ABSTRACT
Introduction
Gingivitis is a prevalent complication in adolescents undergoing fixed orthodontic treatments. However, changes in the supragingival microbiome associated with gingivitis and the impact of Candida albicans remain elusive. Therefore, we investigated supragingival microbiome discrepancy and C. albicans colonization in adolescent orthodontic patients with gingivitis.
Methods
Dental plaques were collected from 30 gingivitis patients and 24 healthy adolescents, all undergoing fixed orthodontic treatment. The supragingival microbiome composition was analyzed using 16S rRNA sequencing. C. albicans colonization was determined using fungal culture and real-time quantitative polymerase chain reaction.
Results
Our analysis revealed significantly heightened microbial diversity in the Gingivitis group. Notably, patients with gingivitis exhibited an enrichment of periodontal pathogens, such as Saccharibacteria (TM7) [G-1], Selenomonas, Actinomyces dentalis, and Selenomonas sputigena. Additionally, 33% of the gingivitis patients tested positive for C. albicans, exhibiting significantly elevated levels of absolute abundance, while all healthy patients tested negative. Significant differences in microbial composition were also noted between C. albicans-positive and -negative samples in the Gingivitis group.
Conclusion
Significant disparities were observed in the supragingival microbiome of adolescent orthodontic patients with and without gingivitis. The presence of C. albicans in the supragingival plaque may alter the microbiome composition and potentially contribute to gingivitis pathogenesis.
KEY MESSAGES
• Adolescent patients undergoing fixed orthodontic treatment, with and without gingivitis, show significant differences in their marginal supragingival plaque microbiomes.
• Adolescent patients with gingivitis exhibit a significantly higher rate of Candida albicans colonization than healthy individuals.
• The colonization of C. albicans alters the composition of the marginal supragingival plaque microbiome in patients with gingivitis.
Introduction
Marginal gingivitis is a common side effect of orthodontic treatment and a major biofilm-related disease that affects treatment effectiveness [Citation1]. Compared to adult patients, adolescents are less diligent in maintaining oral hygiene, leading to a higher incidence of marginal gingivitis [Citation2]. Orthodontics-related gingivitis affects up to 56.8% of adolescents [Citation3]. When the periodontal tissue is in an inflammatory state, the formation and activity of osteoclasts increase during tooth movement, which can worsen alveolar bone loss, posing risks to periodontal tissue and dental health [Citation4].
Dental plaque and its byproducts that accumulate at the gingival margin are key factors in developing marginal gingivitis. Consequently, there is a growing focus on exploring changes in dental plaque associated with marginal gingivitis during fixed orthodontic treatment. Previous studies using bacterial culture techniques have shown a substantial increase in Gram-negative bacteria in the dental plaque of patients with orthodontic gingivitis [Citation5]. Recent advancements in high-throughput techniques have enabled the exploration of changes in the gingival microbiome during orthodontic treatment [Citation6–8]. Previous research on gingival inflammation during orthodontic treatment primarily examined subgingival plaque or saliva. Studies have shown that orthodontic treatment may increase plaque adhesion and alter the subgingival microbiome [Citation9], leading to mild gingival inflammation [Citation10]. It has been confirmed that microbial diversity in subgingival plaque typically increases in patients with fixed orthodontic treatment [Citation11]. Specifically, Tannerella forsythia and Prevotella intermedia have been found to increase significantly in subgingival plaque, indicating an increased risk of periodontal infection during orthodontic treatment [Citation12]. In saliva samples, the microbial compositions of orthodontic patients differed considerably from those of healthy individuals, with orthodontic patients exhibiting higher microbial diversity [Citation7]. While most studies have focused on subgingival plaque or saliva, a few have examined changes in supragingival marginal plaque during orthodontic treatment [Citation13]. These studies found an increase in anaerobic bacteria; however, participants did not exhibit typical symptoms of gingivitis [Citation13]. Research on microbiome changes in orthodontic patients with typical gingivitis, particularly among adolescents, who represent a significant portion of orthodontic patients, remains limited.
In addition to bacteria, fungi are important constituents of the oral microbiome and can contribute to the pathogenesis of various diseases [Citation14–16]. In particular, Candida is closely linked to oral diseases. There are over 150 Candida species, with approximately 20 identified as human pathogens that can interact with bacteria to cause disease [Citation17]. In the context of periodontal disease, research on Candida albicans has mainly focused on patients with periodontitis, and a notable positive correlation between C. albicans and the development of periodontal disease has been reported [Citation18]. C. albicans has been found in the subgingival plaques of approximately 29.8% of individuals with chronic periodontitis [Citation19]. Furthermore, observations have indicated that C. albicans hyphae can infiltrate the periodontal connective tissue [Citation20], interact with subgingival bacterial pathogens, or induce proinflammatory cytokine production, thereby leading to the loss of periodontal attachment and aggravating periodontal disease [Citation18]. C. albicans can also create an anoxic microenvironment in biofilms, supporting the growth of anaerobic bacteria [Citation21]. For example, C. albicans biofilm can protect the anaerobic bacterium Porphyromonas gingivalis from the aerobic environment [Citation21]. However, most studies have primarily investigated the presence of C. albicans in subgingival plaques of individuals diagnosed with chronic periodontitis; however, only a few studies have detected C. albicans in supragingival plaques of adult patients with gingivitis [Citation22]. Moreover, the enrichment of C. albicans in adolescent patients with marginal gingivitis during orthodontic treatment and its potential relationship with bacterial abundance require further exploration.
Therefore, we investigated whether the composition of the supragingival microbiome of healthy adolescent patients undergoing orthodontic treatment differs from that of patients with gingivitis through 16S rRNA sequencing. Additionally, we utilized fungal culture, rDNA internal transcribed spacer identification (ITS) and real-time quantitative polymerase chain reaction (qPCR) to investigate whether C. albicans enrichment occurs in gingivitis and to explore the impact of C. albicans aggregation on bacterial composition, thereby providing further evidence concerning the physiological and ecological significance of the supragingival plaque microbiome in the development of orthodontic gingivitis. The null hypotheses for this research were as follows: (1) There is no statistically significant difference observed in the supragingival microbial composition between orthodontic patients with gingivitis and healthy individuals; (2) C. albicans is not enriched in adolescents with gingivitis; and (3) C. albicans enrichment does not affect the supragingival plaque microbiome composition.
Materials and methods
Recruitment of patients
This study obtained ethical approval from the Ethics Committee of the Beijing Stomatological Hospital. All participants included in this study obtained the consent of the patients and their parents and signed informed consent for the study. Adolescents aged 11–18 years undergoing fixed orthodontic treatment were enrolled from Beijing Stomatological Hospital, involving 30 patients with gingivitis (the Gingivitis group) and 24 patients without gingivitis as the control group (the Periodontal healthy group). In the Gingivitis group, participants had gingivitis in both anterior teeth and premolars with an average gingival index (GI) ≥ 1, attachment loss ≤1 mm, and DMFS index < 10 [Citation23,Citation24]. The inclusion criteria for the Periodontal healthy group were periodontal health, GI < 0.5, PD ≤3 mm, DMFS index < 10, and no periodontal attachment loss.
Dental plaque collection
Participants were instructed to abstain from oral hygiene activities for at least 12 h before sample collection and to avoid eating or drinking for 2 h prior to sampling. Using a periodontal curette, supragingival plaque along the gingival margin of the anterior teeth and premolars was collected.
The plaque samples were then placed into 1 mL of sterilized TE buffer and transported to the laboratory on ice. The collected plaque was divided into two parts. One aliquot underwent bacterial 16S rRNA sequencing analysis and quantification of C. albicans, while the other aliquot was preserved in 1× TE buffer (containing 20% glycerol) for subsequent Candida culture identification and ITS sequencing analysis. All samples were stored at −80°C for future use.
Genomic DNA extraction, illumina MiSeq sequencing, and processing
Genomic DNA was extracted from 54 samples using the FastDNA Spin Kit (MP Biomedicals, USA) following the manufacturer’s instructions. The quality and concentration of DNA samples were assessed using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The V1-V3 region of 16S rRNA fragments was amplified with primers 27F and 533 R, which were 5’ -AGAGTTTGATCCTGGCTCAG-3’ and 5’- TTACCGCGGCTGCTGGCAC-3’ respectively. The PCR protocol comprised an initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturation at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final extension step at 72°C for 10 min. The amplicons were paired-end sequenced using the Illumina MiSeq platform with PE300. The raw sequencing reads were deposited in the NCBI Sequence Read Archive database under the accession number SRP483771.
After demultiplexing, we used FLASH (v1.2.7) to merge the obtained sequences [Citation25] and Fastp (v0.19.6) for quality filtering [Citation26]. The parameters were set as follows: (1) Trim low-quality bases from the end of reads with a minimum quality threshold of 20; use sliding window trimming with a 50 bp window; discard reads under 50 bp in length or containing N bases. (2) Pairwise reads with overlapping PE reads were merged into a sequence, and the minimum overlap length was 10 bp. (3) The maximum mismatch ratio in the overlap region of the splicing sequence was 0.2. (4) Samples were identified by barcode and primer sequences at both ends, allowing zero mismatches for barcodes and up to two for primers. Using the DADA2 [Citation27] plug-in within the Qiime2 pipeline, sequences were denoised under the following conditions [Citation28]: discard reads with a length less than or equal to zero; remove sequences with a total abundance below ten across all samples or less than two in any single sample. The MaxEE was set at two, and the truncQ at zero. Post-denoising, amplicon sequence variants were identified and assigned taxonomic classifications using the Qiime2 naïve Bayes consensus taxonomic classifier and the Human Oral Microbiome Database (HOMD) (v15.2) [Citation29].
Culture, identification, and quantification of C. albicans
Dental plaque samples and C. albicans (SC5314) used to establish standard curves were inoculated onto CHROMagar selective culture medium (Becton Dickinson & Co., USA) and incubated at 37°C for 72 h [Citation30]. C. albicans-positive colonies, which appear as green colonies on CHROMagar Candida-selective medium [Citation31], were then collected using a sterile inoculation ring and transferred to 2 mL sterile centrifuge tubes for subsequent DNA extraction and analysis of the rDNA ITS sequences [Citation31].
The total genomic DNA was extracted using Epicenter MasterPure DNA extraction kits (Lucigen Corporation, USA), following the manufacturer’s instructions. The total volume of the PCR amplification mixture was 50 μL, including enzyme-free water (17 μL), 2 × Taq PCR Mastermix (25 μL) (KT201, Tiangen Biotechnologies, China), template DNA (4 μL), and ITS primers (2 μL). The amplification of the extracted DNA was performed using primers ITS4 and ITS5, which were 5’ - TCCTCCGCTTATTGATATGC − 3’ and 5’ - GGAAGTAAAAGTCGTAACAAGG − 3’ respectively. The sample denaturation was conducted at 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s, and a final extension step at 72°C for 10 min [Citation32].
The amplified products were sequenced by the Beijing Genomics Institution to identify fungal strains. The PCR products were initially detected on a 1.0% agarose gel using 3 μL of the PCR products and subsequently purified following the standard operating procedure for magnetic bead purification. This process leverages the principle that magnetic beads can absorb or release charged substances – DNA is adsorbed in a high-salt, low-pH solution, facilitating DNA separation and purification. The purified PCR products were sequenced using an ABI 3730 sequencer (Applied Biosystems, Inc, USA). BioEdit and Cexpress software were used for sequence assembly and correction. The obtained sequences were then subjected to comparative analysis against homologous sequences available using the BLAST software in the GenBank database. Strains showing high sequence similarity were selected for further study. The raw sequencing reads have been deposited in the NCBI GenBank database under the accession numbers PP563750-PP563769.
To quantify the abundance of C. albicans in the samples, we performed a real-time quantitative polymerase chain reaction. We extracted and purified the total genomic DNA from the C. albicans standard strain and another aliquot of the supragingival plaque, as described in section 2.3. We used C. albicans-specific primers CALB1 (5’ - TTTATCAACTTGTCACACCAGA − 3’) and CALB2 (5’ - ATCCCGCCTTACCACTACCG − 3’) for the qPCR reactions, which were conducted on a Bio-Rad CFX Connect (BioRad, Hercules, CA, USA). The protocol included denaturation at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. We determined the quantification of C. albicans in each sample by converting DNA concentrations to colony-forming units (CFU/mL) using a standard curve.
Statistical analysis
Four diversity indices – ACE, Chao1, Simpson, and Shannon – were selected to evaluate the alpha diversity in this study. Beta diversity was analyzed through principal coordinate analysis (PCoA) using the Bray-Curtis distance matrix, and the ADONIS test was applied to assess statistical distinctions. Both alpha and beta diversity analyses were performed on the Majorbio Cloud Platform. The Wilcoxon rank-sum test was used to assess differences in taxonomic composition between groups, whereas the Kruskal-Wallis test was used to compare the absolute abundance of C. albicans across different groups. A two-tailed p value of < 0.05 was considered statistically significant.
Results
Significant differences in the marginal supragingival plaque microbiome between the gingivitis and periodontal healthy groups
This study compared the differences in supragingival microbiota between adolescents undergoing fixed orthodontic treatment with typical gingivitis and those with periodontally healthy status (). All patients in the Gingivitis group met the diagnostic criteria for moderate gingivitis, as indicated by GI values ranging from 1.1 to 1.9 [Citation23]. Sequencing analysis yielded 4,335,107 raw reads from the 54 supragingival plaque samples. Subsequent quality filtration yielded 4,083,635 optimized sequences, with an average length of 482 base pairs. Denoising the optimized sequences using the DADA2 plug-in in the Qiime2 pipeline yielded 1,148,723 sequences. Finally, 12 phyla, 29 classes, 48 orders, 79 families, 144 genera, and 482 species were detected through high-throughput sequencing. The alpha diversity of the supragingival microbiome of the two groups was analyzed to evaluate differences in species richness and diversity (). The Shannon index was significantly higher in the Gingivitis group than in the Periodontal healthy group (p < 0.05), indicating a significantly higher diversity of the periodontal microbiome in the Gingivitis group. This result corresponded with the previous findings, showing that inflamed periodontal tissue is associated with increased microbial diversity and a more complex microbiome community structure [Citation10]. The PCoA of the supragingival microbiome composition indicated that the beta diversity of the Gingivitis and Periodontal healthy groups was significantly different from each other (p < 0.05) ().
Figure 1. Comparison of alpha and beta diversities of microbial communities between the Gingivitis group and Periodontal healthy group. (a) Four indices, ACE, Chao1, Simpson, and Shannon, were selected to compare the alpha diversity (*p < 0.05). (b) Principal Coordinate Analysis (PCoA) was used to analyze beta diversity, and the ADONIS test was applied to evaluate statistical distinctions.
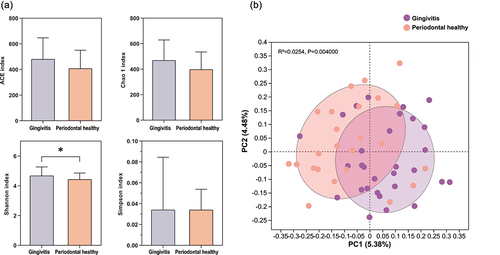
Table 1. Mean (standard deviation) of basic information and clinical indicators.
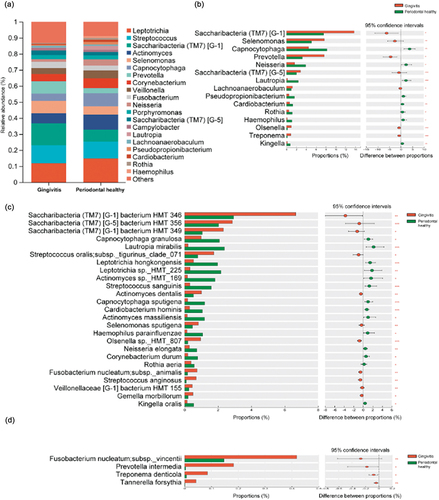
We further explored the differences in bacterial abundance in the supragingival microbiome between the two groups by identifying the core microbiota with a relative abundance of more than 1.0% at the genus level. The results showed that the core genus compositions of the two groups were similar. As shown in , the ten dominant genera included Leptotrichia, Streptococcus, Saccharibacteria (TM7) [G-1], Actinomyces, Selenomonas, Capnocytophaga, Prevotella, Corynebacterium, Veillonella, and Fusobacterium. The Gingivitis group exhibited a high relative abundance of Saccharibacteria (TM7) [G-1], Selenomonas, and Prevotella; however, Capnocytophaga, Neisseria, Lautropia, and Rothia exhibited significant enrichment in the Periodontal healthy group (p < 0.05) (). Several TM7 species like Saccharibacteria (TM7) [G-1] bacterium HMT 346 and Saccharibacteria (TM7) [G-5] bacterium HMT 356; bacteria of the Streptococcus genus, such as Streptococcus oralis subsp._tigurinus_clade_071, and Streptococcus anginosus; as well as bacteria of other genera, such as Actinomyces dentalis and Selenomonas sputigena, also exhibited significant enrichment in the Gingivitis group at the species level (p < 0.05) (). Furthermore, dominant bacteria found in gingivitis patients reported by previous studies, such as P. intermedia and Fusobacterium nucleatum, and bacteria typically detected in chronic periodontitis, such as T. forsythia and Treponema denticola, were also enriched in the Gingivitis group (). However, Capnocytophaga granulosa, Lautropia mirabilis, Streptococcus sanguinis, Actinomyces sp. HMT_169, Capnocytophaga sputigena, Cardiobacterium hominis, Actinomyces massiliensis, Neisseria elongata, Rothia aeria, and Corynebacterium durum were significantly enriched in the Periodontal healthy group (p < 0.05) ().
Figure 2. Species composition analysis between Gingivitis group and Periodontal healthy group. (a) Microbial composition at the genus level between the Gingivitis group and Periodontal healthy group. (b) The genera with significant differences between the Gingivitis group and the Periodontal healthy group (taking the top 1–15 bacteria ranged by relative abundance). (c) The species with significant differences between the Gingivitis group and the Periodontal healthy group (taking the top 1–25 bacteria ranged by relative abundance). (d) Periodontal pathogens with low relative abundance but significant differences between the Gingivitis group and Periodontal healthy group (*p < 0.05, *p < 0.01, **p < 0.001).
The gingivitis group exhibits a high C. albicans colonization rate and load
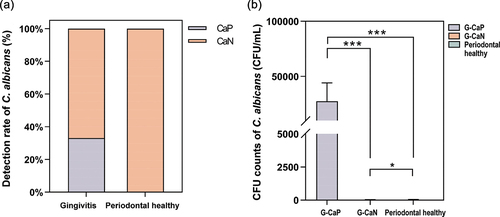
C. albicans colonization was initially identified by inoculating the plaque sample on CHROMagar Candida-selective culture medium, and positive colonies were confirmed through ITS sequencing. No C. albicans-positive colonies emerged on CHROMagar Candida-selective medium inoculated with samples from the Periodontal healthy group; however, ten out of the 30 samples from the Gingivitis group yielded green colonies. ITS sequencing results confirmed that all ten Candida-positive samples in the Gingivitis group were C. albicans, indicating a colonization rate of up to 33% in the Gingivitis group (). The detection rate of C. albicans was higher in the Gingivitis group than in the Periodontal healthy group, suggesting that C. albicans colonization is more likely in patients with gingivitis than in healthy patients. Based on the detection rates of C. albicans, we classified the Gingivitis group into two subgroups: Gingivitis-C. albicans positive (G-CaP, n = 10) and Gingivitis-C. albicans negative (G-CaN, n = 20). As shown in , the abundance of C. albicans in the G-CaP group was significantly higher than that in the G-CaN group and Periodontal healthy groups (p < 0.01).
Figure 3. (a) the difference of Candida albicans detection rate between Gingivitis group and Periodontal healthy group. CaP: C. albicans-positive; CaN: C. albicans-negative. (b) The CFU counts of C. albicans in G-CaP, G-CaN, and Periodontal healthy groups (NS: no significance, *p < 0.05, *p < 0.01, **p < 0.001).
C. albicans influences the supragingival microbiome composition in patients with gingivitis
To understand the effect of C. albicans on the supragingival microbiome of gingivitis plaque, we further performed the taxonomic analysis between the G-CaP (n = 10) and G-CaN (n = 20) groups. No statistical differences were observed in alpha diversity between the two subgroups (p > 0.05) (); however, the PCoA of the supragingival microbiome composition showed that the beta diversity of the two groups was significantly different from each other (p < 0.05), indicating that the microbial community composition of the G-CaP group was significantly different from that of G-CaN samples (). Further investigation of the differences in bacterial abundance in the supragingival microbiome of the G-CaP and G-CaN groups indicated that the core genera of the two groups were similar; however, the G-CaP group exhibited a greater abundance of Leptotrichia, Selenomonas, and Prevotella than the G-CaN group (). Furthermore, the difference in taxonomic composition showed that Campylobacter was more abundant in the G-CaP group, whereas Saccharibacteria (TM7) [G-5] and Treponema were more abundant in the G-CaN group (p < 0.05) ().
Figure 4. Comparison of alpha and beta diversities of microbial communities between the G-CaP and G-CaN group. (a) Four indices, ACE, Chao1, Simpson, and Shannon, were selected to compare the alpha diversity (*p < 0.05). (b) Principal Coordinate Analysis (PCoA) was used to analyze beta diversity, and the ADONIS test was applied to evaluate statistical distinctions.
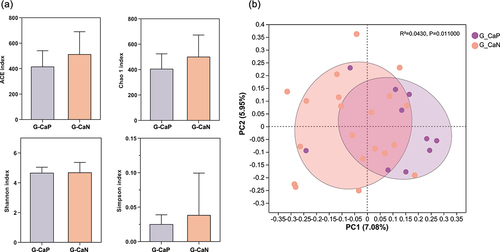
Figure 5. Species composition analysis between G-CaP and G-CaN group. (a) Microbial composition at the genus level between the G-CaP and G-CaN group. (b) The genera with significant differences between the G-CaP and G-CaN group (taking the top 1–15 bacteria ranged by relative abundance). (c) The species with significant differences between the G-CaP and G-CaN group (taking the top 1–25 bacteria ranged by relative abundance). (d) Periodontal pathogens with low relative abundance but significant differences between the G-CaP and G-CaN group (*p < 0.05, *p < 0.01, **p < 0.001).
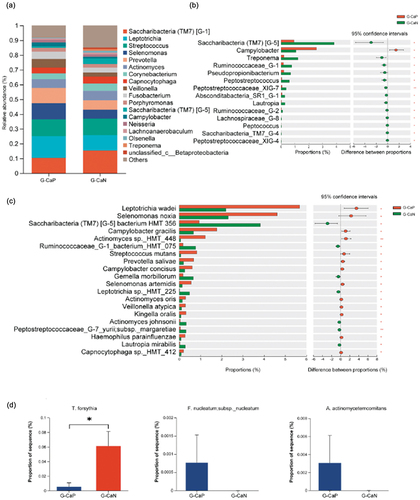
Species-level bacteria related to periodontal diseases, such as S. noxia, Campylobacter gracilis, Actinomyces sp._HMT_448, Campylobacter concisus, Prevotella salivae, Actinomyces oris, and Veillonella atypica, were enriched in the G-CaP group (p < 0.05) (). However, the abundance of TM7 species HMT-356, Gemella morbillorum, Leptotrichia sp._HMT_225, and L. mirabilis were significantly increased in the G-CaN group (p < 0.05). Additionally, although their abundance was relatively low, some periodontal pathogens are more commonly detected in subgingival plaque. For example, F. nucleatum subsp. nucleatum and Aggregatibacter actinomycetemcomitans were only detected in the G-CaP group ().
Discussion
Gingivitis is a common complication of orthodontic treatment that affects the periodontal tissue. In order to clarify the role of the oral microbiome in the pathogenesis of adolescent gingivitis, we investigated the disparities in the oral microbiome between adolescent orthodontic patients with and without gingivitis, as well as the impact of C. albicans for the first time. Our results showed significant differences in the supragingival microbiome between adolescents with gingivitis and those with a healthy status. Furthermore, all three invalid hypotheses were rejected.
During orthodontic treatment, the accumulation of dental plaque near the gingival margin and direct stimulation by orthodontic appliances can lead to gingivitis. Dental plaque and its byproducts at the gingival margin are pivotal in developing marginal gingivitis. Fixed orthodontic appliances, such as brackets and bands, complicate oral hygiene and aggravate plaque buildup [Citation33]. Additionally, the metal components in these appliances, such as nickel alloys, can be toxic to oral bacteria and may prompt gingival overgrowth by increasing epithelial cell proliferation due to continuous low-dose nickel release [Citation34]. Furthermore, nickel ions exert toxic effects on oral bacteria, contrasting with other metal like iron, which are essential for the metabolism of bacteria such as P. intermedia [Citation35,Citation36]. Therefore, beyond promoting plaque accumulation, orthodontic appliances may also disturb the ecological balance of the supragingival microbiome. Furthermore, orthodontic forces can elevate matrix metalloproteinase 8 levels, potentially contributing to gingival hyperplasia in orthodontic patients [Citation37]. Additionally, during certain movements like intrusion or tipping, plaque can migrate into subgingival areas, intensifying periodontal inflammation [Citation12]. Therefore, given that dental plaque is the primary factor in developing marginal gingivitis, gingivitis associated with orthodontic treatment exhibits distinct characteristics compared to gingivitis without orthodontic intervention. This specificity is also confirmed by the differences in microorganisms found in orthodontic versus non-orthodontic gingivitis. Research has shown significant variations in the periodontal pathogens present in subgingival plaque between patients with fixed orthodontic appliances and those without. Notably, the presence of T. forsythia, T. denticola, and P. nigrescens is markedly elevated in orthodontic patients, suggesting that changes associated with fixed orthodontic appliances may influence the prevalence of these periodontal pathogens in subgingival plaques [Citation38]. Another study revealed an 85% detection rate of A. actinomycetemcomitans in the subgingival plaque of adolescents undergoing orthodontic treatment, which significantly exceeded the 15% in adolescents without such treatment. Additionally, these patients exhibited a heightened gingival bleeding index [Citation39]. Moreover, in adolescents undergoing orthodontic treatment, factors such as poor oral hygiene, and hormonal changes contribute to a more pronounced response to dental plaque and an increased risk of periodontal hyperplasia. Studies indicate that adolescents with gingivitis are more prone to gingival bleeding and may experience attachment loss in adulthood compared to periodontal healthy adolescents [Citation40].
High-throughput sequencing suggested that patients with gingivitis exhibited significantly greater alpha diversity in their oral microbiome than healthy individuals, along with distinct clustering characteristics. These findings correspond to previous studies showing that periodontitis patients have significantly higher microbial diversity than healthy individuals [Citation10,Citation41]. In this study, we found that TM7 species HMT-346 and 356 were enriched in patients with gingivitis, consistent with previous studies [Citation42,Citation43]. Saccharibacteria (TM7) is commonly found in human oral, skin, and gut microbiomes [Citation44,Citation45] and is typically dominant in inflammatory environments, particularly in periodontal-related diseases, such as periodontitis [Citation46] and gingivitis [Citation13]. Although Saccharibacteria (TM7) is generally considered a potentially pathogenic bacterium that could initiate or aggravate periodontitis and is categorized as a pathogenic red complex [Citation47], some studies suggest it may have a protective effect by attenuating the pathogenicity of other bacteria [Citation33]. Therefore, the precise role of Saccharibacteria (TM7) in gingivitis pathogenesis is yet to be elucidated. We found that Selenomonas, specifically S. sputigena, was enriched in patients with gingivitis [Citation48]. The increased abundance of S. sputigena in dental plaque is closely associated with periodontal disease. S. sputigena adheres to gingival keratinocytes and induces the expression and secretion of cytokines and chemokines related to inflammation and leukocyte recruitment. Interaction between S. sputigena and the host may lead to bacteria-induced inflammation and tissue destruction, resulting in the progression of gingivitis [Citation48]. Patients with chronic periodontitis exhibit significantly higher detection rates of A. dentalis than those of A. naeslundii or A. oris [Citation49]. Our study findings support this observation, showing that A. dentalis is more abundant in the Gingivitis group, suggesting its potential role in altering dental plaque composition between healthy and periodontal disease states [Citation49]. Additionally, we observed a high accumulation of S. anginosus and S. oralis subsp._tigurinus_clade_071 in patients with gingivitis, consistent with previous research [Citation1,Citation50]. However, further studies are required to elucidate their role in gingivitis pathogenesis.
In adolescent patients undergoing orthodontic treatment, elevated hormone levels can impact the oral microbiome and exacerbate gingival hyperplasia, leading to increased gingival sulcus depth. There is increasing evidence that periodontal tissue response is regulated by hormones such as androgen, estrogen, and progesterone. Several factors influence the incidence and severity of gingivitis in adolescents, including dental plaque biofilm, dental caries, oral respiration, and tooth crowding. Notably, steroid hormone levels are a remarkable modifier of plaque-induced gingivitis in adolescents [Citation51]. Endocrine changes enhance the response of gingival tissue to local irritants like plaque. Despite this, dental plaque remains the primary cause of gingivitis in adolescents, and eliminating local plaque stimulation is crucial for treatment [Citation51]. It has been reported that maintaining oral hygiene is more critical for gingival health than the increase in steroid hormone levels among adolescents [Citation52]. Moreover, reducing gingivitis in adolescents involves removing dental plaque clinically and reducing pathogen abundance rather than regulating hormones. Our study confirmed significant differences in the marginal supragingival plaque microbiomes between adolescent patients with and without gingivitis undergoing fixed orthodontic treatment. These results indicate that while hormone levels affect periodontal tissue, the microbiomes of the Gingivitis and Periodontal healthy groups significantly differ. This study enrolled adolescents aged 11–18 undergoing fixed orthodontic treatment. The average age of the Gingivitis group (14.17 ± 1.60) was similar to that of the Periodontal healthy group (14.58 ± 1.52), suggesting comparable hormone levels, which could similarly impact the oral microbiome. However, as hormone levels vary with age, gender, and individual differences, the average age serves only as a preliminary reference, and represents a limitation of this study.
Currently, research on the relationship between hormones and gingivitis in adolescents mostly focuses on the correlation between hormones and clinical symptoms of gingivitis, with few studies examining the relationship between hormones and oral microbiomes. Previous longitudinal studies have compared pre-adolescent and adolescent orthodontic treatments to assess the impact of hormone levels on clinical and microbiological parameters. These studies reported a statistically significant increase in gingival inflammation and abundance of P. intermedia compared to baseline values, which may be associated with increased systemic hormone levels [Citation53]. The results of another study indicated that the abundance of C. rectus was positively correlated with estradiol levels [Citation54]. Additionally, estradiol levels in saliva were associated with the abundance of C. gingivalis, Peptostreptococcus micros, T. denticola, and T. forsythia [Citation55]. Among these bacteria, P. intermedia, T. denticola and T. forsythia were relatively abundant in the Gingivitis group compared to the Periodontal healthy group, although their relative abundances were low. For the core genera and species with high abundance in this study, there is no existing literature on the hormonal effects on these microorganisms. Conversely, another study suggested that changes in pubertal hormones do not promote the colonization of periodontitis pathogens [Citation56]. Therefore, the relationship between hormones and periodontal pathogens warrants further investigation.
As the most detected fungus in the oral cavity [Citation57], C. albicans can aggregate within subgingival biofilms of periodontitis patients, exhibiting a significantly positive correlation with periodontal disease and playing a role in the initiation and progression of periodontitis [Citation18,Citation58]. Previous studies have primarily focused on subgingival plaque in adults, while C. albicans colonization rates in the supragingival plaque of adolescents with gingivitis undergoing orthodontic treatment have not been extensively investigated. Our results suggest that a high C. albicans colonization occurs in the supragingival plaque of adolescent patients with gingivitis. Moreover, C. albicans enrichment influences the microbial composition of gingival plaques.
Co-aggregation of C. albicans and A. oris has been previously reported. These exhibit growth synergism and can form a strong dual-species biofilm, thereby increasing the total biofilm biomass [Citation59,Citation60]. In addition, although the relative abundance was low, we detected F. nucleatum and A. actinomycetemcomitans only in the G-CaP group. F. nucleatum can aggregate with C. albicans by binding to mannose receptors on the surface of C. albicans [Citation18,Citation61]. A. actinomycetemcomitans and C. albicans exhibit symbiosis and increased virulence, which aggravates periodontal tissue destruction [Citation62]. Furthermore, the invasion of C. albicans hyphae has been detected in the gingival connective tissue of patients with periodontal disease, which is also related to the enrichment of A. actinomycetemcomitans [Citation63,Citation64]. Other bacteria enriched in the G-CaP group in this study, such as C. concisus, C. gracilis, S. noxia, and V. atypica, are presumed to be involved in the occurrence and development of periodontal disease. For example, C. gracilis is enriched in the plaque of patients with refractory periodontitis [Citation65]. C. concisus, which belongs to the green complex, is primarily associated with the early formation of subgingival biofilms linked to early-onset periodontitis [Citation66]. S. noxia can exist within supragingival or subgingival biofilms [Citation67] and may contribute to the transition of periodontal tissues from a healthy to a diseased state [Citation68]. V. atypica possesses a multivalent hemagglutinin that promotes adhesion to P. gingivalis and oral buccal cells [Citation69], making it more likely to be enriched in refractory periodontitis [Citation65]. However, further investigation is needed to understand the interactions between the bacteria and C. albicans.
This study confirmed that the occurrence of gingivitis during orthodontic treatment in adolescents impacts the microbial ecology of the oral microbiome, leading to changes in both the bacterial microbiota and C. albicans. By monitoring changes in biomarkers reflecting periodontal status, it is expected to be utilized for diagnosing and predicting gingivitis in orthodontic patients, enabling the implementation of preventive measures. However, one of the limitations of this study was the relatively small sample size, which should be increased in future research. Additionally, the interactions between C. albicans and various periodontal pathogens warrant further investigation to elucidate their mechanisms in the onset and progression of gingivitis. Moreover, this study is cross-sectional. Future research should involve designing a cohort study to explore the relationship between hormonal changes, clinical symptoms, and changes in bacterial and fungal abundance. This could provide a foundation for reducing the incidence of gingivitis among adolescent orthodontic patients.
Conclusions
The study identified significant differences in both microbial diversity and composition within the supragingival microbiome between adolescent orthodontic patients with gingivitis and those in a healthy state. Additionally, we observed that C. albicans was more prevalent in patients with gingivitis, impacting the microbiome composition of supragingival plaque in adolescent patients undergoing orthodontic treatment. This suggests a potential role for C. albicans in the pathogenesis of gingivitis.
Ethical approval
This study was approved by the Ethics Committee of the Beijing Stomatological Hospital (CMUSH-IRB-KJ-YJ-2022-11).
Author contributions
Hao Yang was responsible for data collection, processing and analysis, and paper writing. Hongyu Gao assisted in data acquisition, processing, and analysis. Xianju Xie provided experimental samples and participated in the research design. Hongmei Wang provided samples to assist in data processing and analysis, and Xiaowei Li provided experimental samples and method selection. Correspondence authors are Yansong Ma and Yuxing Bai. Yansong Ma was responsible for the planning and design of the whole study to ensure the integrity and accuracy of the study. Yuxing Bai designed research methods, and supervised and coordinated the research process, and provided economic and technical support.
Acknowledgments
We are very appreciative of Professor Dongming Li from the Dermatology Department of Peking University for providing the theoretical basis and relevant experimental methods for ITS identification in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Tanner ACR, Sonis AL, Lif Holgerson P, et al. White-spot lesions and gingivitis microbiotas in orthodontic patients. J Dent Res. 2012;91(9):853–13. doi: 10.1177/0022034512455031
- Eid HA, Assiri HAM, Kandyala R, et al. Gingival enlargement in different age groups during fixed orthodontic treatment. J Int Oral Health. 2014;6(1):1–4.
- Zanatta FB, Ardenghi TM, Antoniazzi RP, et al. Association between gingivitis and anterior gingival enlargement in subjects undergoing fixed orthodontic treatment. Dental Press J Orthod. 2014;19(3):59–66. doi: 10.1590/2176-9451.19.3.059-066.oar
- Kirschneck C, Fanghänel J, Wahlmann U, et al. Interactive effects of periodontitis and orthodontic tooth movement on dental root resorption, tooth movement velocity and alveolar bone loss in a rat model. Anat Anz. 2017;210:32–43. doi: 10.1016/j.aanat.2016.10.004
- van Gastel J, Quirynen M, Teughels W, et al. Longitudinal changes in microbiology and clinical periodontal parameters after removal of fixed orthodontic appliances. Eur J Orthodnt. 2010;33(1):15–21. doi: 10.1093/ejo/cjq032
- Sadeq A, Risk JM, Pender N, et al. Evaluation of the co-existence of the red fluorescent plaque bacteria P. gingivalis with S. gordonii and S. mutans in white spot lesion formation during orthodontic treatment. Photodiagn Photodyn. 2015;12(2):232–237. doi: 10.1016/j.pdpdt.2015.03.001
- Sun F, Ahmed A, Wang L, et al. Comparison of oral microbiota in orthodontic patients and healthy individuals. Microb Pathog. 2018;123:473–477. doi: 10.1016/j.micpath.2018.08.011
- Campobasso A, Lo Muzio E, Battista G, et al. Taxonomic analysis of oral microbiome during Orthodontic treatment. Int J Dent. 2021;2021:1–12. doi: 10.1155/2021/8275181
- Guo R, Zheng Y, Zhang L, et al. Salivary microbiome and periodontal status of patients with periodontitis during the initial stage of orthodontic treatment. Am J Orthod Dentofacial Orthop. 2021;159(5):644–652. doi: 10.1016/j.ajodo.2019.11.026
- Guo R, Liu H, Li X, et al. Subgingival microbial changes during the first 3 months of fixed appliance treatment in female adult patients. Curr Microbiol. 2018;76(2):213–221. doi: 10.1007/s00284-018-1610-1
- Chen I, Chung J, Vella R, et al. Alterations in subgingival microbiota during full-fixed appliance orthodontic treatment—A prospective study. Orthod Craniofac Res. 2021;25(2):260–268. doi: 10.1111/ocr.12534
- Palone M, Preite C, Lombardo L. Microbiota changes in the periodontium in response to orthodontic forces. Semin Orthod. 2024;30(2):135–140. doi: 10.1053/j.sodo.2023.10.001.
- Kado I, Hisatsune J, Tsuruda K, et al. The impact of fixed orthodontic appliances on oral microbiome dynamics in Japanese patients. Sci Rep. 2020;10(1):21989. doi: 10.1038/s41598-020-78971-2
- Dilhari A, Weerasekera MM, Siriwardhana A, et al. Candida infection in oral leukoplakia: an unperceived public health problem. Acta Odontol Scand. 2016;74(7):565–569. doi: 10.1080/00016357.2016.1220018
- Mohd Bakri M, Mohd Hussaini H, Rachel Holmes A, et al. Revisiting the association between candidal infection and carcinoma, particularly oral squamous cell carcinoma. J Oral Microbiol. 2010;2(1):5780–5785. doi: 10.3402/jom.v2i0.5780
- He H, Xia X, Yang H, et al. A pilot study: a possible implication of Candida as an etiologically endogenous pathogen for oral lichen planus. BMC Oral Health. 2020;20(1):72–79. doi: 10.1186/s12903-020-1042-8
- Xu H, Sobue T, Thompson A, et al. Streptococcal co‐infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2013;16(2):214–231. doi: 10.1111/cmi.12216
- Suresh Unniachan A, Krishnavilasom Jayakumari N, Sethuraman S. Association between Candida species and periodontal disease: A systematic review. Curr Med Mycol. 2020;6(2):63–68. doi: 10.18502/cmm.6.2.3420
- Monroy-Pérez E, Rodríguez-Bedolla RM, Garzón J, et al. Marked virulence and azole resistance in Candida albicans isolated from patients with periodontal disease. Microb Pathog. 2020;148:104436. doi: 10.1016/j.micpath.2020.104436
- Järvensivu A, Hietanen J, Rautemaa R, et al. Candida yeasts in chronic periodontitis tissues and subgingival microbial biofilms in vivo. Oral Dis. 2004;10(2):106–112. doi: 10.1046/j.1354-523X.2003.00978.x
- Bartnicka D, Karkowska-Kuleta J, Zawrotniak M, et al. Adhesive protein-mediated cross-talk between Candida albicans and porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci Rep. 2019;9(1):4376–4388. doi: 10.1038/s41598-019-40771-8
- Wawrzyk-Bochenek I, Łobacz M, Wilczyński S, et al. Evaluation of the tooth surface after irradiation with diode laser applied for removal of dental microorganisms from teeth of patients with gingivitis, using X-ray photoelectron (XPS) and optical profilometry (OP). J Clin Med. 2022;11(22):6840–6853. doi: 10.3390/jcm11226840
- Peycheva S, Apostolova E, Gardjeva P, et al. Effect of Bulgarian propolis on the oral microflora in adolescents with plaque-induced gingivitis. Rev Bras Farmacogn. 2019;29(3):271–277. doi: 10.1016/j.bjp.2018.11.001
- Margolis HC, Moreno EC. Composition of pooled plaque fluid from caries-free and caries-positive individuals following sucrose exposure. J Dent Res. 1992;71(11):1776–1784. doi: 10.1177/00220345920710110301
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507
- Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9
- Chen T, Yu WH, Izard J, et al. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013–baq13. doi: 10.1093/database/baq013
- Yang H, Ma Y, Xie X, et al. Candida albicans enriched in orthodontic derived white spot lesions and shaped focal supragingival bacteriome. Front Microbiol. 2023;14:1084850. doi: 10.3389/fmicb.2023.1084850
- Thiyahuddin NM, Lamping E, Rich AM, et al. Yeast species in the oral cavities of older people: a comparison between people living in their own homes and those in rest homes. J Fungi. 2019;5(2):30–39. doi: 10.3390/jof5020030
- Nasehi A, Kadir JB, Esfahani MN, et al. Leaf spot on lettuce (lactuca sativa) caused by stemphylium solani, a new disease in malaysia. Plant Dis. 2013;97(5):689. doi:10.1094/PDIS-10-12-0902-PDN.
- Abbate GM, Caria MP, Montanari P, et al. Parodontale Gesundheit von Teenagern mit herausnehmbaren Alignern und festsitzenden kieferorthopädischen Apparaturen. J Orofac Orthop. 2015;76(3):240–250. doi: 10.1007/s00056-015-0285-5
- Gursoy UK, Sokucu O, Uitto VJ, et al. The role of nickel accumulation and epithelial cell proliferation in orthodontic treatment-induced gingival overgrowth. Eur J Orthod. 2007;29(6):555–558. doi: 10.1093/ejo/cjm074
- Thornberg MJ, Riolo CS, Bayirli B, et al. Periodontal pathogen levels in adolescents before, during, and after fixed orthodontic appliance therapy. Am J Orthod Dentofacial Orthop. 2009;135(1):95–98. doi: 10.1016/j.ajodo.2007.02.057
- ŽivkovićSandić M, Popović B, Čarkić J, et al. Changes in subgingival microflora after placement and removal of fixed orthodontic appliances. Srp Arh Celok Lek. 2014;142(5–6):301–305. doi: 10.2298/SARH1406301Z
- Surlin P, Rauten AM, Mogoantă L, et al. Correlations between the gingival crevicular fluid MMP8 levels and gingival overgrowth in patients with fixed orthodontic devices. Rom J Morphol Embryol. 2010;51(3):515–519.
- Lee SM, Yoo SY, Kim HS, et al. Prevalence of putative periodontopathogens in subgingival dental plaques from gingivitis lesions in Korean orthodontic patients. J Microbiol. 2005;43(3):260–265.
- Paolantonio M, di Girolamo G, Pedrazzoll V, et al. Occurrence of actinobacillus actinomycetemcomitans in patients wearing orthodontic appliances. J Clin Periodontol. 2005;23(2):112–118. doi: 10.1111/j.1600-051X.1996.tb00543.x
- Mombelli A, Rutar A, Lang NP. Correlation of the periodontal status 6 years after puberty with clinical and microbiological conditions during puberty. J Clin Periodontol. 2005;22(4):300–305. doi: 10.1111/j.1600-051X.1995.tb00152.x
- Lu H, Zhao Y, Feng X, et al. Microbiome in maintained periodontitis and its shift over a single maintenance interval of 3 months. J Clin Periodontol. 2019;46(11):1094–1104. doi: 10.1111/jcpe.13177
- Chipashvili O, Utter DR, Bedree JK, et al. Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe. 2021;29(11):1649–1662.e7. doi: 10.1016/j.chom.2021.09.009
- McLean JS, Bor B, Kerns KA, et al. Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep. 2020;32(3):107939. doi: 10.1016/j.celrep.2020.107939
- Camanocha A, Dewhirst FE. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol. 2014;8(1):6–28. doi: 10.3402/jom.v6.25468
- Zhu Q, Dupont CL, Jones MB, et al. Visualization-assisted binning of metagenome assemblies reveals potential new pathogenic profiles in idiopathic travelers’ diarrhea. Microbiome. 2018;6(1):201–220. doi: 10.1186/s40168-018-0579-0
- Nie J, Utter DR, Kerns KA, et al. Strain-level variation and diverse host bacterial responses in episymbiotic Saccharibacteria. mSystems. 2022;7(2):e0148821. doi: 10.1128/msystems.01488-21
- Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7(5):1016–1025. doi: 10.1038/ismej.2012.174
- Hawkes CG, Hinson AN, Vashishta A, et al. Selenomonas sputigena interactions with gingival epithelial cells that promote inflammation. Infect Immun. 2023;91(2):e0031922. doi: 10.1128/iai.00319-22
- Vielkind P, Jentsch H, Eschrich K, et al. Prevalence of actinomyces spp. In patients with chronic periodontitis. Int J Med Microbiol. 2015;305(7):682–688. doi: 10.1016/j.ijmm.2015.08.018
- Moore WE, Holdeman LV, Smibert RM, et al. Bacteriology of experimental gingivitis in young adult humans. Infect Immun. 1982;38(2):651–667. doi: 10.1128/iai.38.2.651-667.1982
- Murakami S, Mealey BL, Mariotti A, et al. Dental plaque-induced gingival conditions. J Periodontol. 2018;89(Suppl 1):S17–S27. doi: 10.1002/JPER.17-0095
- Tiainen L, Asikainen S, Saxén L. Puberty‐associated gingivitis. Oral Epidemiol. 1992;20(2):87–89. doi: 10.1111/j.1600-0528.1992.tb00683.x
- Nakagawa S, Fujii H, Machida Y, et al. A longitudinal study from prepuberty to puberty of gingivitis. J Clin Periodont. 2005;21(10):658–665. doi: 10.1111/j.1600-051X.1994.tb00783.x
- Yokoyama M, Hinode D, Yoshioka M, et al. Relationship between Campylobacter rectus and periodontal status during pregnancy. Oral Microb Immun. 2007;23(1):55–59. doi: 10.1111/j.1399-302X.2007.00391.x
- Mitova N, Rashkova MR, Popova CL. Saliva diagnostics of sex hormones and subgingival microflora in children in puberty. Biotechnol Biotec Eq. 2019;35(1):408–414. doi: 10.1080/13102818.2019.1688190
- Yanover L, Ellen RP. A clinical and microbiologic examination of gingival disease in parapubescent females. J periodont. 1986;57(9):562–567. doi: 10.1902/jop.1986.57.9.562
- Cuesta AI, Jewtuchowicz V, Brusca MI, et al. Prevalence of Staphylococcus spp and Candida spp in the oral cavity and periodontal pockets of periodontal disease patients. Acta Odontol Latinoam. 2010;23(1):20–26.
- Canabarro A, Valle C, Farias MR, et al. Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J Periodont Res. 2012;48(4):428–432. doi: 10.1111/jre.12022
- Cavalcanti IMG, Del Bel Cury AA, Jenkinson HF, et al. Interactions between Streptococcus oralis, Actinomyces oris, and Candida albicans in the development of multispecies oral microbial biofilms on salivary pellicle. Mol Oral Microb. 2016;32(1):60–73. doi: 10.1111/omi.12154
- Reardon-Robinson ME, Wu C, Mishra A, et al. Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc Natl Acad Sci. 2014;111(10):3835–3840. doi: 10.1073/pnas.1321417111
- Jabra-Rizk MA, Falkler WA Jr, Merz WG, et al. Coaggregation of Candida dubliniensis with Fusobacterium nucleatum. J Clin Microbiol. 1999;37(5):1464–1468. doi: 10.1128/JCM.37.5.1464-1468.1999
- Brusca MI, Rosa A, Albaina O, et al. The impact of oral contraceptives on women’s periodontal health and the subgingival occurrence of aggressive periodontopathogens and candida species. J Periodontol. 2010;81(7):1010–1018. doi: 10.1902/jop.2010.090575
- Reynaud AH, Nygaard‐Østby B, Bøygard GK, et al. Yeasts in periodontal pockets. J Clin Periodont. 2001;28(9):860–864. doi: 10.1034/j.1600-051x.2001.028009860.x
- Rubio NA, Puia S, Toranzo S, et al. Invasión fúngica en tejido conectivo en pacientes con enfermedad gingivo-periodontal. Rev Iberoam Micol. 2015;32(1):20–24. doi: 10.1016/j.riam.2012.07.002
- Colombo APV, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J periodont. 2009;80(9):1421–1432. doi: 10.1902/jop.2009.090185
- Kamma JJ, Nakou M, Baehni PC. Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodontol Res. 1999;34(1):25–33. doi: 10.1111/j.1600-0765.1999.tb02218.x
- McDaniel J, McDaniel S, Samiano BJ, et al. Microbial screening reveals oral site-specific locations of the periodontal pathogen selenomonas noxia. Curr Issues Mol Biol. 2021;43(1):353–364. doi: 10.3390/cimb43010029
- Cruz P, Mehretu AM, Buttner MP, et al. Development of a polymerase chain reaction assay for the rapid detection of the oral pathogenic bacterium, Selenomonas noxia. BMC Oral Health. 2015;15(1):1–8. doi: 10.1186/s12903-015-0071-1
- Zhou P, Liu J, Merritt J, et al. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis , and human oral buccal cells. Mol Oral Microb. 2015;30(4):269–279. doi: 10.1111/omi.12091
