ABSTRACT
Background
Gingipains are important virulence factors present in Porphyromonas gingivalis. Arginine-specific gingipains (RgpA and RgpB) are critically associated with increased proteolytic activity and immune system dysfunction, including neutrophilic activity. In this study, we assessed the impact of gingipains (RgpA and RgpB) on neutrophil function.
Methods
Peripheral blood samples were obtained; neutrophils were isolated and incubated with P. gingivalis A7436, W50, and the double RgpA/RgpB double knockout mutant E8 at MOI 20 for 2 hours. Neutrophil viability was assessed by Sytox staining. Phagocytic capacity and apoptosis were measured by flow cytometry. Superoxide release was measured by superoxide dismutase and cytochrome c reduction assay. Gene expression of TLR2, p47-phox, p67-phox, and P2 × 7was measured by qPCR. Inflammatory cytokine and chemokine production was measured by IL-1β, IL-8, RANTES, and TNF-α in cell supernatants.
Results
Neutrophil TLR2 gene expression was reduced in the absence of RgpA/RgpB (p < 0.05), while superoxide production was not significantly impacted. RgpA/RgpB−/− significantly impaired neutrophil phagocytic function (p < 0.05) and increased TNF-α production when compared with the wild-type control (p < 0.05). Neutrophil apoptosis was not altered when exposed to RgpA/RgpB−/− E8 (p > 0.05).
Conclusion
These data suggest that arginine-specific gingipains (RgpA/RgpB) can modulate neutrophil responses against P. gingivalis infection.
Summary of key findings
P. gingivalis-derived arginine-specific gingipains impaired the phagocytic and apoptotic function in neutrophils.
Introduction
Periodontitis is a chronic multifactorial inflammatory disease associated with a dysbiotic biofilm causing tooth loss [Citation1]. Porphyromonas gingivalis is a Gram-negative, non-motile, proteolytic, obligatory anaerobic species that forms black-pigmented colonies on blood agar plates and requires iron under the form of hemin to grow [Citation2–5]. P. gingivalis dysregulates innate immunity pathways in a susceptible host and facilitates an increase in overall community biomass [Citation6]. P. gingivalis and host interaction is mediated by virulence factors that activate cell receptors, eliciting the immune response [Citation7]. Gingipains are among the most important virulence factors of P. gingivalis [Citation8,Citation9]. The proteolytic activity of the gingipains is aimed at bacterial access nutrients [Citation10], stimulation of host’s cytokines [Citation11–13], and regulation of receptors [Citation14,Citation15] and components of the complement system [Citation16]. The gingipain family comprises three related cysteine proteases that hydrolyze peptide bonds at the carbonyl groups of arginine (Arg-Xaa) and lysine residues (Lys-Xaa) [Citation17]. The Lys-specific gingipain (Kgp), encoded by the Kgp gene [Citation18], degrades host proteins and tissues, aids in immune evasion, enhances bacterial adherence to host cells and tissues, and facilitates iron acquisition and the maturation of other virulence factors [Citation19–21]. RgpA and RgpB, encoded by the RgpA and RgpB genes, are Arg-Xaa gingipains, that participate in many of the pathological effects of gingipains associated with periodontal destruction, such as increased collagenase activity [Citation22,Citation23], activation of matrix metalloproteinase 2 [Citation24], increased RANKL/OPG ratio [Citation13], and alveolar bone loss [Citation25,Citation26] are activated by the Arg-Xaa group [Citation27].
Neutrophils are the most prevalent cells in the gingival crevicular fluid in periodontitis, playing a pivotal role in periodontal health maintenance and the defense against microorganisms and virulence factors [Citation28]. Neutrophil numbers in the periodontal tissues are maintained with a gradient of interleukin (IL)-8 produced by the gingival epithelium and other neutrophils. This equilibrium can be disrupted when IL-8 is digested by Rgps [Citation11] and P. gingivalis serine phosphatase [Citation29]. Rgps also down-regulate and degrade intercellular adhesion molecule (ICAM)-1, preventing a transient neutrophil migration into gingival tissues, allowing the development of mature pathogenic biofilm, and inciting inflammation [Citation30]. Consequently, inflammation leads to heightened recruitment and activity of neutrophil functions such as superoxide production, phagocytosis, and cytokine release, contributing to periodontal destruction [Citation31].
Although neutrophils initially follow the IL-8 gradient, in infected or inflamed sites, they move towards chemo-attractants derived from bacteria or local activation of the complement system [Citation32]. Interestingly, RgpA regulates the cleavage of C3 into C3b and C5 into C5a, both of which are potent chemotactic factors favoring leukocytic infiltration [Citation10,Citation33]. Neutrophils are ‘tagged’ by Rgps to confuse the recognition signals of phagocytosis by macrophages, which in turn results not only in the enhancement of the dying cells clearance but also stimulation of the uptake of healthy and functional neutrophils by macrophages [Citation34], indicating that P. gingivalis has developed a pathway for subverting the host defense dependent on neutrophils [Citation34,Citation35].
Since neutrophils and P. gingivalis play a key role in the pathogenesis of periodontal disease, mapping specific neutrophil functions affected by Arg-gingipains from P. gingivalis is crucial for understanding the disease. In this study, we tested the hypothesis that RgpA and RgpB regulate the oxidative stress response, viability, and phagocytic and apoptotic functions in neutrophils.
Materials and methods
Ethical compliance
Informed consent was obtained from all subjects, and ethical approval was granted by the Forsyth Institutional Review Board (protocol number 11–03) before collecting patient blood samples at the Forsyth Institute Center for Clinical and Translational Research.
P. gingivalis culture
The study design is shown in . P. gingivalis wild-type strains A7436 and W50 were cultured in blood agar plates and individually stored in an anaerobic chamber (5% CO2, 10% H2, and 85% N2 atmosphere) at 37°C. The colonies were re-plated every three days and 24 hours before each experiment; bacterial suspensions were prepared in BHI broth* from the primary culture at their log phase of growth. Concentrations were determined by an optical density of 600 nm# corresponding to 1 × 109 bacteria/mL. To test the role of Rgp’s, RgpA/RgpB double mutant E8 was obtained from Dr. J. Aduse-Opoku (Queen Mary’s School of Medicine and Dentistry, London, UK) and grown as above.
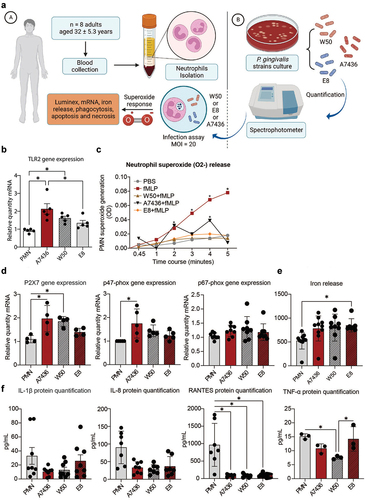
Figure 1. (a) Human neutrophil isolation and experimental workflow with P. gingivalisstrains. (b) TLR2 gene expression. (c) Time course of the fMLP-induced O2- production by human neutrophils (0.5 × 106) in response to P. gingivalis strains compared with fMLP stimulation alone. PBS was used as a negative control (n = 8). *: p < 0.05 statistical difference when compared with baseline PBS. (d) Gene expression of P2X7, p47-phox, and p67-phox in response to different P. gingivalis strains. *: p < 0.05 statistical difference when compared with the control group. #: p < 0.05: statistical difference when compared with W50 group. (e) Iron release optical density (565 nm). *: p < 0.05 statistical difference when compared with the control group. (f) Levels of IL-1β, IL-8, RANTES, and TNF-α (pg/mL) * p < 0.05 statistical difference from PMN. # p < 0.05 statistical difference from E8. Panel a created with BioRender. One-way ANOVA followed by Tukey’s posthoc test was performed for graphs on B, D, E and F. Two-way ANOVA followed by Tukey’s posthoc test was performed on C.
Neutrophil isolation
Donors were nonsmokers and did not present any systemic or periodontal disease. Their average age was 32 (±5.3) years, with a gender distribution of 3 females and 5 males. Approximately 50 mL of peripheral venous blood samples were collected. The samples were placed into vacutainer tubes containing 25 units/ml heparin. Polymorphonuclear neutrophils were isolated by gradient centrifugation as described [Citation36].
Superoxide assay
Neutrophils (0.5 × 105) were infected with P. gingivalis A7436, W50 or E8 at a multiplicity of infection (MOI) of 20. Superoxide production (O [Citation2-] production was monitored in a spectrophotometer (VMax, SpectraMax 340PC, Molecular Devices, Sunnyvale, CA) at 37°C by reducing Cytochrome C at an absorbance of 550 nm. Superoxide dismutase was used as a negative control, and N-fMLP‖ (N-formyl-methionine-leucine-phenylalanine) at 1 µM was used as a positive control. Superoxide production was evaluated linearly and expressed in O [Citation2]/min/cell.
Viability, phagocytosis, and apoptosis
Porphyromonas gingivalis strains isolates were cultured overnight in Brain Heart Infusion (BHI) broth (NutriSelect® Plus, Millipore, Burlington, MA). Post-cultivation, the isolates were subjected to three successive washes with sterile phosphate-buffered saline (PBS) and resuspended to a final concentration of 1 × 10^9 cells/mL in PBS. This concentration was quantified using a spectrophotometer (BioPhotometer, Eppendorf, Hamburg, Germany) at an optical density of 660 nm. For opsonization, P. gingivalis were incubated with human serum (H4522, Millipore, Burlington, MA) at 37°C for 30 minutes under shaking conditions and labeled with fluorescein isothiocyanate (FITC; 100 mg/mL in 1× PBS) to allow detection upon phagocytosis by PMNs. Subsequently, the labeled P. gingivalis were incubated with neutrophils at a MOI of 20 in triplicate at 37°C. Apoptotic cell detection was performed using the CellEvent™ Caspase-3/7 Green Flow Cytometry Assay Kit (ThermoFisher Scientific, Waltham, MA) following the manufacturer’s protocol. Additionally, Sytox® (ThermoFisher Scientific, Waltham, MA) was utilized to identify damaged cells. After a 2-hour incubation period, samples were washed with PBS and resuspended in 500 μL of PBS for flow cytometric analysis. The analysis was conducted using a FACScan flow cytometer (BD Bioscience, San Jose, CA) with excitation at 525 nm and detection using a 530/30 nm bandpass filter.
Expression of TLR-2, p47-phox and P2X7
Following infection, polymorphonuclear leukocytes (PMNs) were lysed using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA (tRNA) was quantified, and 1 µg was used for reverse transcription (RT) with a high-capacity cDNA RT kit (Applied Biosystems, Foster City, CA, USA) in 30 µL reactions. The RT conditions were set to 25°C for 10 minutes, 37°C for 60 minutes, and 95°C for 5 minutes. Quantitative real-time PCR (qPCR) was performed using TaqMan fast advanced master mix (Applied Biosystems) with pre-designed probes targeting TLR-2, P2X7, p47-phox, p67-phox, and β-actin (TaqMan, Applied Biosystems, Thermo Fisher Scientific, Waltham, MA). The NCBI Reference Sequences utilized were TLR-2 (NM_001318787.2), P2×7(NM_002562.5), p47-phox (NR_110044.1), p67-phox (NM_000433.3), and β-actin (BNM_001101.3). For each qPCR reaction, 10 ng of cDNA (50 ng/µL) were used in a 20 µL volume, performed in triplicate using a standard 96-well format. The reactions were carried out in a StepOnePlus real-time PCR system (Applied Biosystems) with the following thermal cycling conditions: 50°C for 2 minutes, 95°C for 20 seconds, followed by 40 cycles of 95°C for 1 second and 60°C for 20 seconds. The cycle threshold (CT) values were determined using StepOnePlus Software v2.3 (Applied Biosystems). Data were analyzed using the 2-ΔΔct method, and TLR-2, P2X7, p47-phox, and p67-phox mRNA levels were calculated by comparing with corresponding control samples running simultaneously, each normalized to β-actin mRNA as endogenous controls.
Cytokine generation
To measure cytokine release by neutrophils in response to gingipains, cells were infected with P. gingivalis A7436, W50, or E8, incubated for 2 hours at 37°C, and a relative humidity of about 95% before collecting the supernatants. All assays were performed on 100 mL of culture supernatant using four multiplex magnetic bead panels on a flexible laser analyzer platform (Luminex 200, Luminex, Austin, TX). IL-1β, IL-8, RANTES, and TNF-α were measured by a human cytokine/chemokine panel (Millipore, MILLIPLEX, Billerica, MA) with no dilution. All assays were performed following the manufacturer’s protocol [Citation37] and analyzed (Bio-Plex Manager, Version 5.0, Bio-Rad, Hercules, CA).
Iron release
To measure the iron (Fe2+) released by neutrophils in response to gingipains, cells (1 × 106) were infected with P. gingivalis A7436, W50, or E8 at MOI = 20. After 2 hours, supernatants were harvested and transferred to a 96-well plate. Iron was evaluated using QuantiChromTM Iron Assay Kit (DIFE-250) following the manufacturer’s protocol (BioAssay Systems, Hayward, CA).
Statistical analyses
All raw data were used in averages of at least three experiments repeated in triplicate. After statistical analysis, data were displayed in mean and standard error. All analyses were performed using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. In the case of non-parametric data, the Kruskal-Wallis test was used, followed by the Dunn-Bonferroni post hoc test in case of statistical significance. A 95% confidence level was attributed before running the statistical analysis.
Results
PMN superoxide generation and TLR2 expression under P. gingivalis challenge
To evaluate the impact of gingipain RgpA/RgpB on human PMN function, freshly isolated neutrophils were exposed to the challenge by P. gingivalis W50 or E8 (MOI = 20). P. gingivalis A7436 was used as a control (). Neutrophil TLR2 gene expression was decreased in the absence of RgpA/RgpB when compared to the A7436 (p < 0.05) (). Superoxide production was depleted to negative control levels (PBS) in the RgpA/RgpB knockdown group. The presence of RgpA/RgpB gingipains resulted in peaks of superoxide production after 2 and 4 minutes of stimulation (p < 0.05) (). There was an upregulation of P2×7and p47-phox gene expressions in the RgpA/RgpB positive strains and, conversely, a non-significant expression in the knockdown group (). Regardless of the presence of RgpA/RgpB gingipain, no impact was found on p67-phox gene expression (p > 0.05) (), suggesting an arginine-specific mechanism of P2×7and p47-phox mediated regulation of superoxide formation in neutrophils in response to gingipains.
RgpA/RgpB knockdown upregulates neutrophil-derived iron release
To check the possibility that RgpA/RgpB gingipains interfere with iron release ability from neutrophils, iron concentration was assessed after neutrophils were incubated with different P. gingivalis strains for 2 hours (). Data revealed a significant increase in iron release by neutrophils in the absence of RgpA/RgpB compared to the PMN control group (p < 0.05), suggesting gingipain-induced negative feedback on the neutrophil iron release.
Lack of RgpA/RgpB upregulates TNF-α and downregulates RANTES in a strain-specific manner
The IL-1β and IL-8 release were not dependent on RgpA/RgpB gingipain expression while RANTES gene expression was significantly decreased in response to all P. gingivalis strains (p < 0.05) (). Interestingly, TNF-α levels in the RgpA/RgpB knockdown E8 were significantly higher when compared to its strain-specific wild-type control (W50), revealing a negative TNF-α feedback response related to RgpA/RgpB absence (p < 0.05) ().
Human neutrophil phagocytic capacity is impaired in the absence of gingipains
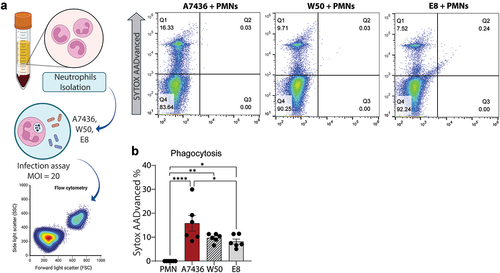
Following a two-hour incubation period, the phagocytic capacity of neutrophils in response to P. gingivalis was assessed. The lack of RgpA/RgpB notably impaired the neutrophil phagocytic potential when comparing the E8 to the A7436 (p < 0.05) (). Although no difference was found between W50 and E8, only the RgpA/RgpB knockdown significantly reduced the percentage of phagocytic events compared to the wild-type controls, suggesting the contribution of gingipains in governing neutrophil phagocytic function ().
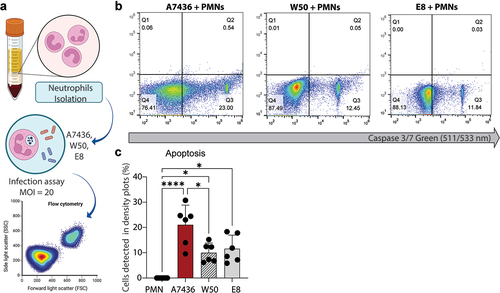
Figure 2. (a) Experimental workflow. (b) Representative flow cytometry density plots. (c) Influence of three different strains of P. gingivalis (A7436, W50, and E8) on PMN phagocytic capacity. *: p < 0.05 statistical difference when compared with the control group. #: p < 0.05: statistical difference when compared with W50 group. Panel A was created with BioRender. One-way ANOVA followed by Tukey’s posthoc test was performed.
The RgpA/RgpB profile does not regulate neutrophil apoptotic cell death
After 2 hours of incubation, apoptotic cell death was visualized in neutrophils exposed to P. gingivalis (). Data revealed no statistical significance, suggesting that apoptotic cell death was not dependent on RgpA/RgpB gingipain status ().
Figure 3. (a) Experimental workflow. (b) Results from caspase-3/7 activity assayed by measuring the fluorescence of AMC; the activity of caspase-3 was expressed in % of relative fluorescence units (RFU), which is translated to PMN apoptosis. (c) Results from Sytox staining of % PMN necrosis. Data are the mean of six independent experiments, and bars are SE. *: p < 0.05 statistical difference. Panel A was created with BioRender. One-way ANOVA followed by Tukey’s posthoc test was performed.
Discussion
P. gingivalis is a keystone etiologic agent associated with dysbiosis and disrupts the host’s innate responses in the periodontium. When neutrophil functions such as migration, peptides/enzyme production, and superoxide release are defective, tissue breakdown may be catalyzed during periodontal disease [Citation38]. Gingipains are one of the principal proteinases exerted by this bacterium. They are associated with immunoglobulin degradation, disruption of the coagulation cascade, iron caption, collagen destruction, cytokines lysing, and destruction of pro-adhesion molecules [Citation39]. RgpA/RgpB gingipains are involved in several mechanisms that shape the neutrophilic response to P. gingivalis infection. Neutrophil extracellular traps (NETs), which are networks with antimicrobial properties, are directly affected by the presence of Rgps, where a proteolytic activation of the protease-activated receptor-2 (PAR-2) takes place, inducing the malformation of NETs lacking bactericidal properties [Citation40]. We investigated whether the absence of gingipains RgpA/RgpB impacted neutrophil function and immune regulation. We found that arginine-specific gingipains (RgpA/RgpB) can modulate neutrophil functions and regulate the survival of P. gingivalis. The RgpA/RgpB absence led to an impaired phagocytic function, suggesting that gingipains play an important role in P. gingivalis survival. Our human data corroborates a murine periodontitis study demonstrating that eliminating RgpA from P. gingivalis increased neutrophil phagocytosis [Citation26]. A plausible mechanistic explanation for these findings is that Arg-gingipains can degrade components of the complement system [Citation16,Citation41,Citation42], leading to a decrease in bacterial opsonization and leukocyte phagocytosis.
Superoxide produced by neutrophils is essential for the elimination of phagocytosed microorganisms [Citation43]. Our data revealed neutrophil superoxide production was not impacted by the absence of arginine-specific gingipains. Neutrophil apoptosis and its subsequent clearance by macrophages are important events during the resolution of inflammation [Citation44]. The inability to effectively complete the efferocytic process fuels a prolonged inflammatory response that impairs resolution and subsequent events to achieve tissue homeostasis [Citation45]. In this study, neutrophil apoptosis was not impacted regardless of the RgpA/RgpB presence, suggesting that although arginine-specific gingipains can increase the number of living neutrophils, they are functionally compromised. These findings corroborate with the previous literature, where a human study found fewer apoptotic neutrophils in gingival tissue sections from periodontitis patients compared to healthy subjects [Citation46]. Further, some studies have demonstrated that P. gingivalis LPS can prolong the lifetime of neutrophils [Citation47] and may lead to diminished secretion of proapoptotic signals, resulting in persistent neutrophil migration with a delay in their physiological clearance [Citation48].
Our data demonstrated that P. gingivalis had no significant impact on PMN apoptosis independent of the presence of arginine-specific gingipains. An increased expression of TLR2 was found in both W50 and E8 groups, corroborating with studies that have demonstrated the TLR2-PI3K stimulus as a way in which P. gingivalis avoids clearance from immune cells, such as neutrophils [Citation49].
Increased number of living neutrophils could result in greater iron concentration in the microenvironment, favoring its scavenging by P. gingivalis and boosting the growth and survival of this microorganism. Iron availability regulates P. gingivalis virulence and influences its growth and survival [Citation50]. We quantified iron concentration to check whether P. gingivalis increased iron secretion in neutrophils. P. gingivalis induced neutrophil iron release regardless of arginine-specific gingipains RgpA/RgpB. Although the E8 group showed a statistically significant upregulation of iron release, the average values are comparable to those of the other strain groups. This suggests that the observed difference may not translate into a biologically relevant effect.
As P. gingivalis can degrade cytokines disrupting the immune system, we measured the IL-1β, IL-8, RANTES, and TNF-α levels. There was no significant impact on IL-1β levels, agreeing with previous research that demonstrated P. gingivalis did not affect IL-1β production by dendritic cells [Citation51]. P. gingivalis can degrade IL-8 and RANTES, essential components of the innate and adaptive immune response, independently of arginine-specific gingipains RgpA/RgpB in line with P. gingivalis proteases degrading IL-8 [Citation52]. P. gingivalis can disturb the cytokine network by eliminating a variety of cytokines [Citation53]. Our results showed that RgpA/RgpB led to TNF-α degradation in line with the literature, showing that P. gingivalis cysteine proteinases can digest TNF-α [Citation54].
Conclusion
Taken together, our data suggested that P. gingivalis can evade neutrophil phagocytosis through gingipains by multiple mechanisms, including the modulation of neutrophils function, apoptosis, and immune regulation, and thus compromise the host’s ability to eliminate infection and sustain chronic periodontal inflammation.
Supplementary 1.tif
Download TIFF Image (101.5 MB)Supplementary 2.tif
Download TIFF Image (106.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/20002297.2024.2376462.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: Consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J Periodontol. [2018 Jun 1];89(S1):S173–9. doi: 10.1002/JPER.17-0721
- Van Winkelhoff AJ, Loos BG, Van Der Reijden WA, et al. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clinic Periodontology. 2002 Nov;29(11):1023–1028. doi: 10.1034/j.1600-051X.2002.291107.x
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38(1):72–122. doi: 10.1111/j.1600-0757.2005.00113.x
- Andrian E, Grenier D, Rouabhia M. In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect Immun. 2004 Aug;72(8):4689–4698. doi: 10.1128/IAI.72.8.4689-4698.2004
- Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013 May;7(5):1016–1025. doi: 10.1038/ismej.2012.174
- Hajishengallis G. Immune evasion strategies of Porphyromonas gingivalis. J Oral Biosci. 2011;53(3):233–240. doi: 10.1016/S1349-0079(11)80006-X
- Mysak J, Podzimek S, Sommerova P, et al. Porphyromonas gingivalis: Major periodontopathic pathogen overview. J Immunol Res [Internet]. 2014 [cited 2021 Mar 12]; Available from 2014:1–8. Available from: https://pubmed.ncbi.nlm.nih.gov/24741603/
- Berker E, Kantarci A, Hasturk H, et al. Blocking proinflammatory cytokine release modulates peripheral blood mononuclear cell response to porphyromonas gingivalis. J Periodontol. 2013 Sep;84(9):1337–1345. doi: 10.1902/jop.2012.120422
- Euzebio Alves VT, da Silva HAB, de França BN, et al. Periodontal treatment downregulates protease-activated receptor 2 in human gingival crevicular fluid cells. Infect Immun. 2013 Dec;81(12):4399–4407. doi: 10.1128/IAI.01107-13
- Grenier D. Inactivation of human serum bactericidal activity by a trypsinlike protease isolated from Porphyromonas gingivalis. Infect Immun. 1992;60(5):1854–1857. doi: 10.1128/iai.60.5.1854-1857.1992
- Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from porphyromonas gingivalis: Implications for pathogenicity of periodontal disease. FEBS Lett. [1998 Dec 4];440(3):282–286. doi: 10.1016/S0014-5793(98)01461-6
- Mȩzyk-Kopeć R, Bzowska M, Potempa J, et al. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect Immun. 2005 Mar;73(3):1506–1514. doi: 10.1128/IAI.73.3.1506-1514.2005
- Belibasakis GN, Bostanci N, Hashim A, et al. Regulation of RANKL and OPG gene expression in human gingival fibroblasts and periodontal ligament cells by porphyromonas gingivalis: A putative role of the arg-gingipains. Microb Pathog. 2007 Jul;43(1):46–53. doi: 10.1016/j.micpath.2007.03.001
- Lourbakos A, Yuan YP, Jenkins AL, et al. Activation of protease-activated receptors by gingipains from porphyromonas gingivalis leads to platelet aggregation: A new trait in microbial pathogenicity. Blood. [2001 Jun 15];97(12):3790–3797. doi: 10.1182/blood.V97.12.3790
- Bostanci N, Thurnheer T, Aduse-Opoku J, et al. Porphyromonas gingivalis regulates TREM-1 in human polymorphonuclear neutrophils via its gingipains. PLOS ONE. [2013 Oct 4];8(10):75784. doi: 10.1371/journal.pone.0075784
- Popadiak K, Potempa J, Riesbeck K, et al. Biphasic effect of gingipains from porphyromonas gingivalis on the human complement system. The J Immunol. [2007 Jun 1];178(11):7242–7250. doi: 10.4049/jimmunol.178.11.7242
- Nakayama K. Domain-specific rearrangement between the two arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol Immunol. 1997;41(3):185–196. doi: 10.1111/j.1348-0421.1997.tb01189.x
- Curtis MA, Kuramitsu HK, Lantz M, et al. Molecular genetics and nomenclature of proteases of porphyromonas gingivalis. J Periodontal Res. 1999;34(8):464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x
- Li N, Collyer CA. Gingipains from porphyromonas gingivalis – complex domain structures confer diverse functions. Eur J Microbiol Immunol (Bp). 2011 Mar;1(1):41–58. doi: 10.1556/EuJMI.1.2011.1.7
- Chow YC, Yam HC, Gunasekaran B, et al. Implications of porphyromonas gingivalis peptidyl arginine deiminase and gingipain R in human health and diseases. Front Cell Infect Microbiol. 2022;12:987683. doi: 10.3389/fcimb.2022.987683
- Grenier D, Roy S, Chandad F, et al. Effect of inactivation of the arg- and/or lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect Immun. 2003 Aug;71(8):4742–4748. doi: 10.1128/IAI.71.8.4742-4748.2003
- Tokuda M, Karunakaran T, Duncan M, et al. Role of arg-gingipain a in virulence of porphyromonas gingivalis. Infect Immun. 1998;66(3):1159–1166. doi: 10.1128/IAI.66.3.1159-1166.1998
- Houle MA, Grenier D, Plamondon P, et al. The collagenase activity of porphyromonas gingivalis is due to arg-gingipain. FEMS Microbiol Lett. [2003 Apr 25];221(2):181–185. doi: 10.1016/S0378-1097(03)00178-2
- Grayson R, Douglas CWI, Heath J, et al. Activation of human matrix metalloproteinase 2 by gingival crevicular fluid and Porphyromonas gingivalis. J Clin Periodontol. 2003 Jun;30(6):542–550. doi: 10.1034/j.1600-051X.2003.00301.x
- Gibson FC, Genco CA, Tuomanen EI. Prevention of porphyromonas gingivalis-induced oral bone loss following immunization with gingipain R1. Infect Immun. 2001;69(12):7959–7963. doi: 10.1128/IAI.69.12.7959-7963.2001
- Wilensky A, Polak D, Houri-Haddad Y, et al. The role of RgpA in the pathogenicity of porphyromonas gingivalis in the murine periodontitis model. J Clin Periodontol. 2013 Oct;40(10):924–932. doi: 10.1111/jcpe.12139
- Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24(1):153–192. doi: 10.1034/j.1600-0757.2000.2240108.x
- Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31(1):55–76. doi: 10.1034/j.1600-0757.2003.03105.x
- Takeuchi H, Hirano T, Whitmore SE, et al. The serine phosphatase serb of porphyromonas gingivalis suppresses il-8 production by dephosphorylation of NF-κB RelA/p65. PLOS Pathog [Internet]. 2013 Apr [cited 2021 Mar 12];9(4). Available from e1003326. https://pubmed.ncbi.nlm.nih.gov/23637609/
- Tada H, Sugawara S, Nemoto E, et al. Proteolysis of ICAM-1 on human oral epithelial cells by gingipains. J Dent Res. [2003 Oct 12];82(10):796–801. doi: 10.1177/154405910308201007
- Kantarci A, Van Dyke TE. Neutrophil-mediated host response to porphyromonas gingivalis. J Int Acad Periodontol. [2002 Oct 1];4(4):119–125.
- Teng TS, Ji AL, Ji XY, et al. Neutrophils and immunity: From bactericidal action to being conquered. J Immunol Res [Internet]. 2017 [cited 2021 Mar 12];2017. Available from: https://pubmed.ncbi.nlm.nih.gov/28299345/1–1410.1155/2017/9671604
- Wingrove JA, DiScipio RG, Chen Z, et al. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from porphyromonas (bacteroides) gingivalis. J Biol Chem. [1992 Sep 15];267(26):18902–18907. doi: 10.1016/S0021-9258(19)37046-2
- Guzik K, Potempa J. Friendly fire against neutrophils: Proteolytic enzymes confuse the recognition of apoptotic cells by macrophages. Biochimie. 2008 Feb;90(2):405–415. doi: 10.1016/j.biochi.2007.09.008
- Guzik K, Bzowska M, Smagur J, et al. A new insight into phagocytosis of apoptotic cells: Proteolytic enzymes divert the recognition and clearance of polymorphonuclear leukocytes by macrophages. Cell Death Differ. 2007 Jan;14(1):171–182. doi: 10.1038/sj.cdd.4401927
- Kalmar JR, Arnold RR, Warbington ML, et al. Superior leukocyte separation with a discontinuous one-step ficoll-hypaque gradient for the isolation of human neutrophils. J Immunol Methods. [1988 Jun 13];110(2):275–281. doi: 10.1016/0022-1759(88)90115-9
- Browne RW, Kantarci A, LaMonte MJ, et al. Performance of multiplex cytokine assays in serum and saliva among community-dwelling postmenopausal women. Glogauer M, editor. PLOS ONE. 2013 Apr 5;8(4):e59498. doi: 10.1371/journal.pone.0059498
- Sochalska M, Potempa J. Manipulation of neutrophils by porphyromonas gingivalis in the development of periodontitis. Front Cell Infect Microbiol. 2017 May 23;7(MAY):197. doi: 10.3389/fcimb.2017.00197
- Gibson FC, Savelli J, Van Dyke TE, et al. Gingipain-specific igg in the sera of patients with periodontal disease is necessary for opsonophagocytosis of Porphyromonas gingivalis. J Periodontol. 2005 Oct;76(10):1629–1636. doi: 10.1902/jop.2005.76.10.1629
- Bryzek D, Ciaston I, Dobosz E, et al. Triggering NETosis via protease-activated receptor (PAR)-2 signaling as a mechanism of hijacking neutrophils function for pathogen benefits. PLOS Pathog [Internet]. 2019 May 1 [cited 2021 Mar 12];15(5). Available from e1007773. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1007773
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1(2):70–87. doi: 10.1159/000181144
- Wang M, Krauss JL, Domon H, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal [Internet]. 2010 Feb 16 [cited 2021 Mar 12];3(109). Available from: https://pubmed.ncbi.nlm.nih.gov/20159852/
- Zeng MY, Miralda I, Armstrong CL, et al. The roles of NADPH oxidase in modulating neutrophil effector responses. Mol Oral Microbiol. 2019 Apr 1;34(2):27–38. doi: 10.1111/omi.12252
- Jäättela M, Candé C, Kroemer G. Lysosomes and mitochondria in the commitment to apoptosis: A potential role for cathepsin D and AIF. Cell Death Differ. 2004 Feb;11(2):135–136. doi: 10.1038/sj.cdd.4401333
- Yagi M, Kantarci A, Iwata T, et al. PDK1 regulates chemotaxis in human neutrophils. J Dent Res. 2009 Dec;88(12):1119–1124. doi: 10.1177/0022034509349402
- Gamonal J, Sanz M, O’Connor A, et al. Delayed neutrophil apoptosis in chronic periodontitis patients. J Clin Periodontol. 2003 Jul;30(7):616–623. doi: 10.1034/j.1600-051X.2003.00350.x
- Murray DA, Wilton JMA. Lipopolysaccharide from the periodontal pathogen porphyromonas gingivalis prevents apoptosis of hl60-derived neutrophils in vitro. Infect Immun. 2003 Dec;71(12):7232–7235. doi: 10.1128/IAI.71.12.7232-7235.2003
- Zaric S, Shelburne C, Darveau R, et al. Impaired immune tolerance to porphyromonas gingivalis lipopolysaccharide promotes neutrophil migration and decreased apoptosis. Infect Immun. 2010 Oct;78(10):4151–4156. doi: 10.1128/IAI.00600-10
- Makkawi H, Hoch S, Burns E, et al. Porphyromonas gingivalis stimulates TLR2-PI3K signaling to escape immune clearance and induce bone resorption independently of MyD88. Front Cell Infect Microbiol [Internet]. 2017 Aug 8 [cited 2021 Mar 12];7(AUG). Available from: https://pubmed.ncbi.nlm.nih.gov/28848717/10.3389/fcimb.2017.00359
- Smalley JW, Olczak T. Heme acquisition mechanisms of porphyromonas gingivalis – strategies used in a polymicrobial community in a heme-limited host environment. Mol Oral Microbiol. 2017 Feb 1;32(1):1–23. doi: 10.1111/omi.12149
- Abdi K, Chen T, Klein BA, et al. Mechanisms by which porphyromonas gingivalis evades innate immunity. PLoS One [Internet]. 2017 Aug 1 [cited 2021 Mar 12];12(8). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5542538/
- Zhang J, Dong H, Kashket S, et al. IL-8 degradation by porphyromonas gingivalis proteases. Microb Pathog. 1999;26(5):275–280. doi: 10.1006/mpat.1998.0277
- Stathopoulou PG, Benakanakere MR, Galicia JC, et al. The host cytokine response to porphyromonas gingivalis is modified by gingipains. Oral Microbiol And Immunol. 2009 Feb;24(1):11–17. doi: 10.1111/j.1399-302X.2008.00467.x
- Calkins CC, Platt K, Potempa J, et al. Inactivation of tumor necrosis factor-α by proteinases (gingipains) from the periodontal pathogen, porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998 Mar 20;273(12):6611–6614. doi: 10.1074/jbc.273.12.6611
