinomycetemcomitans by producing the terminal electron acceptor H2O2, which changes the growth mode of A. actinomycetemcomitans from anaerobic to aerobic, increasing its persistence. This interaction is referred to as ‘cross respiration’, implying that influencing the S. gordonii antibacterial regimen helps combat such co-infections [Citation70].
Quorum sensing is a communication method to coordinate a response in a population employed by bacteria, and the study of genetic competence is a model pathway to explore intercellular communication, especially in Streptococcus. Genetic competence is required for obtaining extracellular DNA and also has a significant impact on the expression of virulence-related features, biofilm formation, and stress tolerance [Citation71]. Extensive research has identified two competence-activating signaling systems, the XIP/ComRS system and CSP/ComCDE the system [Citation72]. Streptococcus mutans, which containing both systems, has become an attractive model to study the two signaling systems. In S. mutans, transposon insertion mutated strains (i.e. comR, oppABCDF, comX, and irvR) have been defined to have great fitness in the mouse oral cavity by Tn-Seq [Citation49]. ComR and the OppABCDF are required for the activation of transcription of comX (sigX), which encodes the alternative sigma factor that controls late competence gene activation, and IrvR is an important regulator for genetic competence. Previous studies have shown that the virulence of S. pneumoniae has been attenuated in a ΔcomX mutant, for loss of induction of the allolytic genes cibAB and cbpD [Citation73]. But Orthologs of cibAB and cbpD are not present in the S. mutans. And the production of ComX has been demonstrated to lead to growth arrest and cell lysis of S. mutans [Citation74], which may account for the fitness enhancements of these mutants. In S. pyogenes, transposon mutagenesis screening identified the ABC transporter PptAB, which plays an important role in short hydrophobic peptide (SHP) pheromone output through the Rgg2/Rgg3 pathway. However, in S. mutans, removal of pptAB only partially disrupted XIP signaling suggesting PptAB is not key to the ComRS signaling pathway and the secretion of XIP may have a secondary secretion pathway [Citation75]. Shields, Robert C et al created a transposon insertion library containing the comX promoter in S. mutans, and novel genes associated with competence development have been identified by Tn-seq, and 20 genes have been identified and characterised in addition to known genes associated with ComX expression. These data also highlight DivIB may be the focus of future studies on the crosstalk between ComRS and ComCDE systems in S. mutans [Citation76]. On the other hand, Tn916 mutagenesis has shown that the sgc gene of S. gordonii inhibits the production of bacteriocin regulated by ComCDE system in S. mutans [Citation77].
Mutant strains in a mutant pool may interact to be complemented, thereby concealing their virulence defects and changing their fitness. Droplet Tn‐Seq has been developed to achieve independent growth by using microfluidic technologies to encapsulate each transposon mutant into a growth medium-in-oil droplet [Citation78]. Through defining single-cell fitness in a genome-wide by dTn-seq, it is possible to further explore interbacterial interactions and bacterial microcolony formation. And combining TIS with other high-throughput technologies, such as RNA-Seq and metabolomics, can reveal competition for environmental resources among microorganisms and important new pathways for microbial community interactions. The combination of Tn-Seq with RNA-Seq has been applied to identify genes that are important for the growth of E. faecium in human serum [Citation79] and explore the interaction between Escherichia coli and microorganisms in cheese environments [Citation80]. The microbial community structure and function depend on complex interactions that are both competitive and beneficial. The increasing complexity of the community leads to changes in the genetic requirements for microbial interactions. The online application ShinyOmics has been developed to allow rapid collaboration in the analysis and exploration of the massive accumulation of bio-omics data [Citation81]. Similar methods and analytical tools should be used to explore the complex and rich interactions among oral microorganisms.
Identifying genes involved in drug resistance
Transposon mutation can be utilized to study the drug sensitivity of different mutants, which helps better understand the development of bacterial resistance. Fluoride exerts significant anticaries effects by inhibiting demineralisation, enhancing remineralisation, and inhibiting bacterial growth, which play important roles in oral health [Citation82]. Streptococcus mutans is the major pathogen causing dental caries, and the widespread use of fluoride might lead to the emergence of bacteria that are resistant to fluoride [Citation83]. Transposon insertion mutants of S. mutans were constructed and a library was screened to identify and characterise genes associated with fluorine tolerance. The results showed that smu.1289c-90c overexpression combined with smu.396 deletion resulted in higher fluorine resistance in the smu.1290c-89c operon encoding fluorine antiporters [Citation84,Citation85]. Screening important genes of the S. mutans transposon mutant library for biofilm-related antibiotic resistance indicated that the dlt gene is associated with gentamicin resistance in S. mutans biofilms. The expression of dlt genes mediates the alanylation of teichoic acids, and the negative charge on the surface of dltA mutants is greater than that of the wild-type, which leads to reduced tolerance to positively charged gentamicin [Citation86].
Drug-bacterial interactions are not only limited to a drug and its direct target but also to drug-induced pressure that seems to resonate through bacteria, resulting in selective pressure. Transposon mutagenesis cannot directly assay drug targets such as DNA replication, cell wall synthesis, or protein synthesis. However, detecting the relative changes in the number of genes inserted into transposons during drug exposure helps to reveal this complex multifactorial process. Transposon mutagenesis has shown that the antimicrobial tolerance of S. mutans biofilms also depends on an oxidative stress response mediated by the SpxA1 protein, which functions as a transcription factor [Citation87]. Screening a transposon mutant library revealed that SMU.244 encodes a homologue of undecaprenyl pyrophosphate phosphatase, which plays important roles in bacitracin resistance and cell wall biosynthesis in S. mutans [Citation88].
Sensitivity profiles constructed using Tn-Seq have shown that the two strains of S. pneumoniae use several genes to resist stress triggered by daptomycin, including genes important for membrane integrity, protein conversion, and potassium uptake. The activity patterns of antibiotics have been partially uncovered by confirming numerous genotype-phenotype relationships, investigating temporal gene expression, and mapping genetic interactions [Citation89]. Viridans group streptococci are important normal bacteria in humans; they are most abundant in the oral cavity and are considered the causative pathogens of infective endocarditis, septicaemia, meningitis, and other serious infections. We predict that TIS will be used to reveal oral streptococcal genes and networks related to drug resistance and develop new therapies for targeting drug-resistant bacteria.
Next frontiers
Transposon mutagenesis and NGS have become the preferred methods for large-scale detection of genotype-phenotype interactions because of their high-throughput capability and sensitivity to small differences in fitness. The functions of most nonessential genetic components in organisms can be explored using this technique under various environmental conditions. TIS also works in conjunction with other modern technologies such as RNA-Seq. Since its development, TIS has been applied to studies of in vitro and in vivo models to explore the fitness of genes, shed new light on studied biological processes, and begin to understand how genotypes influence pathogenicity at the genome level.
Although transposon sequencing has some applicability, it still has some limitations. Traditional TIS is mainly used to study the functions of nonessential genes and identify essential genes. However, libraries with large numbers of mutants have a bottleneck effect; insertion mutants may be randomly lost during selective growth for reasons unrelated to fitness, especially in animal models. Liu et al. designed single-guide RNA (sgRNA) sequences targeting core genes identified by Tn-Seq and developed an IPTG-induced CRISPR interference (CRISPRi) system for functional studies of essential S. pneumoniae D39V genes in vitro [Citation90]. Bosch et al. developed a CRISPRi platform for S. thermophilus to provide a genome-level assessment of gene vulnerability, which links the degree of gene inhibition to its effect on fitness [Citation91]. Liu et al. recently developed a doxycycline-induced CRISPRi system and constructed a pooled CRISPRi library that targets almost all operons of S. pneumoniae D39V and can be easily combined with Illumina sequencing (CRISPRi-Seq) [Citation92]. By selecting a sgRNA for each operon, CRISPRi-Seq was used to assess bottlenecks and identify pneumococcal genes that are important in a murine pneumonia model. Genome-wide CRISPR screening can be used to systematically investigate gene functions. However, an sgRNA library is large, and its synthesis is expensive. Jiang et al. used the CRISPR-CAS adaptation mechanism of S. pyogenes to develop CRISPR adaptation-mediated library manufacturing, which transforms bacterial cells into ‘factories’ that generate hundreds of thousands of CRISPR RNAs, covering 95% of all targeted genomic sites [Citation93]. However, this method also produces numerous mutants resulting in bottlenecks. Moreover, when operons contain multiple essential genes, the CRISPRi system results in polarity effects that inhibit the expression of downstream genes. The insertion of transposons within operons in Tn-Seq, owing to the lack of transcriptional terminators, allows for read-through transcription and, thus, minimises polarity effects. With the development of TIS, the functions of essential genes can be studied using transposon libraries with outwards promoters that promote gene overexpression [Citation94]. Along with technical and analytical development, the bottleneck of saturated libraries between different conditions gradually decreases. In general, TIS is simple to operate and inexpensive and is still a powerful tool for high-throughput quantitative studies of microbial genotypes influencing their phenotypes. The CRISPRi system is indispensable for the functional study of essential genes in microorganisms.
Conclusions and perspectives
Transposon mutagenesis could lead to a better understanding of microbial interactions by providing a better annotation for more types of oral Streptococcus phenotypes. Studies on gene function should be further advanced using other basic experimental methods, such as biochemical studies and microscopy, to understand the specific mechanisms of gene function. Furthermore, TIS can also be combined with other methods, such as RNA sequencing, microfluidics, and CRISPRi, to explore microbial interactions in vitro and in vivo and discover new and important toxicity properties that will deepen our understanding of oral health homeostasis and disease dysregulation. As TIS technology has become more advanced and other types of data are combined for analysis, data analysis tools and visualisation have become increasingly important. We predict that transposon mutations and NGS technologies will continue to be developed for applications to address diverse and complex biological questions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Xu X, He J, Xue J, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol. 2015;17:699–13.
- Ajdić D, McShan WM, McLaughlin RE, et al. Genome sequence of streptococcus mutans ua159, a cariogenic dental pathogen. Proc Nat Acad Sci. 2002;99:14434–14439.
- Rudney JD, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171.
- Whiley RA, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216.
- Ni L, Li J, Shi J, et al. New classification of oral streptococcus. Chinese journal of conservative dentistry. 2002;12:507–509.
- Liu S, Zhu W, Zhi Q, et al. Analysis of sucrose-induced small RNAs in streptococcus mutans in the presence of different sucrose concentrations. Appl Microbiol Biotechnol. 2017;101:5739–5748.
- Bottner A, He RY, Sarbu A, et al. Streptococcus mutans isolated from children with severe-early childhood caries form higher levels of persisters. Arch Oral Biol. 2020;110:1–7.
- Kreth J, Merritt J, Shi W, et al. Competition and coexistence between streptococcus mutans and streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–7203.
- Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025.
- Park O-J, Kwon Y, Park C, et al. Streptococcus gordonii: pathogenesis and host response to its cell wall components. Microorganisms. 2020;8:1–22.
- de Paz LC, Svensäter G, Dahlén G, et al. Streptococci from root canals in teeth with apical periodontitis receiving endodontic treatment. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology. 2005;100:232–241.
- Mosailova N, Truong J, Dietrich T, et al. Streptococcus gordonii: a rare cause of infective endocarditis. Case Rep Infect Dis. 2019;2019:1–2.
- Kim SL, Gordon SM, Shrestha NK. Distribution of streptococcal groups causing infective endocarditis: a descriptive study. Diagn Microbiol Infect Dis. 2018;91:269–272.
- Yumoto H, Hirota K, Hirao K, et al. The pathogenic factors from oral streptococci for systemic diseases. Int J Mol Sci. 2019;20:1–18.
- Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144.
- Burton JP, Drummond BK, Chilcott CN, et al. Influence of the probiotic streptococcus salivarius strain m18 on indices of dental health in children: a randomized double-blind, placebo-controlled trial. J Med Microbiol. 2013;62:875–884.
- Choi KH, Kim KJ. Applications of transposon-based gene delivery system in bacteria. J Microbiol Biotechnol. 2009;19:217–228.
- Goodman AL, McNulty NP, Zhao Y, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289.
- Gawronski JD, Wong SM, Giannoukos G, et al. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for haemophilus genes required in the lung. Proceedings of the National Academy of Sciences 2009, 106, 16422–16427,
- Van Opijnen T, Bodi KL, Tn-seq: CA. High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772.
- Langridge GC, Phan M-D, Turner DJ, et al. Simultaneous assay of every salmonella typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316.
- Wei L, Li M, Xia F, et al. Phosphate transport system mediates the resistance of enterococcus faecalis to multidrug. Microbiol Res. 2021;249,:126772.
- Hubbard TP, Chao MC, Abel S, et al. Genetic analysis of vibrio parahaemolyticus intestinal colonization. Proceedings of the National Academy of Sciences of the USA 2016, 113, 6283–6288,
- Wang Y, Liu G, Zhang J, et al. Wbap is required for swarm motility and intramacrophage multiplication of salmonella enteritidis spic mutant by glucose use ability. Microbiol Res. 2021;126686. DOI:10.1016/j.micres.2020.126686
- Yin K, Zhang J, Ma J, et al. Mvin mediates the regulation of environmental osmotic pressure on esrb to control the virulence in the marine fish pathogen edwardsiella piscicida. Microbiol Res. 2020;126528. 10.1016/j.micres.2020.126528
- Su Y, Xu Y, Li Q, et al. The essential genome of ralstonia solanacearum. Microbiol Res. 2020;126500. 10.1016/j.micres.2020.126500
- Walterson AM, Smith DD, Stavrinides J. Identification of a pantoea biosynthetic cluster that directs the synthesis of an antimicrobial natural product. PloS one. 2014;9:e96208.
- Shields RC, Jensen PA. The bare necessities: uncovering essential and condition‐critical genes with transposon sequencing. Mol Oral Microbiol. 2019;34:39–50.
- Chao MC, Abel S, Davis BM, et al. The design and analysis of transposon insertion sequencing experiments. Nature Rev Microbiol. 2016;14:119–128.
- Cain AK, Barquist L, Goodman AL, et al. A decade of advances in transposon-insertion sequencing. Nat Rev Genet. 2020;21:526–540.
- Shapiro JA. Molecular model for the transposition and replication of bacteriophage mu and other transposable elements. Proc Natl Acad Sci U S A. 1933–1937;1979(76). 10.1073/pnas.76.4.1933
- Spatafora G, Rohrer K, Barnard D, et al. A streptococcus mutans mutant that synthesizes elevated levels of intracellular polysaccharide is hypercariogenic in vivo. Infect Immun. 1995;63:2556–2563.
- Gutierrez JA, Crowley PJ, Brown DP, et al. Insertional mutagenesis and recovery of interrupted genes of streptococcus mutans by using transposon tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175.
- Boyd DA, Cvitkovitch DG, Bleiweis AS, et al. Defects in d-alanyl-lipoteichoic acid synthesis in streptococcus mutans results in acid sensitivity. J Bacteriol. 2000;182:6055–6065.
- Roberts AP, Mullany P. A modular master on the move: the tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258.
- Slater JD, Allen AG, May JP, et al. Mutagenesis of streptococcus equi and streptococcus suis by transposon tn917. Vet Microbiol. 2003;93:197–206.
- Thibessard A, Fernandez A, Gintz B, et al. Transposition of pgh9: is s1 is random and efficient in streptococcus thermophilus cnrz368. Can J Microbiol. 2002;48:473–478.
- Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the himar1 mariner transposon in vitro. Genetics. 1998;149:179–187.
- Green B, Bouchier C, Fairhead C, et al. Insertion site preference of mu, tn5, and tn7 transposons. Mob DNA. 2012;3:1–6.
- van Opijnen T, Lazinski DW, Camilli A. Genome‐wide fitness and genetic interactions determined by tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Curr Protoc Mol Biol. 2014;106:7–16.
- Kwon YM, Ricke SC, Mandal RK. Transposon sequencing: methods and expanding applications. Appl Microbiol Biotechnol. 2016;100:31–43.
- van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nature Rev Microbiol. 2013;11:435–442.
- Lampe DJ, Churchill M, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479.
- Nilsson M, Givskov M, Tolker-Nielsen T. Transposon mutagenesis in streptococcus species. In: Ricke SC, Park SH, Davis ML editors. Microbial transposon mutagenesis. New Jersey: Humana Press; 2019. 39–49.
- Hooven TA, Catomeris AJ, Akabas LH, et al. The essential genome of streptococcus agalactiae. BMC Genomics. 2016;17:1–12.
- Hutchison III, Peterson CA, Gill SN, et al. Global transposon mutagenesis and a minimal mycoplasma genome. Science. 1999;286:2165–2169.
- Le Breton Y, Belew AT, Valdes KM, et al. Essential genes in the core genome of the human pathogen streptococcus pyogenes. Sci Rep. 2015;5:1–13.
- Xu P, Ge X, Chen L, et al. Genome-wide essential gene identification in streptococcus sanguinis. Sci Rep. 2011;1:1–9.
- Shields RC, Zeng L, Culp DJ, et al. Genomewide identification of essential genes and fitness determinants of streptococcus mutans ua159. Msphere. 2018;3:1–14.
- Cornejo OE, Lefébure T, Bitar PD, et al. Evolutionary and population genomics of the cavity causing bacteria streptococcus mutans. Mol Biol Evol. 2013;30:881–893.
- Jr Q, Grayhack RG, Faustoferri EJ, et al. Functional profiling in streptococcus mutans: construction and examination of a genomic collection of gene deletion mutants. Mol Oral Microbiol. 2015;30:474–495.
- Verhagen LM, de Jonge MI, Burghout P, et al. Genome-wide identification of genes essential for the survival of streptococcus pneumoniae in human saliva. PloS One. 2014;9:1119–1129.
- Gaspar P, Al-Bayati FA, Andrew PW, et al. Lactate dehydrogenase is the key enzyme for pneumococcal pyruvate metabolism and pneumococcal survival in blood. Infect Immun. 2014;82:5099–5109.
- Hillman JD, Chen A, Duncan M, et al. Evidence that l-(+)-lactate dehydrogenase deficiency is lethal in streptococcus mutans. Infect Immun. 1994;62:60–64.
- Nobbs AH, Jenkinson HF, Everett DB. Generic determinants of streptococcus colonization and infection. Infect Genet Evol. 2015;33:361–370.
- Krzyściak W, Jurczak A, Kościelniak D, et al. The virulence of streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33:499–515.
- Idone V, Brendtro S, Gillespie R, et al. Effect of an orphan response regulator on streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect Immun. 2003;71:4351–4360.
- Harris GS, Michalek SM. Curtiss, r.R. Cloning of a locus involved in streptococcus mutans intracellular polysaccharide accumulation and virulence testing of an intracellular polysaccharide-deficient mutant. Infect Immun. 1992;60:3175–3185.
- Caufield PW, Shah GR, Hollingshead SK. Use of transposon tn916 to inactivate and isolate a mutacin-associated gene from streptococcus mutans. Infect Immun. 1990;58:4126–4135.
- Król JE, Biswas S, King C. Biswas, I. Smu. 746-smu. 747, a putative membrane permease complex, is involved in aciduricity, acidogenesis, and biofilm formation in streptococcus mutans. J Bacteriol. 2014;196:129–139.
- Gutierrez JA, Crowley PJ, Cvitkovitch DG, et al. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kda subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology. 1999;145:357–366.
- Paik S, Senty L, Das S, et al. Identification of virulence determinants for endocarditis in streptococcus sanguinis by signature-tagged mutagenesis. Infect Immun. 2005;73:6064–6074.
- Zhu B, Green SP, Ge X, et al. Genome‐wide identification of streptococcus sanguinis fitness genes in human serum and discovery of potential selective drug targets. Mol Microbiol. 2021;115:658–671.
- Zhu L, Charbonneau AR, Waller AS, et al. Novel genes required for the fitness of streptococcus pyogenes in human saliva. Msphere. 2017;2:1–34.
- Edgar RJ, van Hensbergen VP, Ruda A, et al. Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides. Nat Chem Biol. 2019;15:463–471.
- Abel S, Abel Zur W, Davis P, et al. M.K. Analysis of bottlenecks in experimental models of infection. PLoS Pathog. 2015;11:1–7.
- Chen L, Ge X, Dou Y, et al. Identification of hydrogen peroxide production-related genes in streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology. 2011;157:13–20.
- Zheng L, Chen Z, Itzek A, et al. Catabolite control protein a controls hydrogen peroxide production and cell death in streptococcus sanguinis. J Bacteriol. 2011;193:516–526.
- Redanz S, Treerat P, Mu R, et al. Pyruvate secretion by oral streptococci modulates hydrogen peroxide dependent antagonism. ISME J. 2020;14:1074–1088.
- Selleck EM, Gilmore MS. Oxygen as a virulence determinant in polymicrobial infections. Mbio. 2016;7:1–3.
- Håvarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in streptococcus pneumoniae. Proceedings of the National Academy of Sciences 1995, 92, 11140–11144,
- Son M, Ahn SJ, Guo Q, et al. Microfluidic study of competence regulation in streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comx. Mol Microbiol. 2012;86:258–272.
- Zhu L, Lin J, Kuang Z, et al. Deletion analysis of streptococcus pneumoniae late competence genes distinguishes virulence determinants that are dependent or independent of competence induction. Mol Microbiol. 2015;97:151–165.
- Wenderska IB, Lukenda N, Cordova M, et al. A novel function for the competence inducing peptide, xip, as a cell death effector of streptococcus mutans. FEMS Microbiol Lett. 2012;336:104–112.
- Chang JC, Federle MJ. Pptab exports rgg quorum-sensing peptides in streptococcus. PLoS One. 2016;11:1–12.
- Shields RC, O’Brien G, Maricic N, et al. Genome-wide screens reveal new gene products that influence genetic competence in streptococcus mutans. J Bacteriol. 2017;200:1–16.
- Wang BY, Kuramitsu HK. Interactions between oral bacteria: inhibition of streptococcus mutans bacteriocin production by streptococcus gordonii. Appl Environ Microbiol. 2005;71:354–362.
- Thibault D, Jensen PA, Wood S, et al. Droplet tn-seq combines microfluidics with tn-seq for identifying complex single-cell phenotypes. Nat Commun. 2019;10:1–13.
- Zhang X, de Maat V, Prieto AMG, et al. Rna-seq and tn-seq reveal fitness determinants of vancomycin-resistant enterococcus faecium during growth in human serum. BMC Genomics. 2017;18:1–12.
- Morin M, Pierce EC, Dutton RJ. Changes in the genetic requirements for microbial interactions with increasing community complexity. Elife. 2018;7:1–43.
- Surujon D, van Opijnen T. Shinyomics: collaborative exploration of omics-data. BMC Bioinformatics. 2020;21:1–8.
- Buzalaf MAR, Pessan JP, Honório HM, et al. Mechanisms of action of fluoride for caries control. Fluoride and the Oral Environment. 2011;22:97–114.
- Men X, Shibata Y, Takeshita T, et al. Identification of anion channels responsible for fluoride resistance in oral streptococci. PloS One. 2016;11:1–12.
- Yu J, Wang Y, Han D, et al. Identification of streptococcus mutans genes involved in fluoride resistance by screening of a transposon mutant library. Mol Oral Microbiol. 2020;35:260–270.
- Liao Y, Yang J, Brandt BW, et al. Genetic loci associated with fluoride resistance in streptococcus mutans. Front Microbiol. 2018;9:1–9.
- Nilsson M, Rybtke M, Givskov M, et al. The dlt genes play a role in antimicrobial tolerance of streptococcus mutans biofilms. Int J Antimicrob Agents. 2016;48:298–304.
- Nilsson M, Jakobsen TH, Givskov M, et al. Oxidative stress response plays a role in antibiotic tolerance of streptococcus mutans biofilms. Microbiology. 2019;165:334–342.
- Jalal N, Tian X-L, Dong G, et al. Identification and characterization of smu. 244 encoding a putative undecaprenyl pyrophosphate phosphatase protein required for cell wall biosynthesis and bacitracin resistance in streptococcus mutans. Microbiology. 1857-1870;2015(161). 10.1099/mic.0.000142
- van Opijnen T, Dedrick S, Bento J. Strain dependent genetic networks for antibiotic-sensitivity in a bacterial pathogen with a large pan-genome. PLoS Pathog. 2016;12:1–29.
- Liu X, Gallay C, Kjos M, et al. High‐throughput crispri phenotyping identifies new essential genes in streptococcus pneumoniae. Mol Syst Biol. 2017;13:1–18.
- Bosch B, DeJesus MA, Poulton NC, et al. Genome-wide gene expression tuning reveals diverse vulnerabilities of m. Tuberculosis. Cell. 2021;184:4579–4592.
- Liu X, Kimmey JM, Matarazzo L, et al. Exploration of bacterial bottlenecks and streptococcus pneumoniae pathogenesis by crispri-seq. Cell Host Microbe. 2021;29:107–120.
- Jiang W, Oikonomou P, Tavazoie S. Comprehensive genome-wide perturbations via crispr adaptation reveal complex genetics of antibiotic sensitivity. Cell. 2020;180:1002–1017.
- Coe KA, Lee W, Stone MC, et al. Multi-strain tn-seq reveals common daptomycin resistance determinants in staphylococcus aureus. PLoS Pathog. 2019;15:1–25.
