ABSTRACT
Background
Curcumin is a multi-functional polyphenol with anti-bacterial and anti-inflammatory effects and may have potential for treatment of periodontal diseases. The present study was conducted to examine the molecular basis of the anti-bacterial effect of curcumin against Porphyromonas gingivalis using metabolome analysis.
Materials and Methods
P. gingivalis were incubated with 10 µg/mL curcumin, and then metabolites were analyzed with CE-TOF/MS. Expression levels of sigma factors were also evaluated using RT-PCR assays. The activities of dipeptidyl peptidases (DPPs) were assessed by examining the degradation reactions of MCA-labeled peptides.
Results
The relative amounts of various glycogenic amino acids were significantly decreased when P. gingivalis was incubated with curcumin. Furthermore, the metabolites on the amino acid degradation pathway, including high-energy compounds such as ATP, various intermediate metabolites of RNA/DNA synthesis, nucleoside sugars and amino sugars were also decreased. Additionally, the expression levels of sigma-54 and sigma-70 were significantly decreased, and the same results as noted following nutrient starvation. Curcumin also significantly suppressed the activities of some DPPs, while the human DPP-4 inhibitors markedly inhibited the growth of P. gingivalis and activities of the DPPs.
Conclusions
Curcumin suppresses the growth of P. gingivalis by inhibiting DPPs and also interferes with nucleic acid synthesis and central metabolic pathways, beginning with amino acid metabolism.
Introduction
Periodontal disease is a major chronic infectious disorder found in humans [Citation1] and known to have an influence on oral as well as systemic health [Citation2] The condition is initiated by dysbiosis caused by an imbalance with host defense systems in subgingival biofilm. Porphyromonas gingivalis is considered to be a keystone pathogen and known to play a central role in initiation of dysbiosis [Citation3], and various functional materials for prevention of periodontal disease have been examined [Citation4].
Curcumin, a major constituent of turmeric rhizomes (Curcuma-longa) [Citation5], has been reported to have a variety of functions, including anti-oxidant [Citation6,Citation7], anti-inflammatory [Citation7–9], anti-tumor [Citation10,Citation11] and anti-bacterial [Citation12,Citation13] effects. Thus, curcumin may have potential for treatment of periodontal diseases. We previously showed that curcumin markedly inhibited the growth of P. gingivalis, and prevented biofilm formation as well as bacterial protease activity at a low concentration [Citation14]. It has also been reported that curcumin clearly limited the expression of inflammatory cytokines (IL-6, IL-1β, TNF-α) and inhibited invasion of human gingival epithelial cells by outer membrane vesicles of P. gingivalis [Citation15]. Furthermore, clinical trials of curcumin used in combination with various materials such as gels, collagen sponges and mouthwash have found clear effects on clinical manifestations of periodontal disease [Citation16–18].
Nevertheless, the mechanism of the antimicrobial action of curcumin is unclear. A previous report presented findings suggesting that curcumin caused damage to the bacterial membrane of Staphylococcus aureus [Citation19], while another study found that curcumin disturbed Bacillus subtilis cell division by inhibiting the activity of GTPase [Citation20]. On the other hand, some reports have suggested that only negligible bactericidal effects against some oral bacterial species such as Aggregatibacter actinomycetemcomitans [Citation14] and Streptococcus mitis [Citation21]. Thus, while curcumin seems to exhibit species-specific antimicrobial activities, the molecular basis remains to be elucidated.
The present study was conducted to investigate the mechanism of the antimicrobial action of curcumin toward P. gingivalis.
Materials and methods
Bacterial strains & culture conditions
Porphyromonas gingivalis ATCC33277 and Streptococcus oralis ATCC6249 were obtained from ATCC (Manassas, USA). KDP136 (P. gingivalis ATCC 33,277 rgpA:erm rgpB:tetQ kgp:cat) was kindly provided by Prof. Koji Nakayama (Department of Microbiology and Oral Infection, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaki, Japan). P. gingivalis was anaerobically cultured (80% N2, 10% CO2, 10% H2) at 37°C in 30 g/L trypticase soy broth (TSB; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) containing 5 mg/L hemin (Sigma-Aldrich, St Louis, MO, USA) and 1 mg/L menadione (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan), while S. oralis was aerobically cultured at 37°C in Todd Hewitt broth (THB; Becton, Dickinson and Company).
Growth assays
P. gingivalis and S. oralis were separately incubated in the presence of curcumin (Fujifilm Wako Pure Chemical Corporation) serially diluted two-fold from 500 to 7.8 µg/mL, or sitagliptin monophosphate (Sigma-Aldrich) or Val-boroPro (Selleck Chemicals, TX, USA) serially diluted two-fold from 10,000 to 625 µg/mL. Those were dissolved with 0.1% dimethyl sulfoxide (DMSO) to OD600 nm ≈ 0.05 and dispensed in 96-well plates at 200 µL/well. Following incubation, the lowest concentration at which no growth (OD600 nm < 0.1) was observed was defined as the minimum inhibitory concentration (MIC).
Intracellular metabolite extraction
The concentration of curcumin was based on a previous report [Citation14]. P. gingivalis cells were cultured anaerobically in a liquid medium containing 0 or 10 µg/mL curcumin dissolved with 0.1% DMSO adjusted to OD600 nm ≈ 0.1 at 37°C for 24 h. The bacterial cells were then collected and suspended in methanol as previously described [Citation22].
CE-TOF/MS measurement conditions
The untargeted metabolome analysis with CE-TOF/MS was performed using an Agilent CE-TOFMS system (Agilent Technologies, Santa Clara, CA) equipped with a fused silica capillary [50 µm (inner diameter) × 80 cm]. For measurements of cationic/anionic metabolites, running buffer, a solution composed of Cation Buffer Solution [H3301–1001; Human Metabolome Technologies (HMT), Tsuruoka, Japan], and Anion buffer solution (H3302–1021) were used, with CE voltage + 27 kV/+ 30 kV, MS ionization ESI positive/negative, MS capillary voltage 4000 V/3500 V, MS scan range 50–1000 m/z and HMT Sheath Liquid (H3301–1020). Identification of metabolites and measurement of relative amounts were performed using MASTER HANDS (version 2.1.0.1, 2.9.0.9; Keio University, Tokyo, Japan) and the HMT metabolite database based on internal standards (HMT).
Determination of gene expression
For the low nutrient condition, the liquid medium was diluted 100-fold with PBS, and then P. gingivalis cells were suspended in low nutrient medium adjusted to OD600 nm ≈ 0.5 and anaerobically incubated at 37°C for 3 h. Next, the cells were suspended in curcumin medium adjusted to OD600 nm ≈ 0.05 and incubated for 9 h, then collected by centrifugation (7500 rpm, 7 min, 4°C). Total RNA was extracted using TRIzol Reagent® (Thermo Fisher Scientific Inc., MA, USA) and an RNeasy kit (QIAGEN N.V., Hilden, Germany) according to the manufacturer’s recommendations, then reverse transcribed to cDNA using iScript master mix (Bio-Rad Laboratories, Inc., CA, USA). Real-time PCR was performed using a KAPA SYBR Fast qPCR kit (NIPPON Genetics Co., Ltd., Tokyo, Japan). Primer sets are shown in . Gene expression levels were compared using a comparative Ct method. Acquired data were normalized to the expression level of the 16s rRNA gene.
Table 1. Primers.
Dipeptidyl peptidase activity
Measurements to determine dipeptidyl peptidase (DPP) activity were performed as previously described [Citation25]. Briefly, P. gingivalis KDP136 cells were washed with PBS followed by centrifugation (7500 rpm, 7 min, 4°C), then suspended to OD600 nm ≈ 0.05 in 50 mM phosphate buffer containing 5 mM EDTA, 100 mM NaCl with or without 10 µg/mL curcumin, 5000 µg/mL sitagliptin monophosphate and 2500 µg/mL Val-boroPro dissolved with 0.1% DMSO. Following suspension, the cells were anaerobically incubated at 37°C for 3 h, then methylcoumarin amide (MCA)-labeled peptides (Gry-Pro-MCA, Lys-Ala-MCA, Met-Leu-MCA, Leu-Asp-MCA: PEPTIDE INSTITUTE.Inc, Osaka, Japan) were added at a final concentration 20 µM, and incubation was performed at 37°C for 3 h. Fluorescence intensity (λex = 380 nm, λem = 460 nm, 1.0 s) was determined using a plate reader ARVO MX (PerkinElmer, Inc), then calculated as free aminomethylcoumarin (AMC) concentration using a calibration curve prepared by use of a regent (PEPTIDE INSTITUTE, Inc.).
Statistical analysis
All obtained data were examined three times using a statistical process. The results, except for those obtained with metabolic analysis, were evaluated with a t-test and Dunnett’s test using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan). Multiple comparisons of metabolomic data were performed with Welch’s t-test. Principal component analysis (PCA) was performed using SIMCA, ver. 17 (Sartorius, Göttingen, Germany).
Results
Effects of curcumin on P. gingivalis metabolomic profiles
First, an untargeted metabolome analysis was performed to examine curcumin-treated P. gingivalis. PC1 and PC2 scores were plotted using principal component analysis (PCA) and are shown in . The curcumin-treated and control groups were separated by the PC1 component. shows a plot of factor loadings for each metabolite. In the PC1 component, several amino acids (ornithine, proline, serine, alanine, glutamic acid, glycine, lysine, arginine, citrulline, tryptophan, phenylalanine), NAD and tripeptide (Ilo-Pro-Pro) showed characteristic fluctuations. Relative peak areas of amino acids are presented in . Proline, ornithine, valine, alanine, serine, lysine, glycine, glutamic acid, threonine and aspartic acid were significantly decreased in P. gingivalis treated with curcumin, whereas phenylalanine and tryptophan were significantly increased. Although the differences were not statistically significant, asparagine, glutamine and β-alanine showed a decreasing trend, and arginine and citrulline an increasing trend. Therefore, we focused on fluctuation of metabolites on the metabolic pathway starting from glycogenic amino acids, with reference to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (https://www.genome.jp/kegg/pathway.html). shows metabolite fluctuations on the glycogenesis and pentose phosphate pathways, as well as the nucleoside-sugar and sugar-amino acid metabolic pathways. When P. gingivalis was treated with curcumin, most of the detected metabolites on these pathways were significantly decreased or showed a decreasing trend. Fluctuations of metabolites on the purine and pyrimidine metabolic pathways, which represent nucleic acid materials and energy currency, are demonstrated in . In addition, curcumin was found to cause a significant decrease in most of the metabolites of the purine and pyrimidine metabolic pathways in P. gingivalis.
Figure 1. Principal component analysis of metabolites detected in P. gingivalis. (a) PC1 and PC2 score plots of results of principal component analysis for the control (green: Control 1 ~ 3) and curcumin-treated (blue: C1 ~ 3) groups. (b) Factor loading for each metabolite contributing to PC1 and PC2.
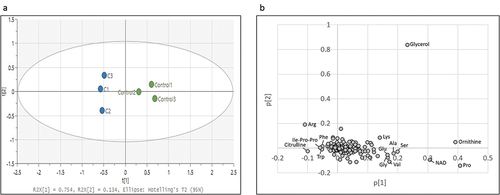
Figure 2. Effects of curcumin on amino acids in P. gingivalis. Corrected peak areas of amino acids detected in P. gingivalis cultured for 24 hours in liquid medium (white) and in TSB with 10 µg/mL curcumin (shaded). Cys was below the detection limit in all samples and excluded.
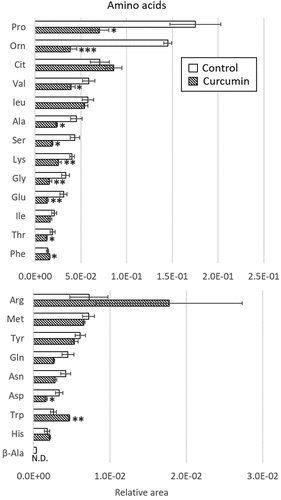
Figure 3. Effects of curcumin on metabolites in (a) glycogenic pathway and TCA cycle, (b) pentose phosphate pathway and (c) sugar amino acids and nucleotide sugars in P. gingivalis. Corrected peak areas of amino acids detected in P. gingivalis incubated for 24 hours in liquid medium (white) and in liquid medium with 10 µg/mL curcumin (shaded) are shown.
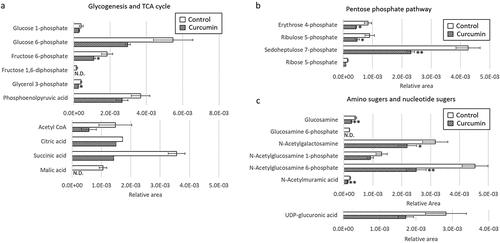
Figure 4. Effects of curcumin on metabolites in (a) purine metabolic pathway and (b) pyrimidine metabolic pathway in P. gingivalis. Corrected peak areas of amino acids detected in P. gingivalis incubated for 24 hours in liquid medium (white) and in liquid medium with 10 µg/mL curcumin (shaded) are shown.
Expression of sigma factors
Findings indicating decreased glycogenic amino acids and downstream metabolites suggested that the bacterial response to nutrient starvation is activated by curcumin. Therefore, the mRNA expression level of the RNA polymerase sigma subunit, which indicates a response to nutrient starvation stress in eubacteria, was examined. Identified in P. gingivalis were a gene encoding sigma-54, termed rpoN (PGN_1202), and a gene encoding sigma-70, termed rpoD (PGN_0638), which are considered to be related to nutrient starvation [Citation25], though their expression levels have not been reported. In the present study, a low nutrient medium diluted with PBS was used, and the expression levels of PGN_0638 and PGN_1202 were determined. As shown in , the expression levels of both were significantly decreased in that low nutrient condition. The expression levels of those genes following stimulation with curcumin were also examined, which revealed a significant decrease in each, the same as noted in the low nutrient condition ().
Figure 5. Effects of curcumin on mRNA expression level of sigma factor in P. gingivalis. (a) mRNA expression levels of PGN_0638(rpoD) and PGN_1202(rpoN) genes in P. gingivalis cultured for three hours in liquid medium (white) or 1/100 diluted liquid medium with PBS (dotted). (b) mRNA expression levels of PGN_0638 and PGN_1202 genes in P. gingivalis cultured for 15 hours in liquid medium (white) or in liquid medium with 10 µg/mL curcumin (shaded). Result of the PGN_0180(fimA) gene with unchanged expression are presented as a control [Citation26]. Expression levels are shown relative to 16srRNA gene.
![Figure 5. Effects of curcumin on mRNA expression level of sigma factor in P. gingivalis. (a) mRNA expression levels of PGN_0638(rpoD) and PGN_1202(rpoN) genes in P. gingivalis cultured for three hours in liquid medium (white) or 1/100 diluted liquid medium with PBS (dotted). (b) mRNA expression levels of PGN_0638 and PGN_1202 genes in P. gingivalis cultured for 15 hours in liquid medium (white) or in liquid medium with 10 µg/mL curcumin (shaded). Result of the PGN_0180(fimA) gene with unchanged expression are presented as a control [Citation26]. Expression levels are shown relative to 16srRNA gene.](/cms/asset/2b81918c-5dd5-4431-b75d-d892a90b6b41/zjom_a_2373040_f0005_b.gif)
Dipeptidyl peptidase activities in P. gingivalis treated with curcumin
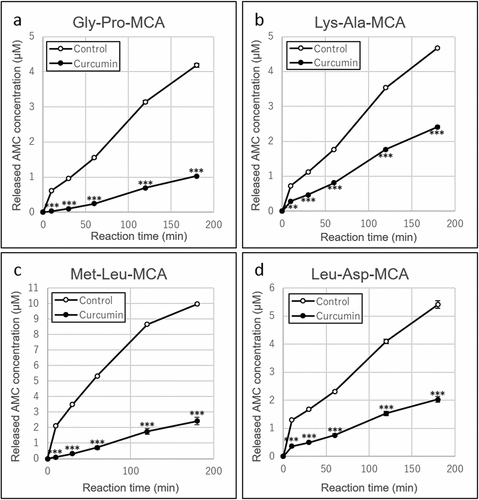
The activities of some DPPs (DPP4, DPP5, DPP7, DPP11) in P. gingivalis, determined by measuring the fluorescence intensity of AMC derived from a decomposition product of MCA-labeled synthetic substrates, were previously reported [Citation27]. In the present study, DPPs in P. gingivalis treated with curcumin were examined. In order to avoid the effects of gingipain, a deletion mutant with knocked out genes coding gingipain of P. gingivalis (KDP136) was used. Following incubation with curcumin, substrates were added, and fluorescence intensity was determined. As shown in , decomposition of Gry-Pro-MCA (substrate of DPP4), Lys-Ala-MCA (substrate of DPP5), Met-Leu-MCA (substrate DPP7) and Leu-Asp-MCA (substrate of DPP11) was inhibited by curcumin.
Figure 6. Inhibitory effects of curcumin on activities of DPPs in P. gingivalis. (a) Gly-Pro-MCA (substrate of DPP4), (b) Lys-Ala-MCA (substrate of DPP5), (c) Met-LeuMCA (substrate of DPP7) and (d) Leu-Asp-MCA (substrate of DPP11) were added to P. gingivalis KDP136 suspensions in reaction buffer without curcumin (white circles) and with 10 µg/mL of curcumin (black circles). Degradation activity was determined based on fluorescence intensity of AMC and converted to concentration by using a calibration curve of the standard product.
DPP-4 inhibitors have inhibitory effect on bacterial growth
Next, whether inhibition of DPP would lead to arrest of P. gingivalis growth was examined. Results showing the MICs of human DPP-4 inhibitors (Sitagliptin monophosphate, Val-boroPro) and curcumin against P. gingivalis and S. oralis are presented in . Both Sitagliptin monophosphate and Val-boroPro markedly inhibited the growth of P. gingivalis at concentrations of 5000 and 2500 µg/mL, whereas those DPP-4 inhibitors had negligible effects on S. oralis. In addition, GryPro-MCA, Lys-Ala-MCA and Met-Leu-MCA degradation were inhibited by the DPP-4 inhibitors at the MIC noted for P. gingivalis ().
Figure 7. Inhibitory effects of DPP-4 inhibitors on activities of DPPs against P. gingivalis. (a) Gly-Pro-MCA, (b) Lys-Ala-MCA, (c) Met-LeuMCA and (d) Leu-Asp-MCA (substrate of DPP11) were added to P. gingivalis KDP136 suspensions in reaction buffer without a DPP-4 inhibitor (white circles), with 5000 µg/mL sitagliptin phosphate (squares), or with 2500 µg/mL Val-boroPro (diamond shapes). Degradation activity was determined based on fluorescence intensity of AMC and converted to concentration using a calibration curve of the standard product.
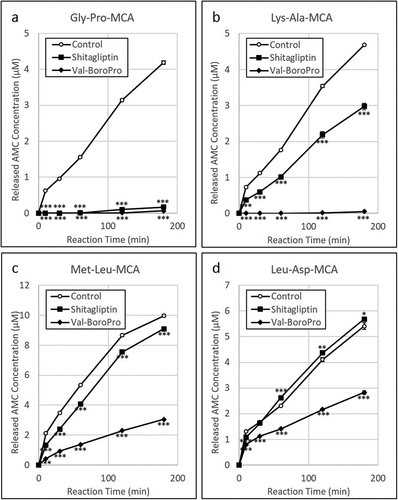
Table 2. Growth inhibitory effects of DPP-4 inhibitor and curcumin on P. gingivalis and S. oralis.
Discussion
The metabolomic profile of P. gingivalis was examined to elucidate the molecular basis of the anti-bacterial effects of curcumin, and the results showed that the metabolism of various amino acids was altered. P. gingivalis is an asaccharolytic bacterium that does not assimilate sugars, such as glucose, lactose, galactose and cellobiose [Citation28], while the pathogen assimilates amino acids obtained from degraded proteins as a carbon source [Citation29]. In particular, glutamic acid and aspartic acid are the starting materials for the energy acquisition pathway, and energy sources are acquired through a metabolic process to synthesize short-chain fatty acids such as acetic acid, propionic acid and butyric acid [Citation30]. It has also been reported that metabolites of the glycogenic pathway are synthesized by glycine, serine, alanine, glutamic acid and aspartic acid as starting materials, after which nucleic acid and polysaccharide are synthesized through the pentose phosphate pathway, and nucleoside sugar and sugar amino acid metabolism [Citation31]. Therefore, a decrease in the supply of amino acids has effects on the growth of P. gingivalis. In the present study, metabolites involved in glycogenesis, the pentose phosphate pathway and the nucleoside-sugar and sugar-amino acid metabolism pathways that start from amino acids were found to be decreased in curcumin-treated P. gingivalis (), while purine and pyrimidine metabolic pathway metabolites synthesized from those metabolites were also decreased. These results suggest that curcumin inhibits bacterial growth by reducing the supply of glycogenic amino acids, which are a nutrient source, thereby preventing synthesis of various downstream metabolites.
Figure 8. Metabolic profile of central metabolism in P. gingivalis treated with curcumin. Fluctuations of mean values of metabolites as compared to the control are visualized (green horizontal arrows: 0.8 < relative value < 1.2, light blue lower arrows: 0.5 < relative value ≤ 0.8, blue lower arrows: relative value ≤ 0.5). Metabolic enzymes are indicated by arrows and EC numbers are shown between each metabolite.
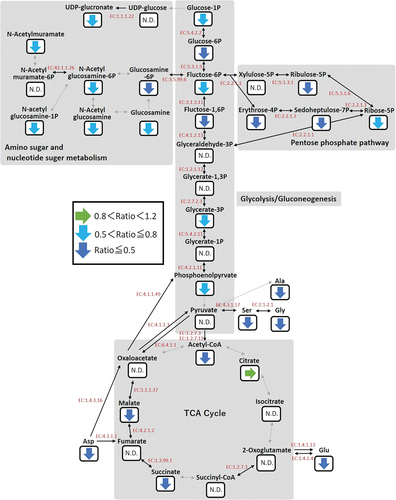
Various bacterial amino acids have been found to be decreased in quantity by curcumin, whereas several amino acids are increased. As for tryptophan, P. gingivalis possesses a gene (PGN_0880) encoding a tryptophanase (EC: 4.1.99.1) that synthesizes indole from tryptophan. Also, enzyme activities have been reported to be controlled by the concentration of pyridoxal-5-phosphate as a cofactor [Citation32]. A decrease in pyridoxal-5’-phosphate by curcumin was noted in the present results. Since the biosynthetic pathway of pyridoxal-5’-phosphate in P. gingivalis is assumed to be via the pentose phosphate pathway [Citation33], tryptophan likely accumulates by reducing the activity of tryptophanase, which is induced by decreases in pyridoxal-5’-phosphate and metabolites of the pentose phosphate pathway caused by curcumin (Figure S1). Metabolomic pathways starting from phenylalanine have not been reported for P. gingivalis. The pathogen expresses a Lys E family transporter that discharges metal ions and neutral amino acids (PGN_0861) [Citation34]; thus, it is possible that curcumin inhibits the amino acid transporter, while some amino acids that can be utilized as secondary metabolites become accumulated in P. gingivalis. As for arginine, it is thought to be a factor in polyamine metabolism, as P. gingivalis has a number of enzymes that synthesize polyamines from arginine. Findings in the present study related to the polyamine metabolic pathway showed that agmatine and putrescine, located downstream of arginine, were increased by curcumin, while N-acetylputrescine was decreased (Figure S2). Diamine N-acetyltransferase (EC: 2.3.1.57) catalyzes the reaction from putrescine and acetyl-CoA to N-acetylputrescine. However, as shown in , curcumin-induced reduction of acetyl-CoA resulted in suppression of N-acetylputrescine synthesis and accumulation of putrescine. Furthermore, it has been reported that plant and bacterial agmatine deiminase (EC: 3.5.3.12) and arginine decarboxylase (EC 4.1.1.19) are inhibited by putrescine and agmatine [Citation35–37]. Therefore, arginine and agmatine in P. gingivalis treated with curcumin become accumulated by feedback inhibition. Recently, polyamines have been reported to be associated with bacterial stress response and pathogenicity [Citation38]. Also, putrescine produced by Fusobacterium nucleatum was shown to enhance biofilm formation by P. gingivalis [Citation39]. On the other hand, P. gingivalis biofilm formation was reported to be inhibited by curcumin [Citation14]. This contradiction may be because of suppression of adhesion factors in P. gingivalis by curcumin [Citation40]. It will be necessary to further examine the effects of polyamine metabolism fluctuation caused by curcumin on stress responses and infectivity.
Results of the present metabolomic analysis indicate that curcumin inhibits the growth of P. gingivalis by nutrient starvation, induced by inhibition of amino acid uptake. Eubacteria respond to various stresses by regulating the expression of the sigma factor, a subunit of RNA polymerase [Citation25]. In E. coli, the expression of ‘sigma-38’ was shown to be increased under nutrient starvation and in response to starvation stress by competing with ‘sigma-70’ [Citation41]. Although P. gingivalis has a sigma-70 gene (rpoD : PGN_0638), it does not possess a gene homologous to sigma-38. Therefore, it is suggested that another sigma factor has a role in starvation stress response. P. gingivalis possesses a sigma-54 gene (rpoN : PGN_1202), and rpoN has been shown to have a relationship to regulation of amino acids, as nitrogen sources, in Escherichia coli as well as other bacteria [Citation25]. Therefore, the mRNA expressions of PGN_0638 and PGN_1202 in P. gingivalis cultured under a low nutrient condition and with curcumin treatment were examined in the present study. Those results showed that the expression level of each was decreased by starvation stress and treatment of curcumin, suggesting that curcumin-treated P. gingivalis may have a condition similar to a low nutrient state in regard to expression level. In contrast to findings of other bacteria previously reported, the expression levels of both sigma-54 and sigma-70 in P. gingivalis were decreased by nutrient starvation. However, PGN_1202 in P. gingivalis is an essential gene, different from other bacteria [Citation42], while another report noted that the amount of sigma-54 in Pseudomonas putida was not changed under a nitrogen starvation stress condition [Citation43]. Although the functions of sigma factors remain largely unknown, results of those previous studies as well as the present suggest that P. gingivalis sigma factors have unique functions.
The present findings indicate that curcumin has effects on enzymatic activities involved in amino acid uptake by P. gingivalis. The pathogen degrades proteins (polypeptides) by gingipain, a protease localized on the outer membrane, and generated oligopeptides are degraded to dipeptides by DPPs, which are localized in periplasm, then dipeptides are degraded to amino acids or directly used for various metabolic activities [Citation44]. We previously reported that curcumin inhibits the activity of gingipain [Citation14], though it is thought that curcumin has effects on other target enzymes, because growth of the KDP136 strain was also inhibited by curcumin in the present study (Figure S3). Therefore, degradation of oligopeptides by DPPs received focus in the present experiments. A previous study found that the activities of some DPPs in P. gingivalis could be measured by the use of MCA-labeled peptides [Citation27], thus DPPs treated with curcumin were examined, which showed inhibition of the activities of DPP4, DPP5, DPP7 and DPP11. A recent report noted that the DPPs examined in the present study are able to recognize amino acids in oligopeptides such as proline (DPP4), hydrophobic amino acids and alanine (DPP5), hydrophobic amino acid (DPP7) and aspartic acid or glutamic acid (DPP11) [Citation44]. Analysis of the metabolomic profile of P. gingivalis after treatment with curcumin showed that all of those amino acids were reduced. In addition, another study reported that curcumin binds to the active site of human DPP-4 and was found to directly inhibit enzyme activity in in silico and in vitro experiments [Citation45]. Since curcumin is a relatively small molecule (molecular weight: 368.4), it is capable of penetration into the periplasm, thus suggesting an ability to inhibit the enzymatic activity of DPPs.
To confirm that inhibition of DPP activities leads to growth inhibition of P. gingivalis, the effects of DPP inhibitors were assessed in the present investigation. A previous study reported findings obtained with some human DPP-4 inhibitors [Citation46]. Sitagliptin monophosphate is considered to be a typical inhibitor and used for diabetes treatment [Citation47], while Val-boroPro has been reported to have inhibitory activities against human DPP-8, DPP-9 and PEP, as well as human DPP-4, and considered to show a broad spectrum of effects [Citation48]. P. gingivalis DPP4 has a structure similar to that of human DPP-4, though it has an approximately 32% amino acid sequence homology, and inhibitory activity with use of inhibitors has also been reported [Citation49]. As a result, the growth inhibition effects of a DPP-4 inhibitor against P. gingivalis and S. oralis, oral indigenous bacteria, were examined. The results showed that sitagliptin monophosphate and Val-boroPro at 5000 and 2500 µg/mL inhibited the growth of P. gingivalis, and both inhibited the growth of S. oralis at 10,000 µg/mL. As for curcumin, P. gingivalis growth was inhibited at 10 µg/mL, while that of S. oralis was inhibited at 125 µg/mL. Therefore, the DPP-4 inhibitors and curcumin showed similar effects on growth inhibition. Furthermore, we examined the effects of DPP-4 inhibitors on the activity of DPPs. The results showed that sitagliptin phosphate significantly reduced the activity of DPP4, DPP5 and DPP7, while Val-boroPro also reduced the activity of DPP4, DPP5, DPP7 and DPP11. These results indicate that these DPP-4 inhibitors suppress the growth of P. gingivalis by inhibiting the activity of DPPs, especially DPP4, DPP5 and DPP7.
Results obtained in this study suggest a novel anti-bacterial mechanism of curcumin. It was previously reported that curcumin reduces the uptake of dipeptides and amino acids possessed by P. gingivalis by inhibiting the activity of gingipain [Citation14], while the present findings showed that they also inhibit the activities of DPPs. Decreases in those induce the fluctuation of metabolites, such as the glycosylation pathway, energy acquisition pathway, pentose phosphate pathway, nucleoside sugars, glyco-amino acids, purine metabolism and pyrimidine metabolisms. Therefore, a decrease in metabolites necessary for proliferation may lead to growth suppression. On the other hand, bacteria such as Streptococcus possess metabolic pathways that start from sugars and do not depend on proteins for energy acquisition [Citation50], thus curcumin is thought to have little effect on growth inhibition. Nevertheless, it is difficult to compare the anti-bacterial effects of curcumin, because the solubilization and purification conditions have been found to differ [Citation51,Citation52]. Accordingly, a future investigation of the selective anti-bacterial effects of curcumin on a variety of bacteria using unified test systems will be necessary for accurate comparisons.
Turmeric, which contains a large amount of curcumin, is used as a spice and has abundant dietary applications, thus is considered to be safe and functional for treatment in the oral cavity. It is expected that details regarding the anti-bacterial effects of curcumin revealed by the present study will contribute to development of oral care agents for prevention of periodontal disease.
Conclusions
Curcumin inhibits P. gingivalis DPPs activity and also interferes with nucleic acid synthesis and central metabolic pathways, beginning with amino acid metabolism, with those fluctuations inducing starvation and growth inhibition. Although additional studies are necessary, the present results provide important details regarding the anti-bacterial mechanism of curcumin.
Authors’ contributions statement
H.M., M.Ku. and A.A. designed the study, main conceptual ideas, and proof outline. M.Ku., M.Ka., and H.M. collected the data. H.M., M.Ku., and A.A. analyzed the data. M.Ka., R.M., and Y.H. aided in interpreting the results. A.A. supervised the project. H.M. wrote the manuscript, with support from M.Ku. and A.A. All authors discussed the results and commented on the manuscript.
FigureS3.jpg
Download JPEG Image (58.6 KB)FigureS2.jpg
Download JPEG Image (87.3 KB)FigureS1.jpg
Download JPEG Image (99 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/20002297.2024.2373040
Additional information
Funding
References
- Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62(1):59–11. doi: 10.1111/j.1600-0757.2012.00457.x
- Bui FQ, Almeida-da-Silva CLC, Huynh B, et al. Association between periodontal pathogens and systemic disease. Biomed J. 2019;42(1):27–35. doi: 10.1016/j.bj.2018.12.001
- Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and Pathobionts. Trends Microbiol. 2016;24(6):477–489. doi: 10.1016/j.tim.2016.02.010
- de Sousa Et, de Araújo Jsm, Pires AC, et al. Local delivery natural products to treat periodontitis: a systematic review and meta-analysis. Clin Oral Investig. 2021;25(7):4599–4619. doi: 10.1007/s00784-021-03774-2
- Fuloria S, Mehta J, Chandel A, et al. A comprehensive review on the therapeutic potential of curcuma longa Linn. In relation to its major active constituent curcumin. Front Pharmacol. 2022;13:820806. doi: 10.3389/fphar.2022.820806
- Motterlini R, Foresti R, Bassi R, et al. Curcumin, an antioxidant and anti-inflammatory agent, induces heme oxygenase-1 and protects endothelial cells against oxidative stress. Free Radic Biol Med. 2000;28(8):1303–1312. doi: 10.1016/s0891-5849(00)00294-x
- Dehzad MJ, Ghalandari H, Nouri M, et al. Antioxidant and anti-inflammatory effects of curcumin/turmeric supplementation in adults: a GRADE-assessed systematic review and dose-response meta-analysis of randomized controlled trials. Cytokine. 2023;164:156144. doi: 10.1016/j.cyto.2023.156144
- Chainani-Wu N. Safety and anti-inflammatory activity of curcumin: a component of tumeric (curcuma longa). J Altern Complement Med. 2003;9(1):161–168. doi: 10.1089/107555303321223035
- Peng Y, Ao M, Dong B, et al. Anti-inflammatory effects of curcumin in the inflammatory diseases: status, limitations and countermeasures. Drug Des Devel Ther. 2021;15:4503–4525. doi: 10.2147/DDDT.S327378
- Norris L, Karmokar A, Howells L, et al. The role of cancer stem cells in the anti-carcinogenicity of curcumin. Mol Nutr Food Res. 2013;57(9):1630–1637. doi: 10.1002/mnfr.201300120
- Mansouri K, Rasoulpoor S, Daneshkhah A, et al. Clinical effects of curcumin in enhancing cancer therapy: A systematic review. BMC Cancer. 2020;20(1):791. doi: 10.1186/s12885-020-07256-8
- Yun DG, Lee DG. Antibacterial activity of curcumin via apoptosis-like response in Escherichia coli. Appl Microbiol Biotechnol. 2016;100(12):5505–5514. doi: 10.1007/s00253-016-7415-x
- Gunes H, Gulen D, Mutlu R, et al. Antibacterial effects of curcumin: an in vitro minimum inhibitory concentration study. Toxicol Ind Health. 2016;32(2):246–250. doi: 10.1177/0748233713498458
- Izui S, Sekine S, Maeda K, et al. Antibacterial activity of curcumin against periodontopathic bacteria. J Periodontol. 2016;87(1):83–90. doi: 10.1902/jop.2015.150260
- Izui S, Sekine S, Murai H, et al. Inhibitory effects of curcumin against cytotoxicity of Porphyromonas gingivalis outer membrane vesicles. Arch Oral Biol. 2021;124:105058. doi: 10.1016/j.archoralbio.2021.105058
- Gottumukkala SN, Koneru S, Mannem S, et al. Effectiveness of sub gingival irrigation of an indigenous 1% curcumin solution on clinical and microbiological parameters in chronic periodontitis patients: a pilot randomized clinical trial. Contemp Clin Dent. 2013;4(2):186–191. doi: 10.4103/0976-237X.114874
- Muglikar S, Patil KC, Shivswami S, et al. Efficacy of curcumin in the treatment of chronic gingivitis: a pilot study. Oral Health Prev Dent. 2013;11(1):81–86. doi: 10.3290/j.ohpd.a29379
- Bhatia M, Urolagin SS, Pentyala KB, et al. Novel therapeutic approach for the treatment of periodontitis by curcumin. J Clin Diagn Res. 2014;8(12):ZC65–9. doi: 10.7860/JCDR/2014/8231.5343
- Tyagi P, Singh M, Kumari H, et al. Bactericidal activity of curcumin I is associated with damaging of bacterial membrane. PLOS ONE. 2015;10(3):e0121313. doi: 10.1371/journal.pone.0121313
- Rai D, Singh JK, Roy N, et al. Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem J. 2008;410(1):147–155. doi: 10.1042/BJ20070891
- Shahzad M, Millhouse E, Culshaw S, et al. Combet E. Selected dietary (poly)phenols inhibit periodontal pathogen growth and biofilm formation. Food Funct. 2015;6(3):719–729. doi: 10.1039/c4fo01087f
- Hashino E, Kuboniwa M, Alghamdi SA, et al. Erythritol alters microstructure and metabolomic profiles of biofilm composed of streptococcus gordonii and porphyromonas gingivalis. Mol Oral Microbiol. 2013;28(6):435–451. doi: 10.1111/omi.12037
- Amano A, Nakagawa I, Kataoka K, et al. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37(5):1426–1430. doi: 10.1128/JCM.37.5.1426-1430.1999
- McClellan DL, Griffen AL, Leys EJ. Age and prevalence of porphyromonas gingivalis in children. J Clin Microbiol. 1996;34(8):2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996
- Wösten MM. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22(3):127–150. doi: 10.1016/S0168-6445(98)00011-4
- James CE, Hasegawa Y, Park Y, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74(7):3834–3844. doi: 10.1128/IAI.01768-05
- Ohara-Nemoto Y, Rouf SM, Naito M, et al. Identification and characterization of prokaryotic dipeptidyl-peptidase 5 from Porphyromonas gingivalis. J Biol Chem. 2014;289(9):5436–5448. doi: 10.1074/jbc.M113.527333
- Yamasaki T, Nagata A, Kiyoshige T, et al. Black-pigmented, asaccharolytic bacteroides species resembling porphyromonas gingivalis (bacteroides gingivalis) from beagle dogs. Oral Microbiol Immunol. 1990;5(6):332–335. doi: 10.1111/j.1399-302x.1990.tb00436.x
- Shah HN, Williams RAD. Utilization of glucose and amino acids by Bacteroides intermedius and Bacteroides gingivalis. Curr Microbiol. 1987;15(5):241–246. doi: 10.1007/BF01589374
- Takahashi N, Sato T, Yamada T. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by porphyromonas gingivalis. J Bacteriol. 2000;182(17):4704–4710. doi: 10.1128/JB.182.17.4704-4710.2000
- Mazumdar V, Snitkin ES, Amar S, et al. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191(1):74–90. doi: 10.1128/JB.01123-08
- Yoshida Y, Sasaki T, Ito S, et al. Identification and molecular characterization of tryptophanase encoded by tnaA in Porphyromonas gingivalis. Microbiology (Reading). 2009;155(Pt 3):968–978. doi: 10.1099/mic.0.024174-0
- Kuboniwa M, Houser JR, Hendrickson EL, et al. Metabolic crosstalk regulates porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2017;2(11):1493–1499. doi: 10.1038/s41564-017-0021-6
- Tsu BV, Saier MH, Rokas A. The LysE superfamily of transport proteins involved in cell physiology and pathogenesis. PLOS ONE. 2015;10(10):e0137184. doi: 10.1371/journal.pone.0137184
- Sindhu RK, Desai HV. Purification and properties of agmatine iminohydrolase from groundnut cotyledons. Phytochemistry. 1979;18(12):1937–1938. doi: 10.1016/S0031-9422(00)82706-5
- Smith TA. Arginine decarboxylase of oat seedlings. Phytochemistry. 1979;18(9):1447–1452. doi: 10.1016/S0031-9422(00)98473-5
- Balasundaram D, Tyagi AK. Modulation of arginine decarboxylase activity from mycobacterium smegmatis. Eur J Biochein. 1989;183(2):339–345. doi: 10.1111/j.1432-1033.1989.tb14934.x
- Gevrekci A. The roles of polyamines in microorganisms. World J Microbiol Biotechnol. 2017;33(11):204. doi: 10.1007/s11274-017-2370-y
- Sakanaka A, Kuboniwa M, Shimma S, et al. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems. 2022;7(4):e0017022. doi: 10.1128/msystems.00170-22
- Kumbar VM, Peram MR, Kugaji MS, et al. Effect of curcumin on growth, biofilm formation and virulence factor gene expression of Porphyromonas gingivalis. Odontology. 2021;109(1):18–28. doi: 10.1007/s10266-020-00514-y
- Nandy P. The role of sigma factor competition in bacterial adaptation under prolonged starvation. Microbiology (Reading). 2022;168(5). doi: 10.1099/mic.0.001195
- Hutcherson JA, Gogeneni H, Yoder-Himes D, et al. Comparison of inherently essential genes of Porphyromonas gingivalis identified in two transposon-sequencing libraries. Mol Oral Microbiol. 2016;31(4):354–364. doi: 10.1111/omi.12135
- Jurado P, Fernandez LA, de Lorenzo V. Sigma 54 levels and physiological control of the Pseudomonas putida Pu promoter. J Bacteriol. 2003;185(11):3379–3383. doi: 10.1128/JB.185.11.3379-3383.2003
- Nemoto TK, Ohara NY. Dipeptidyl-peptidases: Key enzymes producing entry forms of extracellular proteins in asaccharolytic periodontopathic bacterium porphyromonas gingivalis. Mol Oral Microbiol. 2021;36(2):145–156. doi: 10.1111/omi.12317
- Huang PK, Lin SR, Chang CH, et al. Natural phenolic compounds potentiate hypoglycemia via inhibition of dipeptidyl peptidase IV. Sci Rep. 2019;9(1):15585. doi: 10.1038/s41598-019-52088-7
- Gallwitz B. Clinical Use of DPP-4 Inhibitors. Front Endocrinol. 2019;10:389. doi: 10.3389/fendo.2019.00389
- Gallwitz B. Sitagliptin: profile of a novel DPP-4 inhibitor for the treatment of type 2 diabetes (update). Drugs Today (Barc). 2007;43(11):801–814. doi: 10.1358/dot.2007.43.11.1157620
- Lankas GR, Leiting B, Roy RS, et al. Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes. 2005;54(10):2988–2994. doi: 10.2337/diabetes.54.10.2988
- Rea D, Van Elzen R, De Winter H, et al. Crystal structure of Porphyromonas gingivalis dipeptidyl peptidase 4 and structure-activity relationships based on inhibitor profiling. Eur J Med Chem. 2017;139:482–491. doi: 10.1016/j.ejmech.2017.08.024
- Takahashi N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J Dent Res. 2015;94(12):1628–1637. doi: 10.1177/0022034515606045
- Zheng D, Huang C, Huang H, et al. Antibacterial mechanism of curcumin: a review. Chem Biodivers. 2020;17(8):e2000171. doi: 10.1002/cbdv.202000171
- Hussain Y, Alam W, Ullah H, et al. Antimicrobial Potential of Curcumin: therapeutic potential and challenges to clinical applications. Antibiotics (Basel). 2022;11(3):322. doi: 10.3390/antibiotics11030322
