ABSTRACT
Circular RNAs (circRNAs) are a novel class of noncoding RNAs present in all eukaryotic cells investigated so far and generated by a special mode of alternative splicing of pre-mRNAs. Thereby, single exons, or multiple adjacent and spliced exons, are released in a circular form. CircRNAs are cell-type specifically expressed, are unusually stable, and can be found in various body fluids such as blood and saliva. Here we analysed circRNAs and the corresponding linear splice isoforms from human platelets, where circRNAs are particularly abundant, compared with other hematopoietic cell types. In addition, we isolated extracellular vesicles from purified and in vitro activated human platelets, using density-gradient centrifugation, followed by RNA-seq analysis for circRNA detection. We could demonstrate that circRNAs are packaged and released within both types of vesicles (microvesicles and exosomes) derived from platelets. Interestingly, we observed a selective release of circRNAs into the vesicles, suggesting a specific sorting mechanism. In sum, circRNAs represent yet another class of extracellular RNAs that circulate in the body and may be involved in signalling pathways. Since platelets are essential for central physiological processes such as haemostasis, wound healing, inflammation and cancer metastasis, these findings should greatly extend the potential of circRNAs as prognostic and diagnostic biomarkers.
Introduction
Thrombocytes – also referred as platelets – belong to the corpuscular components of mammalian blood and are essential for central physiological processes such as haemostasis (see, for review, [Citation1]). Platelets are peculiar in that they are, first, relatively short-lived (~7 days half-life); second, they are the smallest cells of blood, and third, although without a nucleus, they exhibit active RNA metabolism and are translationally active [Citation2–Citation5]. During thrombopoiesis, anucleated platelets are derived from megakaryocytes in the bone marrow and are released into the bloodstream. In the absence of de novo RNA synthesis, platelets mirror the particular expression status of the megakaryocyte at the specific time point they are generated. To orchestrate all their tasks, platelets release – in addition to chemokines and cytokines – two major types of extracellular vesicles (EVs) into the bloodstream, which can be internalised by different target cells, such as endothelial cells [Citation6,Citation7]. In fact, platelets release massive amounts of EVs, reflected by that approximately 25% of all EVs in the circulating plasma are platelet-derived [Citation8,Citation9]. Therefore, the functional relevance of EVs and their molecular content, in particular miRNAs, have become an important new focus of current research.
Recently, transcriptomic approaches uncovered circular RNAs (circRNAs) as a novel class of noncoding RNAs, functionally still largely undefined [Citation10–Citation14]. They are characterised by a special splice junction, in which the 5’- and 3’-ends of a single exon, or of several adjacent exons, are joined through an alternative mRNA splicing mechanism also referred to as backsplicing (see Starke et al. [Citation15] and references therein). As a result of their circular configuration and their resistance to exoribonucleolytic degradation, circRNAs are unusually stable. Several studies established that circRNAs are cell-type specifically expressed, relatively abundant in the brain, and also exist in body fluids such as human blood [Citation16–Citation19]. In particular, the platelet fraction shows a high proportion of circRNAs, attributed to general degradation of linear RNAs during the lifetime of platelets [Citation20].
In this study, we characterised circRNAs in human platelets, as well as in isolated EVs (microvesicles and exosomes) derived from activated platelets. Our analysis of circRNAs and their corresponding linear spliced isoforms revealed that, compared to other hematopoietic cell types, circRNAs are particularly enriched in platelets. In addition, by activating platelets and thereby inducing the release of EVs, we showed that circRNAs are selectively packaged and released within vesicles, suggesting a specific sorting mechanism.
Materials and methods
Platelets
Apheresis platelets from normal donors were obtained from the Institute for Clinical Immunology and Transfusion Medicine, Justus Liebig University of Giessen. Fresh apheresis leukoreduced platelets from healthy single donors were isolated, using a Trima Accel Automated Blood Collection System equipped with a leukocyte reduction chamber (Terumo; Eschborn, Germany). The average platelet yield was around 1.2 × 109 platelets/mL with a leukocyte contamination of less than 2 × 105 cells/mL. Aliquots of 10 mL platelet suspension were used as starting material for this study. The ethics committee of the Justus Liebig University of Giessen approved the study, and a written informed consent was obtained from all donors.
Platelets were washed twice in Phillips buffer (96.5 mM NaCl, 87.5 mM glucose, 1.1 mM EDTA, 8.5 mM Tris-Cl pH 7.4), pelleted by centrifugation (15 min at 800 xg, at room temperature) and resuspended in Phillips buffer. For activation, Phillips buffer was supplemented with 1 mM MgCl2, 2 mM CaCl2, and 3 mM KCl, and platelets were activated for 60 min at 37°C with 10 µM TRAP-6 (Sigma-Aldrich).
RNA isolation, Northern blotting, RT-PCR
Total RNA was extracted using TRIzol (Ambion). 200 ng of total RNA were used for cDNA synthesis (qScript cDNA Flex Synthesis Kit, Quanta). For RT-PCR, primers were designed to flank the circRNA-specific junction (circular isoforms) or the linear splice junction of downstream exons (linear isoforms). PCR products were analysed by 2% agarose gel electrophoresis. Primer were designed using the program primer3 version 4.0.0 (http://www.bioinfo.ut.ee/primer3/). All oligonucleotides can be found in Supplementary Table S4.
Real-time PCR was carried out using PerfeCTa SYBR Green FastMix (Quanta) on an Eppendorf realplex2 thermocycler. The relative expression levels of circular versus linear isoforms in platelets were calculated by the ΔΔCt-method, with each target normalised to ACTB (referring to Figure 1(c)). Biological replicates were used to calculate standard deviations. The relative levels of circular versus linear isoforms in microvesicles and exosomes versus platelets were determined (2−ΔΔCt) and normalised to the geometric mean of two housekeeping genes (ACTB, B2M) (referring to ).
Figure 1. High abundance of circRNAs in human platelets. (a) Relative expression of circRNA vs. linear isoforms, based on the ratio of total circRNA-junction/total linear-junction reads, comparing whole blood and different human hematopoietic cell types. (b) The read counts (log2) of corresponding circular and linear isoforms (Y-axis, circRNA-junction; X-axis, linear-junction) are plotted for the 100 most abundant circRNAs from each sample. Genes with predominant circular isoforms are plotted in red, with predominant linear isoforms in black. (c) RT-PCR validation of circRNAs in human platelets. 10 corresponding circular and linear isoforms were detected by semi-quantitative RT-PCR, using total RNA from purified resting and activated platelets (-/+TRAP) and either convergent (linear) or divergent (circular) primer pairs, with ß-actin (ACTB) as a loading control (left panel). M, markers (500, 400, 300, 200, 100 bp). The right panel shows the ratio of circular/linear isoforms for the same set of 10 targets, as determined by RT-qPCR in activated (+TRAP, Y-axis) and resting (-TRAP, X-axis) platelets.
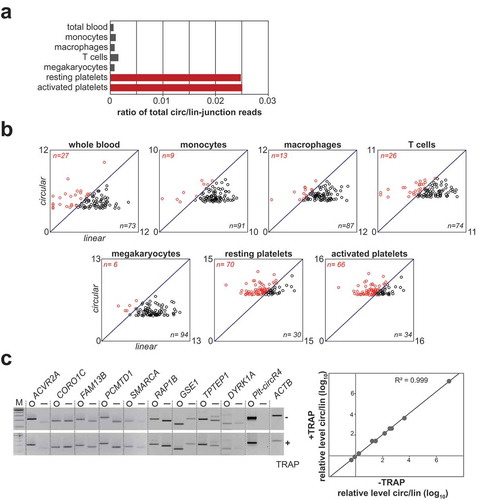
Figure 2. Platelet-specific circRNA Plt-circR4. (a) Plt-circR4 is exclusively expressed in platelets. Plt-circR4 was detected by semi-quantitative RT-PCR, comparing platelets and cell lines indicated. (b) Exon-intron structure of Plt-circR4 genomic locus and its expression. RNA-seq read coverage is displayed above the four exons, and the read counts for the circRNA-junction of exon 3 and the linear splice junctions are indicated below. The region around the Hsmar1 transposon is shown enlarged, including its terminal inverted repeats (arrows). The bottom panel shows RT-PCR detection of circular and linear isoforms, using total RNA from platelets after mock or RNase R treatment. (c) Glyoxal Northern blot analysis reveals exact size of Plt-circR4. Total platelet RNA was analysed by glyoxal agarose gel electrophoresis and Northern blotting. (d) Evidence for circularity of Plt-circR4 by combined Northern/RNase H analysis. The experimental strategy is outlined on the right, including expected fragment sizes after oligonucleotide-directed RNase H cleavage. Total RNA from platelets was treated by RNase H, using either a single antisense oligonucleotide (oligo #1 or #2) or both in combination (oligo #1/2). Control reactions were done in the absence of oligonucleotide and with/without RNase H. RNA was analysed by denaturing polyacrylamide gel electrophoresis and Northern blotting. The mobilities of RNase H-cleaved species are indicated by arrows.
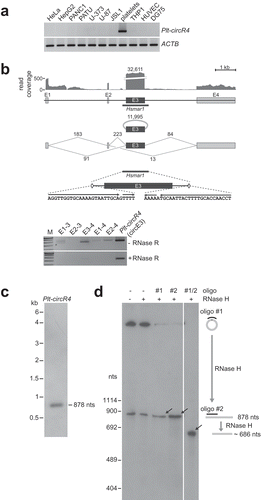
Figure 3. CircRNA-protein complexes in human platelets. Sedimentation profiles of circRNPs. Platelet extract and corresponding free RNA (prepared from the same extract) were fractionated by glycerol gradient centrifugation (fractions #1–12 from top to bottom), followed by RT-PCR analysis across the gradient for five abundant circRNAs (RAP1B, GSE1, CORO1C, PCMTD1 and Plt-circR4) and for one linear mRNA control (RAP1B). The positions of ribosomal RNAs are marked by arrowheads (5S, 18S and 28S). M, markers (300, 200, 100 bp); lane I, 10% platelet extract input. The right panel shows quantification of the gradient fractionation of extract vs. RNA.
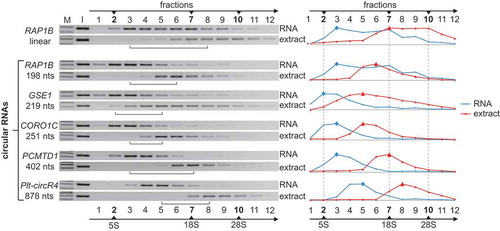
Figure 4. Isolation of platelet-derived EVs. (a) Flow chart of the purification of platelet-derived microvesicles and exosomes, based on differential centrifugation (for details, see Materials and methods). (b) Electron microscopy of purified vesicles. Purified exosomes and microvesicles were counterstained by uranyl acetate. In addition, exosomes were stained with 10 nm gold-conjugated anti-CD63 antibody (in two different magnifications). Scale bar, 100 nm. (c) Characterisation of exosome preparation by Western blot. Exosomes were purified by sucrose-density centrifugation, and each fraction (#1–12, from top to bottom) was subjected to Western blot analysis, using the exosomal marker CD63, with calnexin as a negative control. Platelet extract was loaded as input control. Sucrose densities (in g/mL) of the individual fractions are indicated at the bottom. (d) CircRNAs cofractionate with exosomes. Total RNA was isolated from each fraction of the sucrose gradient (#1–12, from top to bottom), followed by RT-PCR, using circRNA-specific primers for GSE1 and NRIP1. M, markers (200, 100 bp). (e) Purified microvesicles and exosomes were analysed by dynamic light scattering, depicting the calculated size distribution.
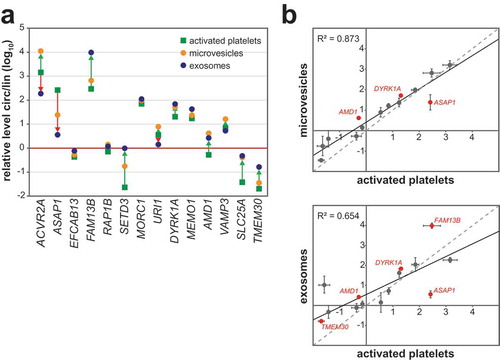
Figure 5. CircRNAs are selectively released into platelet-derived EVs. (a) The relative levels of corresponding circular and linear isoforms are shown, as determined by RT-qPCR for 14 targets, comparing activated platelets, microvesicles, and exosomes. Only mean values of two replicates are given (for replicate data, see panel b) (b) The relative circ/lin levels (as in panel a) are plotted for microvesicles and exosomes (Y-axis), each compared with activated platelets (X-axis). The fitted regression line and coefficient of determination R2 are indicated. CircRNAs preferentially released into EVs or preferentially retained in platelets are plotted in red.
RNase R treatment was carried out using 2 µg total RNA treated with or without RNase R (2.5 U/μg, Epicentre) for 20 min at 37°C. RNA was phenolised, ethanol-precipitated, and 20% were used for RT-PCR as described above.
For glyoxal Northern blot 5 µg of total RNA from platelets was mixed with glyoxal loading dye (Ambion) and incubated at 50°C for 30 min. RNA was separated by agarose gel electrophoresis (1.2% in 1x MOPS buffer), using the 0.1–2 kb RNA ladder (Thermo Fisher Scientific). The gel was transferred onto a nylon membrane by semidry blotting. RNA was crosslinked by ultraviolet (UV) light and probed with single-stranded RNA probe (DIG RNA Labeling Mix; Roche), which covered the circ-junction. Hybridisation was performed in NorthernMax buffer (Thermo Fisher Scientific), and washes of the blot and probe detection with alkaline phosphatase-conjugated anti-DIG-Fab fragments were done as described in the Roche manual (Roche). RNase H assays were performed as described previously [Citation15]. Subsequent Northern hybridisation analysis on denaturing polyacrylamide gels was done as described above.
RNA-seq and data analysis
For high-throughput sequencing, total RNA from platelets and microvesicles was depleted of ribosomal RNA, using the Ribo-Zero Gold rRNA Removal Kit (Epicenter, MRZG126), followed by library preparation (NEBNext Ultra Directional RNA Library Prep Kit, NEB, E7420, according to the manufacturer’s instructions). For library preparation of RNA isolated from exosomes the SMARTer® Stranded RNA-Seq Kit (Takara-Clontech) was used, according to the manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq 2500 (paired-end, 2 × 130 bp, platelets), or on Illumina NextSeq 500 (single-read, 150 bp, microvesicles and exosomes). RNA-seq data were deposited in the Sequence Read Archive (SRP118609) from NCBI.
Sequence reads were aligned to the human genome sequence (hg19 assembly) using STAR, an ultrafast universal RNA-seq aligner with chimeric alignment options [Citation21]. Comprehensive gene annotation from GENCODE Version 19 (http://www.gencodegenes.org) was used for gene annotation. From the chimeric mapped reads, circRNA-specific junction reads were selected, applying these four criteria:
Sequence read maps to the same chromosome and the same strand, with the two segments mapping to the genomic region in reverse order.
The overhang spanning the circRNA junction is ≥12 nts.
The difference between the alignment score of the chimeric mapped read (using column 14: AS of Standard SAM attributes) and the score of linear alignment with genomic sequences or annotated transcripts must be greater than 2.
Both 5′ and 3′ splice sites are either annotated or conform to canonical splice sites. The circRNA-specific junction reads were used to estimate the abundance of circRNAs. The linear-junction read counts were used to estimate the abundance of the corresponding linear isoform and were determined by halving the total number of up- and downstream linear junction reads that share 5’ and 3’ ends of the circular exon(s).
Glycerol gradient sedimentation analysis
Extracts of TRAP-6 activated platelets were prepared in gradient-lysis buffer [5 mM HEPES-KOH pH 7.5, 1.5 mM KCl, 1.5 mM MgCl2, 0.5% Triton X-100, 0.5% deoxycholate, 5% glycerol, 1 mM DTT, RNaseOut and HALT protease inhibitor cocktail (Thermo Fisher Scientific)]. In parallel, total RNA was extracted from an equal amount of activated platelets to be used for the control gradient (free RNA). Extracts/free RNA were loaded onto 10–30% (v/v, 12 mL) linear glycerol gradients [20 mM HEPES-KOH pH 8.0, 150 mM KCl, 1.5 mM MgCl2, 1 mM DTT, HALT protease inhibitor cocktail (Thermo Fisher Scientific)], prepared with the Gradient MasterTM 108 (BioComp) and subjected to ultracentrifugation (16 h at 182,000 xg, at 4°C). After centrifugation, the gradient was fractionated manually into 12 fractions of 1 mL each. RNA was isolated by phenol/chloroform extraction (Roth) from 750 µL of each fraction and from 10% of the input, followed by RT-PCR (as described above). PCR products were analysed on 2% agarose gels and quantified (GeneTools software; Syngene). As size markers, the sedimentation of ribosomal RNAs (present in HeLa total RNA) was analysed (Agilent 2100 Bioanalyzer; RNA 6000 Nano Kit, Agilent).
Isolation of platelet-derived microvesicles and exosomes
A total of approximately 1.8 × 109 platelets in 5 mL Phillips buffer were activated TRAP-6. Activated platelets and cell debris were pelleted by subsequent centrifugation steps (10 min at 1000 xg, at room temperature; 5 min at 2500 xg, at 4°C; and 10 min at 5000 xg, at 4°C), each time transferring the supernatant into a new tube. To enrich for microvesicles, after the last centrifugation the supernatant was transferred and centrifuged for 60 min at 12,000 xg, at 4°C. The microvesicle pellet was resuspended in 50 µl 1x PBS and analysed by electron microscopy, Western blot analysis and dynamic light scattering. For exosomes, the supernatant after microvesicle enrichment was concentrated to 500 µL with Amicon Ultra-15 10K Centrifugal Filter Device (Merck Milipore) using a Heraeus MultifugeTM X1R centrifuge at 4000 xg at 4°C. The concentrate was recovered with a reverse spin at 1000 xg for 2 min. Subsequently, concentrate was applied on top of a linear sucrose density gradient (10–50%) prepared with the Gradient MasterTM 108 (BioComp) and subjected to ultracentrifugation for 18 h at 4°C (100,000 xg). Exosome-containing fractions were pooled, diluted fivefold with 1x PBS, and exosomes were pelleted by ultracentrifugation (2 h at 100,000 xg, at 4°C).
Electron microscopy
Electron microscopy was performed on EV pellets stored at −80°C. For whole-mount analysis, EV suspensions (5 µL each) were incubated for 1 min on formvar and carbon-coated glow-discharged copper grids and subsequently stained with 2% uranyl acetate. Immunogold labelling was performed as described [Citation22–Citation24]. After blocking with 1% BSA in phosphate-buffered saline, grids were incubated with a 1:50 dilution of primary mouse anti-CD63 antibody (Thermo Fisher Scientific; TS63) in PBS containing 0.1% BSA for 1 h at room temperature. Extensive washes in the same buffer were followed by incubation with rabbit-anti-mouse antibody (Jackson ImmunoResearch) for 30 min, and after repeated washes 5-nm Protein A-gold (UMC Utrecht, the Netherlands) was applied for 1 h. After subsequent thorough washing, preparations were fixed using 1% glutaraldehyde in PBS, washed again and finally stained with 2% uranyl acetate. For controls, the primary antibody was omitted, resulting in complete loss of labelling (Supplementary figure S1). Preparations were inspected in an EM912 AB transmission electron microscope (Zeiss) at 120 kV under zero-loss conditions at slight underfocus. Images were recorded using a 2k x 2k slow-scan CCD camera (TRS, Germany) and the iTEM software package (Olympus-SIS).
SDS-PAGE and Western blotting
Platelets as well as microvesicle and exosome preparations containing 1 µg of total protein were lysed with 1x SDS sample buffer and run on a 12% SDS protein gel. Proteins were transferred onto a nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% (w/v) milk powder (Roth) in phosphate-buffered saline supplemented with 0.05% Tween (PBS-T) and probed with a 1:500 dilution of primary mouse anti-CD63 (Thermo Fisher Scientific; TS63) and a 1:20,000 dilution of anti-calnexin (Santa Cruz; AF18) for 60 min at room temperature in 3% milk in PBS-T buffer, followed by incubation with the secondary antibody (anti-mouse-IgG-HRP; Sigma-Aldrich) in 5% milk in PBS-T for 45 min at room temperature. After extensive washing, the blots were incubated for 1 min at room temperature with ECL Western Blotting Substrate (Roche) and visualised with Amersham HyperfilmTM ECL (GE Healthcare).
Dynamic light scattering (DLS)
DLS measurements were performed with a Laser Spectroscatter 201 (RiNA GmbH). Samples were diluted 1:1000 in 1x PBS. 3 × 10 measurement runs were performed, with standard settings (refractive index = 1.331, viscosity = 0.89, temperature = 25°C).
Results and discussion
Circular RNAs are highly abundant in platelets
Since circular RNAs constitute promising new biomarker candidates [Citation25], we examined their expression in human hematopoietic cells, based on available RNA-seq data from ribosomal-RNA-depleted total RNA (European Nucleotide Archive and NCBI Sequence Read Archive): whole blood (SRP029990), monocytes (ERP005301), macrophages (ERX326139), T cells (ERX326142) and megakaryocytes (ERX326138). In addition, we generated our own data based on two RNA-seq libraries of ribosomal RNA-depleted total RNA from human platelets, comparing resting state and after activation by TRAP-6 (Thrombin Receptor Activator Peptide-6). CircRNA abundance was determined based on the read counts of circRNA-specific back-splice junctions (for alignment statistics, see Supplementary Table S1).
First, we compared in whole blood and in the individual cell types the ratios of the total number of circRNA-junction reads to the uniquely-mapped, linear junction reads ()). Clearly, in all cases except for platelets this ratio is relatively low, in the order of 0.001. In contrast, in platelets this is very different, with the circ/lin ratio around 0.025, indicating that circRNAs are particularly abundant in platelets.
To analyse this in more detail, we selected the one hundred most abundant circRNAs from each sample; for each target, the circRNA-junction read counts were compared with the corresponding linear junction read counts to estimate the ratio between circular and linear isoforms (), Supplementary Table S2). Among the selected circRNAs, about 95% are derived from the exons of protein-coding genes. Compared with monocytes, macrophages, T cells and megakaryocytes, circular isoforms in whole blood are more abundant. Strikingly, in platelets, a large set of circular isoforms (70% in resting, 66% in activated platelets) are predominantly expressed, and 95% of these abundant circRNAs are identical in resting and activated platelets. These results are consistent with the previous study by Alhasan et al. [Citation20], which showed that circRNAs are highly enriched in platelets and, in addition, that for some genes only the circular isoform was detected; especially the enrichment of back-splice junctions detected is clearly in line with our finding, when comparing the circular/linear-junction reads in the different hematopoietic cell types (see )).
For validation, 10 of the 100 most abundant predicted circRNAs in platelets were selected. We monitored their expression by semi-quantitative RT-PCR and quantitative RT-qPCR, comparing resting and TRAP-6-activated platelets, using either divergent primer pairs, which specifically detect the circular junction, or primer pairs, which detect a canonical linear splice junction downstream of the circularising exons ()). Consistent with our RNA-seq approach, most of the RNAs tested exist predominantly in their circular configuration, and we observed no significant difference between resting and activated platelets.
Plt-circR4, a platelet-specific circRNA
Next we focused on the most abundant circRNA (called Plt-circR4 in the following) in platelets and screened its expression in 10 different cell lines. We conclude that Plt-circR4 appears to be exclusively expressed in platelets ()).
Regarding the genomic structure of Plt-circR4, we identified, based on RNA-seq data, a novel transcript with 4 exons, which mapped to the minus strand of chromosome 4 (); for exon positions, see Supplementary Table S3). In addition to the predominant circular form of exon 3, a very minor proportion of linear-spliced isoforms could be detected. Around exon 3, a DNA mariner transposon (Hsmar1) was recognised by RepeatMasker (http://www.repeatmasker.org) [Citation26]. This class of transposons encodes a mariner transposase that is flanked by inverted terminal repeats [Citation27], which we identified 140 bp upstream of the 5’ end and 195 bp downstream of the 3’ end of the Plt-circR4 exon. We note that inverted repeat elements can modulate RNA circularisation [Citation11,Citation14].
To conclusively prove the circular configuration of Plt-circR4, we used a combination of approaches. RT-PCR validations alone can result in false-positive results, because potential trans-spliced mRNA species would produce the same products detectable by the divergent primer pairs. First we used exoribonuclease treatment by RNase R, followed by RT-PCR, to check for circularity ()). Only Plt-circR4 (circE3) was RNase R resistant, in contrast to the corresponding linear isoforms, as detected by primer combinations covering all identified splice variants.
Second, we applied denaturing glyoxal-agarose electrophoresis and Northern blot hybridisation to determine the exact size of the circular RNA, using a probe specifically for the back-splice junction ()). One single band was detected for Plt-circR4 at approximately 900 nts, consistent with the expected size (878 nts) of Plt-circR4.
Finally, to unequivocally demonstrate circular configuration, we combined Northern blot analysis after polyacrylamide gel electrophoresis with RNase H cleavage assays, as described previously [Citation15] ()). Note that in polyacrylamide gel electrophoresis circular RNAs display a characteristically slow migration, compared to linear RNA markers, in contrast to agarose gel electrophoresis, where circRNA mobility corresponds to linear markers. For example, in case of Plt-circR4 the major band detected runs near the top of this polyacrylamide gel, above the 1114 nts marker. In addition, a minor band was consistently detected, running at the size expected for the linearised form of Plt-circR4 (878 nts). This may represent a linear degradation product of the circRNA, or an alternative processing isoform or intermediate.
To linearise the Plt-circR4 circRNA, total RNA from platelets was treated with RNase H, directed by either one antisense DNA oligonucleotide (#1, #2) or both of them together (#1/2); oligo #1 targets a sequence spanning the circular splice junction of Plt-circR4, oligo #2 a specific exon sequence. When we used oligonucleotide #1 or #2 alone, the band for the presumptive circular isoform disappeared, and the band of the linear form increased (arrow), consistent with the expected characteristic shift in gel mobility from a slowly-migrating circular form to the expected linear form. This was further supported by using both oligonucleotides together (#1/2), which resulted in a product at approximately 686 nts (arrow), the longer one of two cleavage products (the shorter one, of 192 nts, ran off this gel). In sum, the combined evidence (different mobility in agarose versus polyacrylamide gels, see and ); RNase R resistance, see ); Northern/RNase H assays, see )) convincingly establish the circular configuration of Plt-circR4 circRNA.
Circular RNAs are organised in circRNPs of distinct sizes in platelets
In a recent study our group could provide biochemical evidence for distinct cytoplasmic circRNA-protein complexes in mammalian cells [Citation28]. Since the anucleated platelets represent a very special system without active transcription, we analysed whether circRNAs in platelets are organised as circRNPs ().
After fractionating extract prepared from activated platelets and the corresponding free RNA by glycerol gradient centrifugation, we monitored by RT-PCR the distribution across the gradient of five abundant platelet circRNAs (RAP1B, GSE1, CORO1C, PCMTD1, and Plt-circR4), as well as the linear mRNA isoform of RAP1B as a control. Whereas circRNAs detected in the free RNA sample were mainly found in fractions #2–5, the corresponding peaks in the extract sample moved faster into the gradient, shifted by two to four fractions. The individual shifts observed for this set of five circRNAs correspond to a hypothetical calculated molecular mass differences between 230 to 740 kDa. However, there was no correlation between circRNA size and the apparent circRNA-circRNP shift in molecular mass, indicating that circRNAs are organised in distinct large RNP complexes in platelets.
Circular RNAs are associated with platelet-derived extracellular vesicles
Previous studies showed that circRNA can be found in cell-free body fluids such as serum and plasma [Citation19]. Since platelets release two major types of EVs, microvesicles and exosomes [Citation6], we next asked whether circRNAs are associated with platelet-derived EVs. Although the association of circRNAs and EVs had been addressed before [Citation29], one has to be aware of that the results of such studies strongly depend on the methodology how vesicles were isolated. Therefore, we took great care in using stringent isolation procedures and criteria for EV preparation ()). Microvesicles and exosomes are generated by different biogenesis routes and can be distinguished by their size and sedimentation behaviour.
To assess specific enrichment of microvesicles and exosomes, we used transmission electron microscopy, which demonstrated high specificity of our isolation procedure ()). In addition, we confirmed the specificity of our exosome purification by immunogold labelling, using antibodies directed against CD63, one of the classical exosomal markers ()).
Moreover, we used the same CD63 antibody in Western blot analysis to assess specific enrichment of exosomes, assaying all 12 fractions after sucrose-density gradient centrifugation ()). CD63 could be clearly detected in fractions #5 to 7, with the major peak in fraction #6, consistent with the expected density of exosomes (1.10–1.12 g/mL). The endoplasmic reticulum marker calnexin served as a negative control. To examine whether circRNAs are associated with EVs, we used RNA isolated from each fraction across the sucrose gradient (#1–12, from top to bottom) and monitored by RT-PCR the distribution of two abundant circRNAs in platelets, GSE1 and NRIP1 ()). Clearly both circRNAs could be detected in the fractions where exosomes are highly enriched, indicating that circRNAs are associated with platelet-derived vesicles. Finally, we determined the relative size distribution of the enriched EVs (microvesicles and exosomes) by dynamic light scattering ()). However, note that without further characterisation this does not allow to distinguish between microvesicles and similar-sized lipoprotein particles or small platelets [Citation30]. In addition, the measured size of the isolated microvesicles appeared on the lower limit of what one expects. Please note that in particular the isolation of these vesicles from blood samples such as apheresis platelets is affected by venipuncture, centrifugation and washing steps, as well as by the presence of lipoprotein particles. Therefore, larger particles sub-types may aggregate and/or precipitate during the first steps of centrifugation. Nevertheless, we chose differential centrifugation to obtain a clean subpopulation of microvesicles. In sum, together with the electron microscopy and Western blot analysis, we conclude that we specifically enriched for either type of platelet-derived vesicles.
Circular RNAs are selectively released in EVs
To test whether circRNAs are selectively released, we monitored by quantitative RT-qPCR the expression of 14 circRNAs selected from the 100 most abundant circRNAs detected in platelets, comparing corresponding circular and linear isoforms. These circRNA candidates were chosen based on, first, being abundant in platelets and, second, that they were either enriched or non-enriched in microvesicles and exosomes. The relative levels of circular/linear isoforms (circ/lin) were determined in platelet-derived microvesicles and exosomes, as well as in their progenitor cells, activated platelets (). Interestingly, we observed a subset of targets (AMD1, DYRK1A for microvesicles; FAM13B, DYRK1A, AMD1, TMEM30 for exosomes), which are preferentially released into EVs, compared with their corresponding linear isoforms. In contrast, the circular isoform of ASAP1 is preferentially retained in platelets. Taken together, these data indicate that circRNAs are selectively released into vesicles. In exosomes, this phenomenon is more pronounced.
Another study proposed that the release of circRNAs in EVs may reflect a kind of clearance mechanism [Citation31]. However, the selective release described here appears to be contrary to the clearance hypothesis. We favour the interpretation that selective release mechanisms of circRNAs represent a novel way of transferring information from donor to recipient cells. As reported in several studies, vesicles from platelets can be internalised by recipient cells such as endothelial cells, one of the main interaction partners of platelets thereby inducing inflammatory responses in the endothelial cell [Citation7].
In addition, we tried to systematically analyse by RNA-seq the selective release of circular RNAs from platelets into vesicles (Supplementary figure S2A). Relying on the circRNA-junction read counts as a measure for circRNA abundance, we identified 1117 circRNAs in microvesicles and 154 in exosomes. Fisher’s exact test was applied to assess statistical significance for each circRNA, using its circRNA-junction read counts in exosomes/microvesicles versus activated platelets and the corresponding total circRNA-junction read counts in each sample. Volcano plots show for each circRNA the ratio of relative abundance (microvesicles versus platelets in the left panel; exosomes versus platelets in the right panel): 32 circRNAs are preferentially released in microvesicles, whereas three are preferentially retained in platelets; and 35 circRNAs are preferentially released in exosomes. These data further support the existence of a selective sorting pathway for circRNAs.
Due to the very low RNA quantities obtained from our exosome preparations, and the corresponding low number of circRNA-junction reads from RNA-seq data, we are cautious in drawing conclusions. These low read numbers might reflect the fact that circRNAs exist in vesicles only in very low copy numbers, as reported for miRNAs in exosomes [Citation32].
Finally, to search for potential sorting determinants, we asked whether there may be a correlation between circRNA size and packaging into vesicles as hypothesised earlier for other classes of RNA [Citation33]. CircRNA size appears to be plausible in this regard, considering that the available packaging volume within a vesicle might be limiting to accommodate RNP complexes of larger circRNA molecules. Therefore, we compared the size of circRNAs which are preferentially released with those which are preferentially retained in platelets (Supplementary figure S2B). Based on the size distributions, we conclude that preferentially released circRNAs are smaller than preferentially retained circRNAs (mean of 283 versus 459 nts for microvesicles; 286 versus 435 nts for exosomes), suggesting that size appears to be an important determinant for selective vesicle export of circRNAs. However, one has to take into account that circRNAs are organised in RNPs as shown in . Although circRNPs appear to be distinct RNP complexes, small circRNAs can in general bind fewer proteins than very large circRNAs; for example, GSE1, one of the shortest circRNAs analysed here, assembles into a relatively small RNP (see ). We note that, in addition to the circRNA size, there are likely other sorting and selection determinants, for example, particular sequence motifs, as shown for miRNAs [Citation34].
As shown in this work and by others [Citation20,Citation35], circRNAs are clearly enriched in platelets compared to other blood cells. Moreover, the ratio between the linear spliced mRNA and the circular isoform is very often shifted towards the circular form, which is more stable than linear RNA [Citation20]. Therefore, Alhasan et al. had explained this phenomenon by differential degradation of cellular platelet RNA [Citation20]. However, the selective RNA release in EVs, as shown in this study, clearly argues against a role of EVs as a general RNA discard pathway. Selective RNA release is also supported by our identification of an abundant, platelet-specific circRNA (Plt-circR4), which is not preferentially released into vesicles. One might consider thrombocytes themselves as a kind of vesicles, which may selectively deliver certain circRNAs and alter their circRNA profile in response to signals from the environment, as recently reported for mRNA profiles of “tumour-educated” platelets [Citation36]. In sum, our study provides new insights into the circRNA biology of platelets and creates starting points for the development of novel biomarkers linked to platelet-associated diseases including cancer as well as cardiovascular disorders.
Supp_Tables_figs.zip
Download Zip (559.6 KB)Supp_Tables.zip
Download Zip (97.7 KB)Acknowledgements
We thank Joachim Misterek for the recruitment of apheresis blood donors, Vladimir Benes and the EMBL GeneCore sequencing team for excellent sequencing support, and the members of our group for discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, to A.B. and Se.S.), the LOEWE programme Medical RNomics (State of Hessen; to A.B.), and the Excellence Cluster Cardiopulmonary System (to Se.S.).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementry matieriels
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Smyth SS, McEver RP, Weyrich AS, et al. Platelet functions beyond hemostasis. J Thromb Haemost. 2009;7(11):1–12.
- Santoso S, Kalb R, Kiefel V, et al. The presence of messenger RNA for HLA class I in human platelets and its capability for protein biosynthesis. Br J Haematol. 1993;84(3):451–456.
- Lindemann S, Tolley ND, Eyre JR, et al. Integrins regulate the intracellular distribution of eukaryotic initiation factor 4E in platelets. A checkpoint for translational control. J Biol Chem. 2001;276(36):33947–33951.
- Denis MM, Tolley ND, Bunting M, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122(3):379–391.
- Rondina MT, Schwertz H, Harris ES, et al. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost. 2011;9(4):748–758.
- Heijnen HF, Schiel AE, Fijnheer R, et al. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94(11):3791–3799.
- Laffont B, Corduan A, Ple H, et al. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122(2):253–261.
- Aatonen MT, Ohman T, Nyman TA, et al. Isolation and characterization of platelet-derived extracellular vesicles. J Extracell Vesicles. 2014;3:24692.
- Arraud N, Linares R, Tan S, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12(5):614–627.
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7(2):e30733.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842.
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211.
- Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111.
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777.
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885.
- You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610.
- Memczak S, Papavasileiou P, Peters O, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS One. 2015;10(10):e0141214.
- Alhasan AA, Izuogu OG, Al-Balool HH, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127(9):e1–e11.
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21.
- Slot JW, Geuze HJ, Gigengack S. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113(1):123–135.
- Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2(10):2480–2491.
- Ramakrishnaiah V, Thumann C, Fofana I, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110(32):13109–13113.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2013-2015. Available from: http://www.repeatmasker.org
- Robertson HM, Zumpano KL. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205(1–2):203–217.
- Schneider T, Hung LH, Schreiner S, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313.
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984.
- van der Pol E, Hoekstra AG, Sturk A, et al. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8(12):2596–2607.
- Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PloS One. 2016;11(2):e0148407.
- Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111(41):14888–14893.
- Hung ME, Leonard JN. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J Extracell Vesicles. 2016;5:31027.
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980.
- Maass PG, Glažar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95(11):1179–1189.
- Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, Multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–676.
