ABSTRACT
In a recently published study, Anna Krichevsky and colleagues raise the important question of whether results of in vitro extracellular RNA (exRNA) studies, including extracellular vesicle (EV) investigations, are confounded by the presence of RNA in cell culture medium components such as foetal bovine serum (FBS). The answer, according to their data, is a resounding “yes”. Even after lengthy ultracentrifugation to remove bovine EVs from FBS, the majority of exRNA in FBS remained. Although technical factors may affect the degree of depletion, residual EVs and exRNA in FBS could influence the conclusions of in vitro studies: certainly, for secreted RNA, and possibly also for cell-associated RNA. In this commentary, we critically examine some of the literature in this field, including a recent study from some of the authors of this piece, in light of the Wei et al. study and explore how cell culture-derived RNAs may affect what we think we know about EV RNAs. These findings hold particular consequence as the field moves towards a deeper understanding of EV–RNA associations and potential functions.
RESPONSIBLE EDITOR Raymond Schiffelers, University Medical Center Utrecht, The Netherlands
Introduction
Research into extracellular vesicles (EVs), including exosomes, microvesicles, and other cell-derived particles with double-leaflet membranes [Citation1], has expanded rapidly in recent years [Citation2], bolstered in part by high-profile reports of EV-mediated ribonucleic acid (RNA) exchange between non-neighbouring cells [Citation3–Citation6]. As in any field in rapid growth phase, rampant enthusiasm for development of novel paradigms in EV-mediated communication has been accompanied by new challenges [Citation7]. A set of recurring concerns involves the relatively low yields of analytes from typical volumes of biological materials in EV studies and how the stoichiometries of in vivo and in vitro studies relate [Citation8,Citation9]. In the case of RNA, although amplification-based quantitative polymerase chain reaction (qPCR) and sequencing assays are highly sensitive, the often underappreciated effects of contaminants inversely correlate with sample input [Citation10,Citation11]. Artefacts arising from contaminants might play a larger role in the EV field than in others.
Figure 1. Ultracentrifugation of FBS does not remove all RNA or EVs, nor does ultracentrifugation of conditioned medium collect only cultured cell-derived RNAs or EVs.
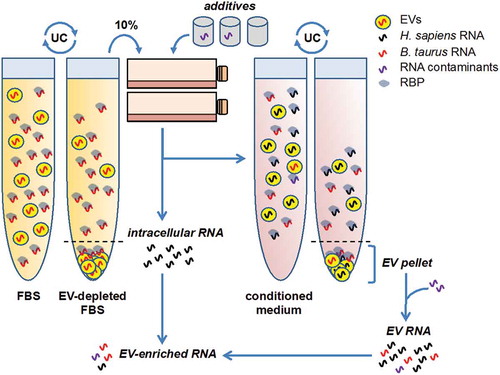
Figure 2. Comparison of ranks of human-mapped bovine miRNAs from ultracentrifuge pellets (EV-enriched) and supernatants (EV-depleted); n = 3. Data are from Wei et al. ([16], GSE78970). Selected apparent pellet- or supernatant-enriched miRNAs are noted, along with miR-122-5p and miR-1246 (the most abundant two mapped miRNAs in both fractions).
![Figure 2. Comparison of ranks of human-mapped bovine miRNAs from ultracentrifuge pellets (EV-enriched) and supernatants (EV-depleted); n = 3. Data are from Wei et al. ([16], GSE78970). Selected apparent pellet- or supernatant-enriched miRNAs are noted, along with miR-122-5p and miR-1246 (the most abundant two mapped miRNAs in both fractions).](/cms/asset/ad5078e4-9cad-4b59-8e13-51da9ad05881/zjev_a_1272832_f0002_b.gif)
The sources of contaminants in EV and extracellular RNA (exRNA) studies are incompletely understood. Serum used in cell culture media, most commonly foetal bovine serum (FBS), is an obvious source of exogenous RNA-containing EVs [Citation12,Citation13], so interpretable isolation of EVs from cell culture requires either serum-free medium or use of vesicle-depleted serum. Because serum contains a variety of soluble growth factors, and optimal serum-free conditions have not yet been established for all cell lines, the majority of reports rely on serum. EV depletion from serum is typically carried out by prolonged (≥18 h), high-speed (~100,000 g) centrifugation of diluted serum [Citation12]. This procedure depletes the majority (75–90%) of FBS EVs, while shorter centrifugation times have even lower depletion efficiency [Citation14,Citation15]. But does even prolonged centrifugation guarantee the absence of serum-born RNAs?
An elegant study by Wei et al. now suggests that ultracentrifugation protocols designed to remove EVs from FBS leave most RNA undepleted, and that this RNA may contribute to misinterpretation of extracellular RNA (including EV) studies [Citation16]. In our opinion, this work is timely and well-presented and gives a quantitative estimate of the impact that contaminating nucleic acids can have on EV and exRNA research. In this case, the warning is focused on cell culture-derived extracellular RNA, and the source of RNA contaminants is FBS. It will also be important to consider other sources of contaminants in high-sensitivity measurements of EV contents and exRNA.
miR-122 and miR-451a as bellwethers
Wei et al. were alerted to the potential problem of FBS contamination after observing high levels of hsa-miR-122 in the conditioned media of glioma cell lines. miR-122 expression is paradigmatic: it is highly expressed in the liver and undetectable or found at only trace amounts elsewhere [Citation17]. The authors contrasted the exciting possibility of extremely efficient miR-122 secretion by glioma cells (given virtually null intracellular detection) with the less-exciting and more parsimonious possibility of an artefact. Together with miR-122, the authors had also found the red blood cell (RBC)-specific miR-451a as a miRNA putatively enriched in EVs from the same cell lines. However, both miRNAs were also detectable at high levels in the non-conditioned media, even in the case of prior serum EV depletion for 24 h. Were these miRNAs medium contaminants instead of EV-enriched miRNAs secreted by glioma cells? Interestingly, while miR-451a detection was completely abolished in serum-free non-conditioned media, detection of miR-122 was not completely dependent on the presence of serum. This observation raises the possibility that some components of serum-free media formulations might also serve as sources of contaminating miRNAs.
Culture-derived miRNAs…that aren’t
To obtain a list of FBS-derived miRNAs that could be misinterpreted as hailing from cultured cells (here: human cells), Wei et al. performed small RNA sequencing of FBS 100,000 g pellets and supernatants after ultracentrifugation for 24 h. (Note that, because miRNAs are highly conserved among mammals, most human miRNAs are 100% identical with their bovine homologues.) The list was headed by miR-122-5p, followed by miR-1246, miR-148a-3p, miR-423-5p, miR-92a-3p, let-7a/b-5p, miR-379-5p, miR-127-3p, miR-320a, and others (see also Supplemental for rankings in supernatant and pellet). The RBC-specific miR-451a, while not among the most abundant in the next-generation sequencing (NGS) analysis, was detected at high levels by qPCR. One might speculate that library preparation method biases and/or mapping strategies affect detection of specific miRNAs; of note, miR-451a is also non-canonically processed, with substantial length polymorphism.
Table 1. Selected experiments reporting EV-enriched microRNAs that may be consistent with cell culture RNA contamination. A variety of methods of FBS EV depletion have been used.
Some of the miRNAs (e.g. miR-122-5p, miR-1246) were present at high levels in the FBS supernatants independent of ultracentrifugation time, but increased their abundance in the 100,000 g pellets as a function of time. This observation is highly relevant for EV RNA research: abundant bovine miRNAs in serum can contaminate initial 100,000 g pellets, but may also remain at high levels in the supernatant to contaminate downstream EV preparations from FBS-containing cell cultures (). These RNAs might be found in carriers including small EVs and protein complexes. In this report, the observation is that most RNA in serum is not easily pelletable, possibly due to its predominant association with ribonucleoprotein complexes rather than EVs [Citation18,Citation19], although this observation is not universal [Citation14,Citation20] and must be considered in light of procedural details (see below). Interestingly, ranks of FBS pellet and supernatant miRNAs were well correlated in the dataset deposited by Wei et al. (GEO accession GSE78970), with a few notable outliers. As just one example, the central nervous system- and neuron-enriched miR-9 appeared to be enriched in EV pellets compared with supernatant (), corresponding to a roughly 50-fold difference in reads per million (GSE78970).
We would add a few more miRNAs to the list of contamination-betraying candidates, even though they did not figure prominently in the Wei et al. sequencing results. miR-486-5p (identical sequence in bovine and human) is enriched in RBCs and platelets and has elsewhere been reported as the most abundant miRNA in serum and serum EVs of cattle [Citation21,Citation22]. miR-144 is part of a bicistronic cluster with miR-451a and expressed at lower, but proportional, levels. miR-150 is a hematopoietic-specific (lymphocyte-dominant) miRNA [Citation23] that should not be observed outside of hematopoietic-related cultures [Citation17]. miR-223-3p is another hematopoietic-specific miRNA, found at high levels in myeloid-lineage cells such as platelets, monocytes/macrophages, and neutrophils [Citation17,Citation24–Citation26]. Although miRs-486, -150, and -223, like miR-451a, were not detected at high levels in the Wei et al. NGS data, these miRNAs have been known to have high expression in serum since the early serum biomarker studies (e.g. [Citation27]). In plasma, expression of miR-150 and miR-223 correlates tightly with lymphocyte and myeloid cell counts, respectively [Citation25] and can vary substantially depending on how plasma or serum is processed [Citation28]. To be sure, blood cell-specific miRNAs have been reported to be present in a wide variety of human tissues (see, for example, [Citation29]). However, as we have discussed elsewhere [Citation9,Citation17], this is due to the ubiquity of blood cells in human tissue; cell specificity becomes apparent when pure cell populations are examined [Citation24]. presents several of the miRNAs from the Wei et al. list and our additions, giving examples of publications in which they were reported to be EV-enriched. Please note that, in assembling this non-exhaustive list, we do not mean to suggest that all of these studies are without merit. Indeed, we have tried to be critical of our own work [Citation30], as well, since it is also subject to the factors we describe.
Methods matter: dilution, fractionation, and beyond
As mentioned above, technical factors may play a role in divergent serum particle and RNA depletion findings between studies. Particle pelleting efficiency is of course lower for viscous, undiluted serum than for serum, e.g. diluted 1:5 with basal medium [Citation12]. We are aware of various serum EV depletion protocols, ranging from no dilution to centrifugation of complete media (often 10% serum). Wei et al. did not report diluting serum prior to ultracentrifugation [Citation16]. Thus, depletion efficiency may have been low despite a relatively long spin (24 h). Other technical questions could be posed for this and most other studies. How was supernatant removed from the ultracentrifuge tube? Due to formation of particle gradients in the tube, supernatant decanted entirely and at once into a new container might show a different degree of depletion compared with supernatant drawn starting from the top of the column by pipette, leaving several millilitres above the pellet. How was residual supernatant removed and the pellet re-suspended or solubilised, and how do these methods affect extracellular RNA results? Although more details are always helpful, and different methods will affect depletion results, we submit that Wei et al. have provided a valuable set of experiments to establish RNA profiles in different fractions of FBS…or, at least, the FBS they used, since technical factors might also affect RNA content of different FBS lots.
Lot-specific effects of FBS in cell culture are well known. Does EV depletion and RNA content also differ by lot? Regarding source, does FBS (also known as foetal calf serum) differ in EV and RNA content from calf serum or adult bovine serum? Does the health of the calf affect these parameters? In human clinical studies, some pay close attention to every detail of obtaining blood, from the needle gauge to when the tourniquet is removed from the donor to the time between draw and processing. The same considerations would apply to obtaining bovine serum, yet these variables are rarely reported. After obtention, what processing steps are followed to remove contaminants from FBS? What are the effects of serum heat inactivation and different heat inactivation protocols? It is clear from an important study of human blood product that processing affects extracellular RNA concentrations (and some miRNAs more than others) [Citation28]. We recommend that these and other factors be considered in more depth in future studies of the influence of serum on cell culture, but they also suggest that avoiding serum is an attractive option where available.
miRNA-specific secretion…or sticky stuff from serum?
One open controversy in EV RNA research is the extent to which RNAs (with miRNAs being the most heavily studied) are sorted into EVs by protein-mediated recognition of defined sequences versus being incorporated non-selectively by pinching off of random bits of cytoplasm. The latter end of the spectrum would suggest that the deciding determinant of miRNA secretion is intracellular concentration in the vicinity of EV formation rather than the presence of specific sequence motifs that destine miRNAs for secretion. An obvious method for choosing the balance point between the two ends of the scale is a comparison of EV and parental cell miRNA profiles. Perhaps an even better comparison would be between EVs and the parental cell cytoplasmic fraction (eliminating nuclear RNAs that are normally less likely to leave the cell) or even membrane fractions (as the sites where vesiculation occurs) [Citation39,Citation40].
An extreme case consists of certain miRNAs, perhaps most prominently miR-451a, that are frequently reported to be undetectable or nearly so in cells yet detected, sometimes in abundance, in EVs harvested from cell culture [Citation13,Citation37]. As another example, miR-223 was recently reported to be efficiently packaged into EVs despite no functional detection in the parent HEK293 cells ([Citation37]; indeed sequencing of two HEK293 lines emphasises the lack of miR-223 in these cells [Sequence Read Archive accessions SRR1240816 and SRR1240817]). A miRNA could be functionally absent in a parent cell while abundant in EVs only if one proposes a set of hypothetical processing, transport, packaging, and secretion mechanisms that are near-instantaneous and 100% efficient. A much simpler explanation is that these microRNAs were never in the cultured cell to begin with, as Wei et al. now point out, but came from the culture medium. In another apparent illustration, Fong et al. [Citation32] reported specific secretion from breast cancer cell lines – but not non-cancerous MCF10A cells – of the liver-restricted and serum-abundant miR-122. Interestingly, all cell lines were reportedly cultured in the American Type Culture Collection (ATCC) “recommended medium”. ATCC recommends culture of MCF10A cells in serum-free medium (no or little miR-122), while 10% FBS is present in most recommended formulations for the other breast cancer cells. In this study, serum was centrifuged without reported dilution [Citation32], much as in Wei et al. [Citation16] and many of the other studies we evaluated ().
Examining wider profiles, several groups have reported substantial differences between cellular and extracellular miRNA fractions [Citation13,Citation35,Citation38,Citation41,Citation42], while we [Citation30] and others [Citation18] have found a strong correlation (r = 0.8) between contents of EVs and cells. Importantly, we switched cells to serum-free media and performed several wash and culture intervals before supernatant collection in an attempt to reduce the effects of FBS contamination [Citation30]. Differences between studies using EV-depleted FBS could arise from laboratory-to-laboratory variation in serum particle depletion (including EVs but possibly other RNA-containing particles), different lots of FBS or other serum, behaviours of diverse cell types under different culture conditions, and even a hypothetical influence of serum EV depletion on RNA sorting mechanisms. Nevertheless, as Wei et al. have noted, FBS enriched miRNAs emerge as a common theme in many (but not all, see for example [Citation41]) EV-cell comparisons.
Recurring themes: FBS-enriched miRNAs are EV-enriched
Now that Wei et al. have demonstrated the outsized role of FBS on EV RNA determinations in culture, we can turn a newly critical eye on this set of literature. Early on, Pigati et al. reported dramatic enrichment of miR-451a and miR-1246 in EVs released by breast cancer cells, concluding that “breast cancer cells release most of their miR-451a and miR-1246 molecules” [Citation13]. To their credit, the authors gave considerable thought to the possibility that FBS might contribute to extracellular RNA profiles, providing a timed release experiment and reporting that they, like another group before them [Citation43] could not amplify miR-451a from serum. Bos taurus miR-451a was noted to differ from human miR-451a by a terminal nucleotide. However, it is now understood that miR-451a is highly abundant in serum and is atypically processed, with length polymorphisms that may interfere with amplification. The results of numerous subsequent studies ([Citation31,Citation33,Citation34,Citation36,Citation37], see also ), including an analysis of several public datasets [Citation33] underscore the influence of FBS (and possibly other contaminants) on interpretation of EV RNA enrichment.
No viral miRNA enrichment in EVs…but possible RNA modifications?
In another particularly noteworthy study, Koppers-Lalic and colleagues sequenced one library each of six B-cell lines and their released vesicles; each batch of EVs was highly related with its parent cell type, consistent with a general non-specific release [Citation35]. However, enriched miRNAs were also observed, included miR-451a, the co-transcribed but less abundant miR-144, and the RBC-enriched miR-486. The most significantly under-represented miRNAs in EVs were miRs-1275 and miR-7974: neither has a homologue in bovine. Also, most of the cell lines were EBV+, and EBV produces miRNAs, yet “not a single viral miRNA was found significantly enriched in exosomes” [Citation35]. Since these RNAs would presumably not be expected in bovine serum (unless perhaps from related bovine viruses), a lack of enrichment is consistent with a non-selective packaging model. On the other hand, one might speculate that viral miRNAs have evolved to avoid motif-specific packaging and export. An intriguing finding of the study was that miRNA modifications, especially non-templated additions, also distinguish EV from cellular miRNAs. This discovery is compelling, but it may be informative to interrogate the presence of these modifications in FBS or other cell culture components.
Placenta-specific miRNAs: are they expressed extra-placentally and specifically exported?
Analysing microarray data from primary T cells and a T cell line, along with their released EVs, Villarroya-Beltri et al. reported two motifs associated with cellular export in EVs (GGAG and CCCU) [Citation38]. The exported miRNAs included FBS miRNAs miR-451a (both studies), miR-122 (Jurkat study), and miR-1246 (primary T-cell study), consistent with Wei et al. [Citation16]. However, several of the reported EV-enriched miRNAs are less likely to have been FBS-derived, such as members of the miR-513 and miR-520 families of the placenta- and primate-specific miRNA cluster on human chromosome 19 (C19MC) [Citation44]. At the same time, T-cell expression of these miRNAs is unexpected. Signal was also quite low, suggesting very low expression levels. Interestingly, many of the C19MC members contain the described export motifs, as do miR-122 and miR-1246. It might be instructive to repeat a motif analysis including only higher-abundance, non-placental, and non FBS-enriched miRNAs from the study. Another publication on EV release by primary human trophoblast cells – which do express C19MC miRNAs – reported no differences of EV and cellular abundance of C19MC miRNAs [Citation45], while a third study, using yet another cell type, concluded that “we were unable to identify any statistically significant primary sequence motifs for miRNAs” secreted in EV subtypes [Citation37]. It appears that the motif-dependence of miRNA export, if any, remains unclear and may vary by cell type.
Long-term persistence of FBS miRNAs in serum-free culture?
In our study of breast cell lines [Citation30], miRNA profiles were generally consistent with non-selective release. Only 8 miRNAs satisfied enrichment criteria, 3 of which (miR-92a-3p, miR-423-5p, and miR-320a) were reported as FBS-enriched by Wei et al. [Citation16]. Furthermore, miR-122-5p showed the highest fold change between EVs and cells, although the difference was not statistically significant because of high variance/low expression. Interestingly, we sought to minimise the influence of FBS in our study by washing and switching cells to serum-free medium for 48 h before a second wash and re-feed with fresh serum-free medium for 48 h before collection. Yet the Wei et al. publication brings caution to our findings. If the few extracellularly enriched miRNAs in our study are indeed explained by carryover of long-lived FBS RNA, they emphasise how pervasive the influence of RNA in culture components can be.
Is serum RNA taken up by cells in culture?
How might serum RNAs persist even after washing and prolonged serum-free culture, as our results seem to indicate? Wei et al. reported that bovine miR-1246 (which is not encoded in the mouse genome) was “consistently and reproducibly detected” in mouse cells cultured in the presence of EV-replete FBS and at lower levels in cells after seven days of EVD culture [Citation16]. Furthermore, analysis of publicly available exRNA sequencing datasets obtained with human cell lines traced up to 17.2% of reads to bovine-specific transcripts [Citation16]. But is this RNA taken up by cells, or is it simply cell-associated and able to diffuse away into the media, where it could be mistaken for secreted or EV-enriched RNA? Considering the abundance of miR-1246 in FBS, the persistence of the majority of serum miR-1246 after EV depletion, and the extremely low levels of cellular miR-1246 detected by qPCR, it is possible that this RNA was not taken up by cells. Rather, miR-1246 carriers – EVs, protein complexes, or both – were associated with the cell surface and incompletely washed away before RNA extraction. We recently cultured primary macrophages, a T-cell line, and a promonocytic line in media prepared with EV replete serum and various EVD sera. Profiling of 48 miRNAs revealed no consistent EV depletion-associated differences across the cell types [Citation46]. One explanation for these results might be that RNA uptake from serum EVs does not occur at high levels for these miRNAs and cell types. To establish genuine and potentially functionally relevant uptake of FBS RNA into cultured cells, careful procedures are needed, e.g. (1) protease treatment of cultured cells along with washing before RNA isolation and (2) LNA in situ hybridisation to demonstrate cytoplasmic vs intraluminal endosomal localisation. Much stronger evidence for uptake is recommended before investigators embark on studies of hypothetical function of exogenous RNA in culture, which we consider to be unwarranted at present.
FBS: tip of a contamination iceberg?
The serum issue exposed by Wei et al. makes considerable sense in retrospect, considering the large EV and RNA concentration differences (up to several orders of magnitude) between serum and cell culture conditioned medium. However, FBS carryover may be just one aspect of widespread confounding of exRNA and exogenous RNA studies. We have previously reported evidence of contamination by surveying exogenous RNAs in publicly available sequencing datasets [Citation47]. In this study, sequences from a plethora of organisms, including bacteria, appeared to be present in human tissues. For example, rRNA fragments from mussels were widespread. Mussels are the primary source of commercially-available glycogen, widely used as a carrier in nucleic acid precipitation. Thus, common lab reagents are genuine sources of nucleic acid contamination (). Even extraction kit spin columns have been reported as a source of contaminants [Citation48], leading to the false discovery of NIH-CQV virus as the etiologic cause of seronegative hepatitis. We were able to find Trypanosoma cruzi-specific reads in human cells when using a bottle of Trizol used for a parasitology project a year earlier, but none when re-sequencing aliquots of the same samples extracted with a new bottle of reagent (Tosar et al., unpublished data). Recombinant enzymes can be contaminated with bacterial RNA. Importantly, the problem of contamination may be most insidious not for exogenous sequences, which are often easily recognised, but instead for sequences from the organism under study. We have reported cross-contamination between sequencing libraries generated in parallel [Citation47], and run-to-run carryover in sequencing platforms is a recognised problem [Citation49].
Ambiguous mapping
Even in the absence of contamination, mis-mapping can affect interpretation of sequencing results. miR-1246 was described by Wei et al. as the second-most abundant miRNA in FBS (and in pellet and supernatant fractions) [Citation16], but the sequence of mature miR-1246 is identical to a sequence within RNU2, a 188-nucleotide small nuclear RNA in human. Numerous copies of RNU2 are found in a variable-length region of human chromosome 17 that also contains BRCA1 [Citation50]. Cross-reactivity of miR-1246 assays with RNU2 fragments was reported in 2013 [Citation51]. In addition to miR-1246, we have found that the following human miRNAs have identity with ncRNA fragments and are often highly represented in sequencing data: miR-4448, miR-3960, miR-1248, miR-1290, miR-574-5p and miR-644b-5p. The existence of these miRNAs is not in dispute, but their quantitation might be confounded by mapping of ncRNA fragments. The presence of longer reads (>25 nt) partially mapping to those sequences is additional evidence of some degree of artefactual mapping.
Towards RNA-free media?
Altogether, as Wei et al. correctly suggest, future work will benefit from the implementation of RNA-free culture conditions. Removing serum is a good start. Certainly, serum introduces many unknowns into cell culture even without considering EVs and extracellular RNA. But removing serum might not be enough. Wei et al. detected some miRNAs (including miR-122-5p) in fresh non-conditioned media even in the absence of FBS. We could speculate that this detection occurred because some components of serum-free media contain impurities including RNA. For example, supplements like transferrin and albumin are biologically sourced and can vary considerably in quality [Citation52]. Growth factors produced from cells grown in serum, if not purified to homogeneity, could carry over bovine RNA or RNA from the cultured cells. Thus, even FBS-free culture conditions may not be truly chemically defined. RNA-free formulations: (1) may need to be developed for specific cell types; (2) should promote adherence, growth, and viability without evident signs of stress; and (3) should not interfere with EV secretion. Gene expression changes between cells grown in vesicle-depleted FBS and in defined RNA-free conditions are likely, but this is not necessarily a problem if the same media are used for different experimental conditions. When RNA-free culture is impossible, full-scale sequencing analysis of fresh media prior to conditioning should be included as a control to establish baseline detection of RNAs. One might also argue for the additional inclusion of what we would call “complete process controls,” in which the non-conditioned medium is passed through all disposables and procedures, including EV depletion, RNA extraction, and more, to be analysed along with the identically treated biological samples.
Summary
FBS-derived RNAs are pervasive and consequential confounding factors in extracellular RNA and EV research, as emphasised recently by Wei et al. Here, we have highlighted examples, including from our own research, in which FBS RNA may have contributed to apparent findings of specific RNA loading into EVs. To be sure, the Wei et al. findings and our analysis here do not suggest that all apparent RNA enrichment in exRNA fractions is artefact. The reportedly extracellular-enriched small RNA family of tRNA fragments [Citation30,Citation31,Citation41], e.g. were found at very low levels in the Wei et al. data, suggesting that their apparently enrichment is genuine. However, we encourage careful surveillance for warning signs of contamination and artefact: detection of cell-specific RNAs out of the expected cellular context, identification of RNAs in EVs but not in the putative parent cells, mapping of short RNA sequences that have 100% identity with other RNA fragments, and mapping of foreign RNAs that could be explained by reagent contamination instead of novel biological processes. Crucially, FBS and other sera are not the only sources of RNA contaminants, and other classes of macromolecules can also be affected. Keeping these observations in mind, experimental diligence and RNA-free or RNA-defined reagents are needed for investigations of EVs and exRNA to mature.
33836-supplementary-table.xlsx
Download MS Excel (39.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental meterial
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol [Internet]. 2013 Feb 20;200(4):1–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23420871
- Kim D-K, Lee J, Kim SR, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015 Mar;31(6):933–939.
- Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006 May;20(5):847–856.
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007 May 9;9(6):654–659.
- Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol [Internet]. 2008 Nov 18;10(12):1470–1476. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19011622
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A [Internet]. 2010 Mar 23;107(14):6328–6333. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20304794
- Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell [Internet]. 2016 Mar 10 [ cited 2016 Oct 9];164(6):1226–1232. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26967288
- Sverdlov ED. Amedeo Avogadro’s cry: what is 1 microg of exosomes? Bioessays [Internet]. 2012 Jul 21;34(10):873–875. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22815202
- Witwer KW, Halushka MK. Towards the promise of microRNAs – enhancing reproducibility and rigor in microRNA research. RNA Biol [Internet]. 2016 Sep 19 [ cited 2016 Oct 6];0. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27645402
- Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol [Internet]. 2014 [ cited 2016 Sep 22];12:87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25387460
- Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One [Internet]. 2014 Jan [ cited 2016 Jan 18];9(10):e110808. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4213012&tool=pmcentrez&rendertype=abstract
- Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol [Internet]. 2006 Jan 30; Chapter 3:Unit 3 22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18228490
- Pigati L, Yaddanapudi SCS, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One [Internet]. 2010 [ cited 2016 Sep 23];5(10):e13515. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20976003
- Shelke GV, Lässer C, Gho YS, et al. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles [Internet]. 2014 [ cited 2016 Sep 23];3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25317276
- Eitan E, Zhang S, Witwer KW, et al. Extracellular vesicle-depleted fetal bovine and human sera have reduced capacity to support cell growth. J Extracell Vesicles [Internet]. 2015;4:26373. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25819213
- Wei Z, Batagov AO, Carter DRF, et al. Fetal bovine serum RNA interferes with the cell culture derived extracellular RNA. Sci Rep [Internet]. 2016 [ cited 2016 Sep 22];6:31175. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27503761
- Kent OA, McCall MN, Cornish TC, et al. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res [Internet]. 2014;42(12):7528–7538. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24875473
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res [Internet]. 2011 May 26;39(16):7223–7233. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21609964
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A [Internet]. 2011 Mar 9;108(12):5003–5008. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21383194
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One [Internet]. 2012 Mar 20;7(3):e30679. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22427800
- Farrell D, Shaughnessy RG, Britton L, et al. The identification of circulating MiRNA in bovine serum and their potential as novel biomarkers of early mycobacterium avium subsp paratuberculosis Infection. PLoS One [Internet]. 2015 [ cited 2016 Oct 9];10(7):e0134310. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26218736
- Zhao K, Liang G, Sun X, et al. Comparative miRNAome analysis revealed different miRNA expression profiles in bovine sera and exosomes. BMC Genomics [Internet]. 2016 [ cited 2016 Oct 9];17(1):630. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27519500
- Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell [Internet]. 2007 Jun [ cited 2016 Nov 23];129(7):1401–1414. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0092867407006046
- Haider BA, Baras AS, McCall MN, et al. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One [Internet]. 2014 Mar 4;9(2):e89565. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24586876
- Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila). 2012 Mar;5(3):492–497.
- Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature [Internet]. 2008 Feb 28 [ cited 2016 Nov 24];451(7182):1125–1129. Available from: http://www.nature.com/doifinder/10.1038/nature06607
- Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem [Internet]. 2010 Oct 15;56(12):1871–1879. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20943850
- Cheng HH, Yi HS, Kim Y, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One [Internet]. 2013 Jun 14;8(6):e64795. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23762257
- Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res [Internet]. 2016 May 5 [ cited 2016 Nov 23];44(8):3865–3877. Available from: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkw116
- Tosar JP, Gambaro F, Sanguinetti J, et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res [Internet]. 2015;43(11):5601–5616. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25940616
- Baglio SR, Rooijers K, Koppers-Lalic D, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther [Internet]. 2015 [ cited 2016 Oct 9];6:127. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26129847
- Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol [Internet]. 2015 Jan 26 [ cited 2016 Nov 24];17(2):183–194. Available from: http://www.nature.com/doifinder/10.1038/ncb3094
- Guduric-Fuchs J, O’Connor A, Camp B, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics [Internet]. 2012 [ cited 2016 Oct 9];13:357. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22849433
- Ji H, Chen M, Greening DW, et al. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One [Internet]. 2014 [ cited 2016 Oct 9];9(10):e110314. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25330373
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep [Internet]. 2014;8(6):1649–1658. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25242326
- Li CCY, Eaton SA, Young PE, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol [Internet]. 2013 Aug [ cited 2016 Oct 9];10(8):1333–1344. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23807490
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife [Internet]. 2016 Aug 25 [ cited 2016 Oct 14];5:5003–5008. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27559612
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun [Internet]. 2013 Dec 21;4:2980. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24356509
- Holden P, Horton WA. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res Notes [Internet]. 2009 [ cited 2016 Nov 22];2(1):243. Available from: http://bmcresnotes.biomedcentral.com/articles/10.1186/1756-0500-2-243
- Graham JM, Rickwood D, editors. Subcellular fractionation: a practical approach [Internet]. Oxford: Oxford University Press; 1997. Available from: https://books.google.com/books?hl=en&id=h4hDKHEPTCcC&oi=fnd&pg=PR17&dq=subcellular+fractionation&ots=LfjjSzYFU5&sig=qUU-kYUjobhOipcwmmoIWz0D5qI#v=onepage&q=subcellularfractionation&f=false
- Nolte-’t Hoen EN, Buermans HP, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res [Internet]. 2012 Jul 24;40:9272–9285. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22821563
- Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res [Internet]. 2010 Jul 10;38(20):7248–7259. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20615901
- Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One [Internet]. 2008 Nov 13;3(11):e3694. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19002258
- Ouyang Y, Mouillet J-F, Coyne CB, et al. Review: placenta-specific microRNAs in exosomes - good things come in nano-packages. Placenta [Internet]. 2014 Feb [ cited 2016 Oct 9];35(Suppl):S69–S73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24280233
- Donker RB, Mouillet JF, Chu T, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod [Internet]. 2012 Aug [ cited 2016 Oct 9];18(8):417–424. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22383544
- Liao Z, Muth DC, Eitan E, et al. Serum extracellular vesicle depletion processes affect release and infectivity of HIV-1 in culture. bioRxiv [Internet]. 2016 Dec 8. Available from: http://biorxiv.org/content/early/2016/12/08/081687.1
- Tosar JP, Rovira C, Naya H, et al. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA [Internet]. 2014 Apr 15;20(6):754–757. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24729469
- Naccache SN, Greninger AL, Lee D, et al. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol [Internet]. 2013 Sep 13;87:11966–11977. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24027301
- Illumina. Reducing run-to-run carryover on the MiSeq using dilute sodium hypochlorite solution tech note.pdf – Google Drive [Internet]; 2013. Available from: https://docs.google.com/file/d/0B383TG7oJh2CTWp6MzZkMHBDd28/view
- Liu X, Barker DF. Evidence for effective suppression of recombination in the chromosome 17q21 segment spanning RNU2-BRCA1. Am J Hum Genet [Internet]. 1999 May [ cited 2016 Oct 9];64(5):1427–1439. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10205276
- Baraniskin A, Nöpel-Dünnebacke S, Ahrens M, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int J Cancer [Internet]. 2013 Jan 15 [ cited 2016 Oct 9];132(2):E48–E57. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22907602
- Chen Y, Stevens B, Chang J, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods [Internet]. 2008 Jun [ cited 2016 Nov 22];171(2):239–247. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0165027008001738
