ABSTRACT
Introduction: A number of arboviruses have previously been isolated from naturally-infected East African bats, however the role of bats in arbovirus maintenance is poorly understood. The aim of this study was to investigate the exposure history of Ugandan bats to a panel of arboviruses.
Materials and methods: Insectivorous and fruit bats were captured from multiple locations throughout Uganda during 2009 and 2011–2013. All serum samples were tested for neutralizing antibodies against West Nile virus (WNV), yellow fever virus (YFV), dengue 2 virus (DENV-2), Zika virus (ZIKV), Babanki virus (BBKV), and Rift Valley fever virus (RVFV) by plaque reduction neutralization test (PRNT). Sera from up to 626 bats were screened for antibodies against each virus.
Results and Discussion: Key findings include the presence of neutralizing antibodies against RVFV in 5/52 (9.6%) of little epauletted fruit bats (Epomophorus labiatus) captured from Kawuku and 3/54 (5.6%) Egyptian rousette bats from Kasokero cave. Antibodies reactive to flaviviruses were widespread across bat taxa and sampling locations.
Conclusion: The data presented demonstrate the widespread exposure of bats in Uganda to arboviruses, and highlight particular virus-bat associations that warrant further investigation.
Introduction
Uganda has a rich history in arbovirology field and laboratory studies beginning in the early 1930’s at the Rockefeller Foundation’s Yellow Fever Research Institute (currently known as the Uganda Virus Research Institute (UVRI)). Notably, researchers at the UVRI are credited with the isolation and discovery of over 20 novel arboviruses from the country including Semliki Forest (SFV), West Nile (WNV), o’nyong-nyong (ONNV), Bunyamwera (BUNV), Bwamba (BWAV), and Zika viruses (ZIKV) [Citation1], as well as the elucidation of the sylvatic transmission cycles for both yellow fever virus (YFV) and ZIKV in Uganda [Citation2–Citation4]. Among the virus ecology studies focused on vertebrate reservoirs, research at UVRI included investigations on bats for the purposes of virus discovery [Citation5,Citation6], surveillance [Citation7,Citation8], and evaluation of pathogenicity and infectivity of potential vertebrate hosts to arthropods [Citation9]. These valuable studies demonstrated a widespread seroprevalence of arboviruses among Ugandan bats [Citation7,Citation8]. However, interpretation of these serological data is challenging due to the cross-reactivity of flavivirus antibodies, providing incomplete field evidence for a potential role of bats in the transmission cycles of particular arboviruses.
During 2008–2010, the CDC Division of Vector-borne Diseases (CDC/DVBD) re-initiated arbovirus surveillance activities in collaboration with UVRI [Citation10]. During these investigations, the sources of a number of blood meals identified from mosquitoes included fruit bats. These mosquito-bat contacts were documented from Maramagambo Forest in Queen Elizabeth National Park, and Semliki Forest in Western Uganda [Citation11]. Additionally, the particular mosquito species which had engorged on fruit bats are known to be associated with a number of medically-important arboviruses: Coquillettidia (Cq.) fuscopennata (Theobald) (YFV, Sindbis, chikungunya viruses), Culex (Cx.) perfuscus Edwards (WNV, Oropouche, Sindbis, Wesselsbron, Usutu, Babanki viruses), Cx. (Cx.) neavei Theobald (WNV, Babanki, Spondweni, Sindbis, Koutango viruses), and Cx. (Cx.) decens group (WNV, chikungunya, Babanki viruses)[Citation10]. Considering this evidence for exposure of bats to bites from mosquitoes associated with arboviruses, an investigation to evaluate the seroprevalence of Ugandan bats to a panel of arboviruses was undertaken. The purpose of this study was to follow-up on earlier field investigations which reported high seroprevalence of known arboviruses, particularly flaviviruses [Citation7,Citation8] in bats. Our goal was to identify bat-virus associations that would warrant further study for their potential ecological significance for a role of bats in arbovirus transmission and maintenance.
Materials and methods
Sample collection
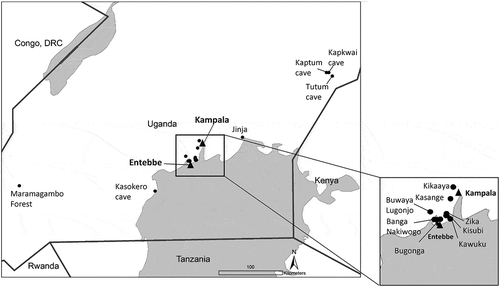
Bats were captured from multiple locations throughout Uganda during 2011–2013 (, .
Table 1. Species and sampling locations for bat sera tested, Uganda 2009, and 2011–2013.
Figure 1. Sampling locations for bats, 2009 – 2013. Capture locations for all bats tested in this study.
Additional samples collected from Egyptian rousette bats captured previously at Maramagambo Forest in 2009 were provided for inclusion in this serosurvey. All bat captures were conducted under the approval of IACUC protocols 1731AMMULX (Maramagambo samples) and 010–015 (all other samples). Bats were captured using harp traps or mist nets, and upon capture, placed individually in holding bags. Blood from bats captured in Maramagambo forest was collected as described by Towner et al. [Citation12]. All other bats were anesthetized with halothane and bled by cardiac puncture, then euthanized by halothane overdose and cervical dislocation. Blood from bats captured in locations other than Maramagambo forest was collected directly into serum separator tubes and centrifuged in the field, and placed immediately in liquid nitrogen dry shippers.
Serological testing
All serum samples were frozen at −80°C until they were tested for neutralizing antibodies against flaviviruses (Flaviviridae: Flavivirus): WNV, YFV, Dengue virus type 2 (DENV-2), ZIKV; Babanki virus (BBKV) (Togaviridae: Alphavirus), and Rift Valley fever virus (RVFV) (Phenuiviridae: Phlebovirus) by plaque reduction neutralization test (PRNT) [Citation13]. While all four dengue serotypes circulate in Africa, we chose to screen samples for DENV-2 due to the frequent involvement of this serotype in epidemics in Africa [Citation14]. Twofold serial dilutions of serum were incubated with 100 plaque-forming units (PFU) of WNV (strain UG2274, Uganda), DENV-2 (strain DakHD10674, Senegal), YFV (strain BA-55, Nigeria), ZIKV (MP766, Uganda), BBKV (46A-186), and RVFV (MP-12). A cutoff of ≥80% reduction in PFU on Vero cells was selected. For flaviviruses, bats with an antibody titer (PRNT80) at least four-fold higher than the other flaviviruses were considered positive for antibody to that virus. Due to the serologic cross-reactivity observed among the flaviviruses, bats with neutralizing antibody titers against multiple flaviviruses for which no fourfold difference in titer was observed or with weak titers PRNT80 ≤ 20, where no four-fold difference could be observed, were considered flavivirus antibody positive with no specific virus identified. For some samples which returned ambiguous flavivirus results, we attempted to detect residual flavivirus RNA from tissue samples collected from those bats in order to positively identify the infecting virus. For these samples, RNA from liver/spleen homogenates was screened using pan-flavivirus primers [Citation15]. For BBKV and RVFV, serum samples with a titer of PRNT80 ≥ 20 were considered seropositive. Because of limited serum volumes, not enough serum was available to test all bats for every virus, so some samples were not tested on the entire panel.
Results
This study reports the results of serological testing conducted on 626 bats captured in Uganda: 323 bats captured from 2011 to 2013, and an additional 303 samples from Egyptian rousette bats sampled during 2009. Neutralizing antibodies against flaviviruses were common. Significant neutralizing antibody titers against WNV were found in 2/8 (25%) African straw-colored fruit bats sampled in Jinja in 2012, as well as in 2/25 (8%) little epauletted fruit bats sampled in Kikaaya and 1/19 (5.3%) from Buwaya Lugonjo ( and ). One of 45 (2.2%) Egyptian rousette bats sampled from Tutum cave had a neutralizing antibody titer against YFV ( and ). One of two little free-tailed bats (50%) and 1/2 (50%) Angolan free-tailed bats sampled from Zika forest in 2011 contained specific antibodies against DENV-2 virus, as well as the single little epauletted fruit bat from Buwaya Lugonjo that same year ( and ). However, 85/626 (13.5%) of bats possessed neutralizing antibodies against an undetermined flavivirus for which a 4-fold difference in antibody titer was not observed for any one of the flaviviruses included in the panel ( and ). Among these bats, all 15/64 of the flavivirus antibody-positive little free-tailed bats captured from Kawuku in 2013 reacted only to DENV-2, however at titers that were not distinguishable from other flaviviruses based on our criteria. presents the endpoint antibody titer of five representative bats for which a specific result was determined, and five representative bats for which serological results were inconclusive based on our criteria.
Table 2. Prevalence of neutralizing antibodies detected in Ugandan bats, 2009 – 2013. Number positive/number tested for each virus in each location and year.
Table 3. Example end-point titers of arbovirus neutralizing antibodies (PRNT80). The top five lines represent bats for which a specific result was determined; the bottom five lines represent bats for which serological results were inconclusive.
A total of 2/432 (0.5%) bats contained specific neutralizing antibodies against BBKV: one Egyptian rousette bat and one little epauletted fruit bat ( and ). Nine of the 303 (3%) Egyptian rousette bats tested from Maramagambo forest exhibited alphavirus neutralizing antibodies, but could not be confirmed as BBKV (). Five of 52 (9.6%) little epauletted fruit bats from Kawuku and 3/54 (5.6%) Egyptian rousette bats from Kasokero cave contained specific neutralizing antibodies against RVFV.
Discussion
Overall, exposure of fruit and insectivorous bat species to flaviviruses was widespread, although prevalence in any one species was often low, suggesting possible incidental exposure (). This common detection of flavivirus antibodies is consistent with previous studies [Citation6–Citation8]. In particular, 23/86 (27%) little free-tailed bats, spanning every collection during the study period, demonstrated neutralizing antibodies against flaviviruses (). We were unfortunately not able to distinguish among the flavivirus infections in the majority of cases, despite using the most specific antibody assay, the PRNT. It is unclear whether these ambiguous results are the result of an infection(s) with one or more of the flaviviruses included in our panel, or with a different flavivirus altogether. Flaviviruses including Entebbe bat virus (ENTV)[Citation5,Citation16], Bukalasa bat virus (BBV) and Dakar bat virus (DBV)[Citation6,Citation8], have been previously isolated from little free-tailed and Angolan free-tailed bats in Uganda, leaving open the possibility that some of the flavivirus antibody-positive bats in this study had been infected with one of those other viruses. We did not assess the serological cross-reactivity between ENTV, BBV, or DBV with the mosquito-borne viruses in our panel because they are classified in different antigenic complexes within the family Flaviviridae and therefore unlikely to cross-react serologically [Citation17]. Egyptian rousette bats also displayed a relatively high flavivirus seroprevalence across all collections, with a total of 53/402 (13%) of Egyptian rousette bats exhibiting flavivirus neutralizing antibodies. Aside from one bat captured at Tutum cave which was considered to be antibody-positive for YFV, we did not generate any convincing evidence indicating which flavivirus was the infecting virus in any other Egyptian rousette bat sampled. Follow-up testing of the liver/spleen homogenates from these seropositive Egyptian rousette bats using pan-flavivirus primers [Citation15] did not result in any positive amplifications (Kading, unpublished data). The tissues from one little free-tailed bat from 2011 which yielded an isolate of ENTV, were flavivirus RNA-positive [Citation16]. Development of specific assays targeting sub-genomic flaviviral RNA (sfRNA), which can be present in high amounts in infected cells, may be one approach to deciphering ambiguous serological results in future investigations [Citation18,Citation19].
Among the alphaviruses, a variant of Sindbis virus, BBKV, is known to cause fever and arthralgia in humans and has been isolated from numerous mosquito species across Africa, including Cx. (Cx.) decens group and Cx. (Cx.) perfuscus Edwards [Citation20,Citation21]. In this study, two bats were seropositive for BBKV: one little epauletted fruit bat from Kikaaya, and one Egyptian rousette bat from Kasokero cave. Mosquitoes collected in Uganda that were documented to have engorged on fruit bats included Cx. perfuscus on straw-colored fruit bats and Egyptian rousette bats in Semliki Forest, Cx. (Cx.) neavei Theobald on Egyptian rousette bats in Semliki Forest and Maramagambo forest, and Cx decens group mosquitoes on Egyptian rousette bats in Maramagambo forest [Citation11]. Further, several isolates of BBKV were obtained from Cx. perfuscus collected from multiple locations in Uganda during this same sampling period [Citation21]. Among the 40 blood meals previously identified from Cx. perfuscus in Uganda [Citation11], 90% were taken from mammalian hosts, and 10% from avian hosts comprising at least 11 different species [Citation11]. The majority of mammalian blood meals (26/36; 72%) came from ungulates, however, blood meals from bats comprised 7.5% of the total blood meals identified from this mosquito species [Citation11]. Whether bats contribute to the transmission cycle of BBKV or are merely incidentally exposed through being fed upon by mosquitoes is unclear.
The involvement of a wild mammal reservoir in the epidemiologic cycle of RVFV is also unknown. Olive et al. [Citation22] reviewed what is currently known about the role of wild mammals in the maintenance of RVFV. As with many vertebrate taxa, the potential involvement of bats in the interepidemic maintenance of RVFV has not been thoroughly investigated. Available experimental data suggest that RVFV is capable of replication in bats, and that infection could be asymptomatic [Citation22]. Experimental inoculation of Schreiber’s long-fingered bats (Miniopterus schreibersii) and Cape serotines (Eptesicus capensis) confirmed that these bats were capable of becoming infected with RVFV without showing signs of clinical illness [Citation22,Citation23]. Further, RVFV has been previously isolated from several bat species including Peters’ lesser epauletted fruit bat (Micropteropus pusillus), the Aba leaf-nosed bat (Hipposideros abae) and Sundevall’s leaf-nosed bat (Hipposideros caffer), Franquet’s epauletted fruit bat (Epomops franqueti), and the common butterfly bat (Glauconycteris argentata) [Citation24–Citation26], demonstrating that bats are naturally exposed to this virus.
RVFV is endemic to Uganda, characterized by low levels of enzootic activity as opposed to the large-scale epidemics experienced by neighboring East African countries [Citation27]. RVFV has been previously isolated from mosquitoes captured on the Entebbe peninsula [Citation28], and a localized outbreak in the Kabale district of Western Uganda in 2016 confirms recent RVFV circulation [Citation29]. In this study, bats from two collections contained several individuals with neutralizing antibodies against RVFV. These collections comprised 3/54 (5.5%) Egyptian rousette bats from Kasokero cave captured in 2013, and 5/52 (9.6%) little epauletted fruit bats from Kawuku, also from 2013. Kasokero cave and Kawuku are both located on the shore of Lake Victoria, and are approximately 80 km apart (). In addition to the bats considered seropositive with an antibody titer of PRNT80 ≥ 20, there were several additional little epauletted fruit bats from Kikaaya (2011) and Buwaya Lugonjo (2011–2012), and one Egyptian rousette bat from Tutum cave (2012) with antibody titers of PRNT80 = 10 for RVFV that were considered inconclusive. If these additional results are indicative of a true infection with RVFV, these observations collectively suggest that exposure of fruit bats in Uganda to RVFV is fairly widespread (Kasokero cave to Tutum cave is >200km). However, it is also possible that at least some of these results are attributable to circulation of a different, unidentified, phlebovirus. Mourya et al. [Citation30] reported a novel bat phlebovirus named Malsoor virus, related to severe fever with thrombocytopenia syndrome virus (SFTSV). Malsoor virus was isolated from a related fruit bat (Rousettus leschenaultia) in Western India [Citation30]. Mossel et al. [Citation21] reported two isolations of Arumowot virus (Phenuiviridae, Phlebovirus) from Culex mosquitoes collected in Jinja, Uganda, in 2012. While Malsoor and Arumowot viruses are both distantly related to RVFV [Citation30], we did not evaluate the potential for cross-reactivity of these samples to other phleboviruses. The ability of Egyptian rousette bats and little epauletted fruit bats to become infected with RVFV and develop viremias high enough to infect feeding mosquitoes is unknown.
An important aspect to the ecology of arbovirus transmission that was not addressed in this study is the influence of geographic location and land-use/land cover on the prevalence of arbovirus antibodies in bats. The bat species captured in this study are all widely distributed in Uganda, but tend to occupy particular habitats such as man-made structures, caves, or tall trees. The agro-ecological conditions associated with the bat collection locations would also influence the presence and abundance of mosquito vectors through the availability of suitable larval habitats and preferred hosts. The co-occurrence of vector, host, and virus with landscape and environmental conditions is important to the emergence and circulation of arboviruses. Bat migration and dispersal patterns also make it difficult to know where they would have become infected. Future investigations should include modelling component to evaluate the co-incidence of vector and host distribution and pathogen detection data in an ecological context.
In conclusion, this study provides novel data on the exposure of multiple species of fruit and insectivorous bats in Uganda to a variety of arboviruses. While the role of bats in arbovirus transmission cycles is still unfolding, we have built upon previous studies [Citation6,Citation8,Citation11,Citation21] to demonstrate ecological associations between bats, arboviruses, and mosquito vectors in Uganda. This study provides additional field evidence of mosquito-bat contact through the detection of neutralizing antibodies in bat populations against arboviruses which were contemporaneously isolated from mosquitoes in Uganda (i.e. WNV, BBKV)[Citation21]. To further define and quantify the contribution of bats to arbovirus transmission cycles, more data are needed on the contact rates between different species of bats with mosquito vectors, as well as the competency of these bat species as amplification hosts for specific viruses [Citation31]. In particular, experimental studies are needed to evaluate the ability of Egyptian rousette bats and little epauletted fruit bats to support replication of BBKV and RVFV and develop viremias high enough to infect feeding mosquitoes. These experimental data would provide important insight into the epidemiological significance and interpretation of the serological data presented in this study.
Acknowledgments
We thank the Field Museum Department of Mammology for donation of voucher material for molecular confirmation of the Rousettus bat species from Tutum cave. Godfrey Kyazze, David Ssekatawa, and Dennis Ssemwogerere served as drivers and managed equipment. Bat sampling was conducted under the permission of the Uganda Wildlife Authority (TDO/7/92/01), and CDC IACUC approval numbers (1731AMMULX; CDC/VSPB, Maramagambo samples)(10-015; CDC/DVBD, all other samples). We also thank Mr. Tom Okello Obong, Dr. Margret Driciru, and the Uganda Wildlife Authority rangers at the python cave.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Notes on contributors
Rebekah C. Kading
Dr. Rebekah C. Kading is currently an Assistant Professor in the Department of Microbiology, Immunology, and Pathology, Colorado State University, where her research program focuses on medical entomology and emerging arboviruses.
Robert M. Kityo
Dr. Robert M. Kityo is a Professor in the College of Natural Sciences, Makerere University, Kampala Uganda. His research specializes in small mammals.
Eric C. Mossel
Dr. Eric C. Mossel is an arbovirologist with the US Centers for Disease Control and Prevention, Division of Vector-borne Diseases in Fort Collins, Colorado. His research interests include arbovirus surveillance, discovery and diagnostic capacity building.
Erin M. Borland
Erin M. Borland is currently a Research Associate in the Department of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO.
Teddie Nakayiki
Teddy Nakayiki is an Entomology Assistant in the Arbovirology, Emerging and Re-Emerging Viral Infections program. She carries out mosquito identification as well as assists with research on bats.
Betty Nalikka
Betty Nalikka is a research scientist in the College of Natural Sciences, Makerere University, Kampala, Uganda.
Luke Nyakarahuka
Dr. Luke Nyakarahuka is qualified in Veterinary Medicine (BVM) and received his Masters in Public Health (MPH) both from Makerere University, Kampala. His PhD topic at Norwegian University of Life Sciences was “Epidemiology of Marburg and Ebola Viruses in Uganda”. Dr. Nyakarahuka is currently employed as an Epidemiologist at Uganda Virus Research Institute, Viral Hemorrhagic Fevers Surveillance Program and Makerere University College of Veterinary Medicine as a lecturer of Epidemiology, Zoonoses and Veterinary Public Health.
Jeremy P. Ledermann
Jeremy P. Ledermann is a microbiologist for the Centers for Disease Control and Prevention, Division of Vector-borne Diseases in Fort Collins, Colorado. He specializes in the biological interactions between arboviruses and their mosquito vector hosts.
Nicholas A. Panella
Nicholas A. Panella is a biologist with the Centers for Disease Control in Fort Collins, CO. He has been researching arthropod borne diseases for over 20 years. His research interests include the ecology of West Nile virus and integrated pest management.
Amy T. Gilbert
Dr. Amy Gilbert is a Research Biologist at the National Wildlife Research Center, which is part of the USDA APHIS Wildlife Services Program. Her current research focuses on ecology and spillover of rabies infections in wildlife, as well as experimental studies regarding the application of novel rabies biologics and technologies to improve vaccination of carnivore wildlife, in efforts to improve national disease management.
Mary B. Crabtree
Mary B. Crabtree, currently retired, was an arbovirologist with the Centers for Disease Control and Prevention, Division of Vector Borne Diseases.
Julian Kerbis Peterhans
Dr. Julian Kerbis Peterhans is a professor at Roosevelt University, Chicago and an Adjunct Curator at the Field Museum (Chicago). He studies carnivore predation and small mammal alpha-taxonomy and biogeography in central Africa.
Jonathan S. Towner
Dr. Jonathan S. Towner is a Virologist at the Centers for Disease Control and Prevention, Viral Special Pathogens Branch in Atlanta, GA.
Brian R. Amman
Dr. Brian R. Amman received his PhD in Zoology from Texas Tech University. He is currently employed as a Disease Ecologist for the Viral Special Pathogens Branch, Virus Host Ecology Unit, at the Centers for Disease Control and Prevention in Atlanta, GA. His research interests include investigating the ecology of, and relationships between, emerging zoonotic viruses and their reservoirs.
Tara K. Sealy
Tara K. Sealy is a microbiologist in the Viral Special Pathogens Branch Virus Host Ecology team.
Stuart T. Nichol
Dr. Stuart T. Nichol is the Chief of the Viral Special Pathogens Branch, Centers for Disease Control and Prevention in Atlanta, GA.
Ann M. Powers
Dr. Ann M. Powers is the Chief of the Alphavirus lab, Division of Vector-borne Diseases, Arbovirus Diseases Branch, Centers for Disease Control and Prevention in Fort Collins, CO.
Julius J. Lutwama
Dr. Julius J. Lutwama is head of the Arbovirology, Emerging and Re-Emerging Viral Infections program, Uganda Virus Research Institute, Entebbe, Uganda.
Barry R. Miller
Barry R. Miller, currently retired, served as the Chief of the Virology Activity, CDC Division of Vector-borne Diseases, Arbovirus Diseases Branch.
References
- Sempala SDK. Institute profile: the Uganda virus research institute. Trends in Microbiol. 2002;10:1—7.
- Haddow AJ, Smithburn KC, Dick GWA, et al. Implication of the mosquito Aedes (Stegomyia) africanus Theobald in the forest cycle of yellow fever in Uganda. Ann Trop Med Parasitol. 1948;42:218—223.
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520.
- Haddow AJ, Williams MC, Woodall JP, et al. Twelve isolations of Zika virus from Aedes (Stegomyia) africanus (Theobald) taken in and above a Uganda forest. Bull World Health Organ. 1964;31:57–69.
- Lumsden WHR, Williams MC, Mason PJ. A virus from insectivorous bats in Uganda. Ann Trop Med Parasitol. 1961;55:389.
- Williams MC, Simpson DIH, Shepherd RC, et al. Virus isolations from bats. East African virus research institute report, July 1963–December 1964. Publication of the East African High Commission. Nairobi, Kenya: Government Printer. 1964; 42.
- Shephard RC, Williams MC. Studies on viruses in east African bats (Chiroptera). 1. Hemagglutination inhibition and circulation of arboviruses. Zoonoses Res. 1964;3:125–139.
- Simpson DIH, Williams MC, O’Sullivan JP, et al. Studies on arboviruses and bats (Chiroptera) in east Africa II. Isolation and haemagglutination-inhibition studies on bats collected in Kenya and throughout Uganda. Ann Trop Med Parasitol. 1968;62:432–440.
- Simpson DI, O’Sullivan JP. Studies on arboviruses and bats (Chiroptera) in east Africa. I. Experimental infection of bats and virus transmission attempts in Aedes (Stegomyia) aegypti (Linnaeus). Ann Trop Med Parasitol. 1968;62:422–431.
- Mutebi JP, Crabtree ME, Kading RC, et al. The mosquitoes of Western Uganda. J Med Entomol. 2012;49:1289–1306.
- Crabtree M, Kading RC, Mutebi J-P, et al. Identification of host blood from engorged mosquitoes collected in western Uganda using cytochrome oxidase I gene sequences. J Wildl Dis. 2013;49:611–626.
- Towner JS, Amman BR, Sealy TK, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536.
- Beaty BJ, Calisher CH, Shope RE. Arboviruses. In: Lennette EH, Lennette DA, Lennette ET, editors. Diagnostic Procedures for Viral, Rickettsial, and Chlamydial Infections. 7th ed. Washington DC: American Public Health Association; 1995. p. 189–212.
- Amarasinghe A, Kuritsky JN, William Letson G, et al. Dengue virus infection in Africa. Emerg Infect Dis. 2011;17:1349–1354.
- Kuno G, Chang G-J-J, Tsuchiya KR, et al. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83.
- Kading RC, Kityo R, Nakayiki T, et al. Detection of Entebbe bat virus after 54 years. Am J Trop Med Hyg. 2015;93:475—477.
- Calisher CH, Karabatsos N, Dalrymple JM, et al. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70:37—43.
- Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005.
- Roby JA, Pijlman GP, Wilusz J, et al. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014;6:404—427.
- Centers for Disease Control and Prevention. Arbovirus Catalog. [cited 2017 Apr 27]. Available from: https://wwwn.cdc.gov/arbocat.
- Mossel EC, Crabtree MB, Mutebi J-P, et al. Arboviruses isolated from mosquitoes collected in Uganda, 2008–2012. J Med Entomol. 2017;54:1403—1409.
- Olive MM, Goodman SM, Reynes JM. The role of wild mammals in the maintenance of Rift Valley fever virus. J Wildl Dis. 2012;48:241—266.
- Oelofsen MJ, Van Der Ryst E. Could bats act as reservoir hosts for Rift Valley fever virus? Onderstepoort J Vet Res. 1999;66:51—54.
- Boiro I, Konstaninov OK, Numerov AD. Isolation of Rift Valley fever virus from bats in the Republic of Guinea. Bull Soc Pathol Exot Filiales. 1987;80:62—67.
- Butenko AM. Arbovirus circulation in the Republic of Guinea. Med Parazitol (Mosk). 1996;2:40—45.
- Calisher CH, Childs JE, Field HE, et al. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545.
- Bird BH, Ksiazek TG, Nichol ST, et al. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234:883—893.
- Weinbren MP, Williams MC, Haddow AJ. Isolation of an atypically reacting strain of Rift Valley fever virus from Lunyo, on the Entebbe Peninsula. East African Virus Research Institute Report, July 1955–June 1956. Publication of the East African High Commission. Nairobi, Kenya: Government Printer. 1956;21—26.
- De St. Maurice A, Nyakarahuka L, Purpura L, et al. Rift Valley fever response — Kabale District, Uganda, March 2016. Morb Mortal Wkly Rep. 2016;65:1200–1201.
- Mourya DT, Yadav PD, Basu A, et al. Malsoor virus, a novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and heartland virus. J Virol. 2014;88:3605—3609.
- Golnar AJ, Turell MJ, LaBeaud AD, et al. Predicting the Mosquito species and vertebrate species involved in the theoretical transmission of Rift Valley fever virus in the USA. PLOS Negl Trop Dis. 2014;8(9):e3163.
