ABSTRACT
Bioinformatics tools are imperative for the in depth analysis of heterogeneous high-throughput data. Most of the software tools are developed by specific laboratories or groups or companies wherein they are designed to perform the required analysis for the group. However, such software tools may fail to capture “what the community needs in a tool”. Here, we describe a novel community-driven approach to build a comprehensive functional enrichment analysis tool. Using the existing FunRich tool as a template, we invited researchers to request additional features and/or changes. Remarkably, with the enthusiastic participation of the community, we were able to implement 90% of the requested features. FunRich enables plugin for extracellular vesicles wherein users can download and analyse data from Vesiclepedia database. By involving researchers early through community needs software development, we believe that comprehensive analysis tools can be developed in various scientific disciplines.
Responsible Editor Peter J. Quesenberry, Brown University, United States
Advances in high-throughput techniques including next generation sequencing, RNA sequencing and proteomics have spurred enormous volume of data [Citation1–Citation3]. Currently, it is amenable to characterise the genome, transcriptome, metabolome and proteome of an organism in a robust manner. These technological developments reduced the amount of sample material, shortened the time to collect raw data and substantially decreased the associated costs [Citation4]. Hence, large scale approaches are now accessible by many research laboratories. To harness the true potential of these heterogeneous high-throughput data, software/bioinformatics tools have become indispensable resources for the ensuing analysis [Citation5]. To match the unprecedented growth in data generation, robust software analysis tools are constantly developed by academic and commercial entities [Citation4]. Most of the software tools are developed by specific laboratories or groups or companies wherein they are designed to perform the required analysis for the group. However, the software tools fail to capture “what the community wants in a tool”. A “community needs software” may overcome these hurdles and aid in the development of a comprehensive data analysis tool.
Here, we report a novel community needs software initiative in the context of functional or gene set enrichment analysis. To achieve this, we initially reached out to the scientific community through editorial [Citation6], conference participations, social networking sites and communicated with researchers in the OMICS community via e-mail. FunRich, an open access standalone functional enrichment analysis tool [Citation7], was used as a template for this purpose. By various means of communication, we invited the researchers to request for additional features/changes in FunRich software through the online forum (http://www.funrich.org/forum). We had enthusiastic participation from many researchers who requested for additional features/changes in the existing software. By September 2016, we had 54 unique requests/changes from users worldwide (Supplementary Table 1). The features were prioritised based on the number of users per request and were implemented in FunRich tool over the last 18 months. Remarkably, 90% of the requested features have been implemented in the new version of FunRich (Version 3). The updated version of FunRich is now freely available for download (http://www.funrich.org/download) both for academic and commercial users.
To gain biological insights, researchers often rely on functional enrichment analysis of large scale data from high-throughput experiments to identify overrepresented classes. Using FunRich, users can perform functional enrichment analysis with minimal or no support from computational and database experts for more than 13,320 species. The database is integrated from heterogeneous genomic and proteomic resources (>6.8 million annotations). The background database in any analysis tool is critical for the analysis and needs to be constantly updated [Citation8]. However, the currently existing functional enrichment analysis tools do not allow the users to control the databases nor to update them in real time [Citation8]. Using the forum, one of the request from the community pertained to “user controlled databases” and “regular update of background databases”. To address this, FunRich now uniquely allows the users to update the background database for 13,320 species from UniProt, Gene Ontology and Reactome in real time (). Additionally, the users can build custom databases with tab delimited files and perform the enrichment analysis irrespective of the organism and the type of dataset (e.g. metabolomics). Hence, these database options allows for longer sustainability of FunRich as a tool to perform functional enrichment analysis.
Figure 1. Features available in FunRich. FunRich is a free standalone functional enrichment analysis tool. Users can obtain customisable graphs, charts, interaction networks and heatmaps. All features of these images including color and font is editable and thus allows for quick representation of analysis. In spite of its ease of use, the images produced are of publication quality and can be directly imported into manuscripts. In addition, users have multiple background database options for more than 13,320 species. One of the features requested by the community is the option to update the background databases in real time. FunRich now allows the users to download data from UniProt, Reactome and Gene Ontology databases in real time. Furthermore, users can perform analyses in the context of biological pathways, gene ontology categories, protein domains, site of expression, cancer signatures, transcription factors, clinical phenotypes, extracellular vesicles, miRNA enrichment, protein interaction network and cross database accession conversion. The custom database option allows users to use any data type for any species thereby allowing for flexibility.
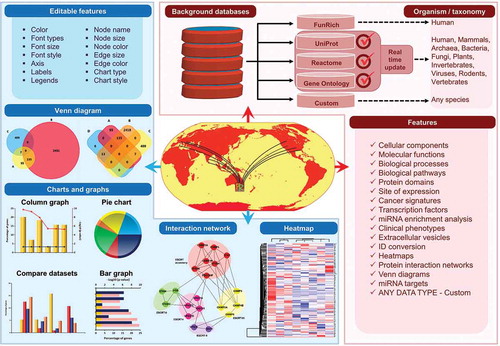
Other popular requests that have been implemented in FunRich include miRNA enrichment analysis (requested by most users – Supplementary Table 1), customisable heat maps, plugin to analyse extracellular vesicle datasets, comparison of oncogenes using COSMIC database and customisable colour for all the publication quality graphs. In miRNA enrichment analysis, users can submit a list of miRNA and identify biological pathways that may be perturbed. Gene set enrichment analysis is normally performed with the number of input genes/proteins and the quantitative data is often ignored. In FunRich, users can upload quantitative data and perform enrichment analysis for gene/protein expression values. For instance, total mRNA/protein abundance of genes involved in Wnt signalling pathway is compared between datasets in addition to number of genes. The quantitative data can also be utilised to generate customisable heat maps. Furthermore, users have complete control on all the graphs where the text and colour can be customised. Based on popular requests, FunRich now allows users to automatically download data from Vesiclepedia, an online compendium that hosts RNA and protein data pertaining to extracellular vesicles including exosomes [Citation9]. The input datasets can be compared with filtered Vesiclepedia data either through enrichment analysis or through Venn diagrams. In addition users can customise the data that can be downloaded from Vesiclepedia by using filters based on extracellular vesicles subtype, sample type, isolation method, cargo type and identification method.
Overall, with the involvement of researchers in the early phase of software development, we have developed a comprehensive tool for functional enrichment analysis. As databases are not regularly updated in most of the functional enrichment analysis tools [Citation8], the community requested features pertaining to automatic database update and custom database feature will allow for the continuous use of FunRich. Though we have completed 90% of the requests from researchers, we are constantly implementing the newer requests. We envision the development of a web-based version of FunRich and implementation of metabolomic analysis in the near future. With the advent of large volumes of data, it is critical to build such comprehensive software tools for data analysis. Based on this fruitful experience, we strongly encourage community driven software development for research purposes so as to build comprehensive software tools and to curtail software duplications.
Supplementary_table_1.pdf
Download PDF (102.8 KB)Acknowledgements
We would like to thank Tejaswee P. Shah for her valuable input in the development of FunRich. We would like to thank Jan Lotvall for helping us in this effort by circulating the email within his group. We thanks many other users who requested for features/changes in FunRich though the e-mail and the online forum. Suresh Mathivanan is supported by Australian Research Council DECRA (DE150101777) and Award U54-DA036134 supported by the NIH Common Fund through the Office of Strategic Coordination/Office of the NIH Director. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
The supplemental material for this article can be accessed here.
Additional information
Funding
References
- Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:1–4.
- Gerstein MB, Rozowsky J, Yan -K-K, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448.
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
- Muir P, Li S, Lou S, et al. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 2016;17:53.
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550.
- Benito-Martin A, Peinado H. FunRich proteomics software analysis, let the fun begin! Proteomics. 2015;15:2555–2556.
- Pathan M, Keerthikumar S, Ang C-S, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601.
- Wadi L, Meyer M, Weiser J, et al. Impact of outdated gene annotations on pathway enrichment analysis. Nat Methods. 2016;13:705–706.
- Kalra H, Simpson RJ, Hong J, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450.
