ABSTRACT
Sample amount is often a limiting factor for multi-parametric analyses that encompass at least three areas of ‘-omics’ research: genomics, transcriptomics and proteomics. Limited sample amounts are also an important consideration when these multi-parametric analyses are performed on extracellular vesicles (EVs), as the amount of EVs (and EV cargo) that can be isolated is often very low. It is well understood that a monophasic solution of phenol and guanidine isothiocyanate (i.e. TRIzol©) can simultaneously isolate DNA, RNA and proteins from biological samples; however, it is most commonly used for the extraction of RNA. Validation of this reagent for the isolation of multiple classes of biological molecules from EVs would provide a widely applicable method for performing multi-parametric analyses of EV material. In this report, we describe a comparison of proteins identified from EVs processed with either TRIzol© or the conventional Laemmli buffer protein-extraction reagents. EVs were isolated from 3 mL of cell-culture supernatant derived from MCF-10A, MCF-7 and MDA-MB-231 cells using the Vn96 EV capture technology. For the TRIzol© extraction protocol, proteins were precipitated with acetone from the organic phase and then re-solubilized in a mixture of 8M urea, 0.2% SDS and 1 M Tris-HCl pH 6.8, followed by dilution in 5× loading buffer prior to fractionation with 1D SDS-PAGE. NanoLC-MS/MS of the trypsin-digested proteins was used to generate proteomic profiles from EV protein samples extracted with each method. Of the identified proteins, 57.7%, 69.2% and 57.0% were common to both extraction methods for EVs from MCF-10A, MCF-7 and MDA-MB-231, respectively. Our results suggest that TRIzol© extraction of proteins from EVs has significant equivalence to the traditional Laemmli method. The advantage of using TRIzol© reagent is the ability to accumulate multi-parametric data (e.g., RNA and protein profiles) on the same limited EV sample while minimizing sample preparation and processing time.
Introduction
Extracellular vesicles (EVs) are a heterogeneous population of lipid membrane-bound nano-scale structures that are released from cells through distinct mechanisms [Citation1]. There are three commonly accepted major EV sub-types, exosomes, microvesicles and apoptotic bodies, which are defined by their cellular origin, size and composition. Exosomes have diameters ranging from 40 nm to 120 nm, and are released into the extracellular media via fusion of cytosolic multi-vesicular bodies with the cell membrane. Microvesicles are typically larger than exosomes, having diameters of 100 nm to 2 μm, and are formed by outward budding of the plasma membrane [Citation2]. Finally, apoptotic bodies are generated during apoptotic cell death and have diameters ranging from 1 μm to 5 μm [Citation3].
EVs incorporate and transport cargoes derived from their parent cells that are relevant for research purposes, as well as for use as diagnostic or prognostic markers in various disease [Citation4]. These include RNA and proteins; however, the average low lumen size of EVs limits the amount of material that they can carry [Citation5,Citation6]. This limited cargo capacity has proven to be a challenge for capitalizing on the potential of exploiting EVs, as it increases the difficulty in identifying low-abundance material, or increases the demands on limited sample amounts. Therefore, an EV isolation method that is highly efficient, amenable to small samples and applicable to multi-parametric analyses is required for further exploitation of the potential for EVs to facilitate these types of applications.
We have recently developed an EV capture method that uses a synthetic peptide called Vn96, which has binding affinity for heat shock proteins that are expressed in high abundance on the surface of EVs[Citation7]. Vn96 is a small (27-mer) peptide, with an amino acid sequence of PSQGKGRGLSLSFRSWGALTLGEFLKL and a molecular weight of 2.9 kDa. Independent validation has shown that the yield of EVs obtained using Vn96 is superior to that of differential ultracentrifugation (UCF) [Citation8]. The use of Vn96 for EV isolation therefore enables the study of much smaller sample volumes than ultracentrifugation methodology, or permits sample enrichment by capturing more EV material, improving the ability to detect and analyse cargo. A comparative proteomic profile of UCF (UCF-EV) and Vn96-isolated (HSP-EV) EVs from HT-29 colorectal cancer cell-conditioned media has been recently described [Citation8]. While the amount of starting material used for ultracentrifugation (UCF) was 30 times greater than that used with Vn96, the authors identified a total of 3085 unique proteins using the Vn96 peptide compared with 3115 proteins using UCF. This report indicated that, ‘the small-scale HSP-EV isolation method captures a significant amount and diversity of the EV proteome to render it useful for clinically relevant comparisons [Citation5]’. More recently, Vn96 has been used to isolate EVs from only 2 mL of mouse B cell-conditioned tissue culture media in vitro, which were subjected to downstream analyses including mass spectrometry and RNA sequencing [Citation9]. A significant research focus within the EV field is to examine and characterize multi-parametric data, such as the complement of RNA and protein carried within an EV population; however, existing methods have been limited by small sample sizes, and the lack of a standardized methodology for extracting these disparate biological entities from a single EV sample.
For proteomics analyses, proteins are commonly extracted from lysed cells using a Laemmli (or similar) buffer that contains 2% sodium dodecyl sulfate (SDS), 10% glycerol and 60 mM Tris-HCl at pH 6.8 [Citation10]. Importantly, both studies described above show that Laemmli buffer is an effective protein-extraction reagent for EVs isolated by either Vn96 or UCF; however, Laemmli-based protein extraction negates the possibility of simultaneously isolating DNA or RNA from the same sample. Therefore, samples are typically sub-divided prior to protein extraction when nucleic acids are also to be examined. Consequently, sample splitting decreases available sample volume for each assay, and ultimately increases the effective cost (in both time and money) of EV isolation protocols and/or reagents. Furthermore, sample splitting decreases the depth and breadth of proteome/transcriptome coverage and increases the difficulty of examining rare or minor components of EVs. The use of TRIzol© reagent enables the extraction of all three molecular entities (DNA, RNA and proteins) from the same biological sample. The use of TRIzol© for the isolation of nucleic acids from cells and EVs has been well established [Citation11]; however, its utility for protein capture is less well investigated and has not been validated in the context of EVs. While work from the group of Edit Buzas [Citation12] demonstrates differential detergent sensitivity of EV sub-populations, a similar effect was not anticipated with the TRIzol© protein-extraction reagent because TRIzol© (or Laemmli) extraction does not depend on this principle. The main barrier to using TRIzol© for protein recovery arises from poor reproducibility and difficulty in the re-solubilization of the protein pellet from the prescribed acetone- or isopropanol-mediated precipitation step [Citation13].
We examined whether the TRIzol© reagent (and, by extension, phenol/guanidine isothiocyanate approaches in general) is amenable for the extraction and analysis of biologically relevant proteins from Vn96-captured EVs, as a means of more easily facilitating multi-parametric analysis of EV contents. We used three human breast cell lines, MCF-10A, MCF-7 and MDA-MB-231, to determine qualitatively if similar biological data could be collected from EV preparations using TRIzol© for protein extraction, when compared with the traditional Laemmli extraction method. We found that these two methods gave similar results in their ability to extract proteins. Overall, the overlapping proteins accounted for 57.7%, 69.2% and 57.0% of the total proteins identified for EVs from MCF-10A, MCF-7 and MDA-MB-231 cells, respectively. The number of overlapping proteins gives an indication of the complementarity of the two protein-extraction methods. Furthermore, when comparing the total number of proteins identified, Laemmli and TRIzol© showed a similar range of detection across samples, ranging from 68.9% to 96.1% of proteins detectable via Laemmli, versus 61.6% to 88.4% detectable by TRIzol©.
Principal component analysis (PCA) on Laemmli and TRIzol© extracted EV proteins was performed to evaluate the similarity of data sets between cell lines and between extraction reagents. Finally, we performed an ontology enrichment analysis to determine whether the common proteins extracted by either Laemmli or TRIzol© contain biologically consistent data that reflect the parent cells from which the EVs were isolated. Together, our findings suggest that TRIzol©-based extraction of EV proteins may be applicable for proteomics analysis or diagnostic marker identification.
Materials and methods
Cell culture
All tissue-culture materials were purchased from Thermo-Fisher Scientific (Mississauga, ON) unless otherwise indicated. MCF-10A cells were grown in DMEM media supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine (Sigma-Aldrich, St. Louis, MO), 1 mM sodium pyruvate, 500 ng/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO), 1 ng/mL insulin and 100 ng/mL cholera toxin (Sigma-Aldrich, St. Louis, MO). MCF-7 and MDA-MB-231 cells were grown in DMEM (high glucose) supplemented with 10% FBS, 2 mM l-glutamine (Sigma-Aldrich, St. Louis, MO) and 1 mM sodium pyruvate. Two separate biological samples of each cell type were grown to 80% confluence in 150 mm tissue culture-treated plates. For the collection of EVs from cell-culture supernatant, cells were washed once in their respective growth media containing all supplements, with the exception of FBS. Cells were then left for 24 h in 20 mL of FBS-free growth media [Citation14]. EV-containing growth media were then removed from the cells and centrifuged at 500 g for 5 min at 4°C to remove cells and large debris. Media was further cleared of debris via centrifugation at 2000 g for 20 min at 4°C and 12,000 g for 45 min, and finally by filtration through a 0.22 μm syringe filter.
EV isolation
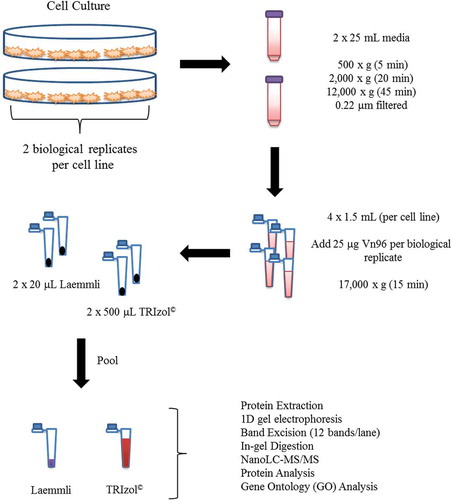
The EV isolation protocol used in this study is shown in . For each biological replicate of MCF-10A, MCF-7 and MDA-MB-231 cells, four aliquots of 1.5 mL of cell culture supernatant were used for EV isolation (eight aliquots total per cell line). Each aliquot was processed for EV isolation using 25 µg of Vn96 (New England Peptide, Gardner, MA) with 2 h of end-over-end rotation at room temperature. Vn96-bound EVs were isolated by centrifugation at 17,000 g for 15 min at room temperature. EV pellets from two Vn96 preparations per biological replicate were re-suspended in 1 mL of 0.1 μm-filtered phosphate buffered saline and centrifuged at 16,000 g for 10 min at room temperature. Four Vn96 pellets (2 per biological replicate for each cell line) were then re-suspended in 20 μL of 2× Laemmli buffer (Bio-Rad, Saint-Laurent, QC, Canada) with the remaining four pellets re-suspended in 500 μL of TRIzol© (Thermo-Fisher Scientific, San Jose, CA) and groups of two pooled together in the same tube, yielding one EV sample per biological replicate for Laemmli or TRIzol© protein extraction as described below.
Protein extraction (TRIzol© and Laemmli methods)
EVs were lysed, and either the proteins were collected by Laemmli extraction or the TRIzol© reagent was used to generate clean fractions of DNA, RNA and protein sequentially. Here, it was found that 8 M urea, 0.2% SDS and 1 M Tris-HCl, pH 6.8, constituted an efficient reagent for returning the protein pellet to solution from the TRIzol© protein-extraction protocol and that it is critical to avoid over-drying of the precipitated protein pellet prior to downstream processing.
SDS-PAGE
Intact protein fractionation by SDS-PAGE ultimately reduces the complexity and amount of proteins (i.e. tryptic peptides) present in each liquid chromatographic sample injection. The process of fixing and washing the gels also removes potentially interfering metabolites, detergents, salts, denaturing agents and lipids from the downstream analysis. In addition, proteins present in high abundance are focused into narrow bands, thereby improving the identification of lower abundance proteins in the sample. We note that the Vn96 peptide does not interfere with our ‘GeLC’ workflow (i.e. SDS-PAGE/nanoLC-MS/MS) owing to its low molecular weight, which allows it to be removed from the samples by the gel electrophoresis step. This experimental design focused on a qualitative analysis of the proteins detected following Vn96 EV isolation and either Laemmli or TRIzol© extraction. The total amount of protein from our Vn96-captured EVs was not quantified prior to 1D SDS-PAGE owing to interference of the abundant Vn96 peptide, added to capture the EVs, with the BCA assay. As such, the contribution of the EV protein cargo would be substantially smaller than the abundance of Vn96 peptide added, and thus not quantifiable in this method. Therefore, in order to maximize the sensitivity of our protein identification by mass spectrometry, equal volumes (consisting of all the Vn96 preparations) were loaded onto the SDS-PAGE in their entirety. In this study, proteins were separated using a 4–20% gradient SDS-PAGE gel (Bio-Rad Laboratories, Mississauga, ON). In preparation for MS analysis, the gels were fixed with 50% methanol and 5% acetic acid, stained with Coomassie EZ-blue and destained overnight in water prior to band excision.
Western blot analyses
EVs were collected, and proteins were extracted as described above for the MCF-10A, MCF-7, MDA-MB-231 cell lines using either Laemmli or TRIzol© reagents. EV samples were divided into two aliquots of equal volume, with one sample prepared for a reducing SDS-PAGE gel using 10% β-mercaptoethanol, and the other for a non-reducing gel (no β-mercaptoethanol), then boiled for 5 min. EV samples were then loaded onto two 4–20% gradient SDS-PAGE gel (Bio-Rad Laboratories) and transferred to 0.45 µm polyvinylidene fluoride (PVDF) membranes. Blocking was performed using 5% (w/v) skim milk in tris-buffered saline with 0.1% Tween-20 (TBST). Anti-human antibodies (from Santa Cruz, Santa Cruz, CA, unless otherwise specified) were prepared in TBST with 5% BSA and incubated overnight at 4°C as follows: 1:200 HSC-70 (SC-7298), 1:1000 Flotillin-1 (Cell Signaling Technology, Danvers, MA; 18,634), 1:200 CD63 (SC-5275), 1:200 CD9 (SC-59,140), 1:1000 CD81 (SC-7637). HSC-70, CD63, CD81 and CD9 were detected using anti-mouse IgG (115-035-003), and Flotillin-1 was detected with anti-rabbit IgG (111-035-003) conjugated to horse radish peroxidase diluted 1:5000 in TBST + 5% BSA.
In-gel digestion
Each sample lane was excised into 12 equal bands approximately 5 mm in height. Gel bands were dehydrated in acetonitrile (ACN) for 5 min, re-swelled in 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate (ABC) and incubated at 56°C for 1 h. Following subsequent dehydration with ACN, the gel bands were treated with 55 mM iodoacetic acid (IAA) in 50 mM ABC in the dark at room temperature for 1 h. Bands were dehydrated once more with ACN and treated with 50 μL of a 10 ng/μL solution of mass spectrometry grade trypsin in 50 mM ABC. Following overnight digestion at 37°C, the gel band supernatant was transferred to a new tube, and the tryptic peptides were extracted three times with 50% acetonitrile containing 5% acetic acid. The combined extracts were concentrated in a vacuum centrifuge to approximately 45 μL and acidified with 5 μL of 10% aqueous formic acid.
C-18 clean-up (solid phase extraction)
Peptide extracts were purified offline using C-18 solid-phase extraction mini spin-filter cartridges (Canadian Life Sciences, Peterborough, ON). Samples were acidified with 2% formic acid in 20% acetonitrile. The cartridges were activated with 50% acetonitrile and then equilibrated with 0.5% formic acid and 5% acetonitrile. Extracted in-gel digests were bound to the C-18 resin, washed with equilibration solution and eluted using 70% acetonitrile. Sample volumes were reduced to approximately 10–15 μL and then diluted to 50 μL in 1% formic acid and stored at −20°C [ref].
LC-MS/MS analysis
Protein tryptic digests were analysed by gradient nanoLC-MS/MS using a hybrid quadrupole-Orbitrap tandem mass spectrometer (Q-Exactive, Thermo-Fisher Scientific, San Jose, CA) interfaced to a Proxeon Easy NanoLC II liquid chromatograph. Each sample was analysed in triplicate by nanoLC-MS/MS. Five microlitre sample volumes were injected onto a narrow bore (20 mm long × 100 μm inner diameter) C-18 pre-column packed with 5 μm Reprosil-Pur resin particles with 300 angstrom channels. Chromatographic separation was achieved on a C-18 column with dimensions of 100 mm × 75 μm i.d., containing 3-μm-diameter Reprosil-Pur particles (Thermo-Fisher Scientific, San Jose, CA). Peptide elution was achieved using a water/acetonitrile gradient solvent system. Solvent A consisted of 0.1% formic acid, while solvent B consisted of 90/9.9/0.1 v/v/v acetonitrile/water/formic acid. A linear acetonitrile gradient was introduced to the C-18 column from 5 to 45% solvent B over 60 min followed by 100% B for 10 min at a flow rate of 300 nL/min. The outlet of the nano-flow emitter (15 μm diameter tip) was biased to 1.9 kV via remote liquid junction and positioned approximately 2 mm from the transfer capillary that was heated to 250°C. The S-lens of the mass spectrometer was maintained at 100 V. Mass spectrometric data were acquired in data-dependent acquisition mode, whereby a full mass scan from 400 to 1500 Thomson (Th) was followed by the acquisition of fragmentation spectra for the 10 most abundant precursor ions with intensities above a signal threshold of 20,000. Precursor ion spectra were collected at a resolution of 70,000 (at m/z 400) and an AGC setting of 1 × 106. Peptide fragmentation was performed using high-energy collision induced dissociation in the HCD cell, and MS/MS spectra were collected in the Orbitrap at a resolution of 17,500 and an AGC setting of 1 × 105. Peptide precursors were selected using a repeat count of 2 and a dynamic exclusion period of 35 s.
Data analysis (Proteome Discoverer 2.0)
Raw data for mass spectrometric protein identification were analysed using Proteome Discoverer 2.0 (Thermo-Fisher Scientific, San Jose, CA) employing the Sequest HT algorithm [Citation15]. A human FASTA database was obtained from the Uniprot web site (uniprot.org) containing both Swissprot and Trembl protein entries (February 2017, 78,782 kb). Searches were performed using the following settings: (a) trypsin enzyme specificity with two allowed missed cleavages, (b) precursor and fragment mass accuracy tolerances of 10 ppm and 0.8 Da, respectively, (c) variable modifications of methionine oxidation (+ 15.994 Da) and lysine acetylation (+42.011 Da), and (d) a fixed modification of cysteine carboxymethylation (+58.005 Da). Proteome Discoverer 2.0 calculates a strict false discovery rate of 0.1% based on the results of a decoy (reverse sequence) database search. Proteins that were identified with identical peptides were grouped to satisfy the principle of parsimony.
PCA
The presence or absence of proteins detected in the biological replicates of Laemmli or TRIzol©-extracted EVs was converted to a numerical score of ‘1’ for present, or ‘0’ for absent. A PCA was then performed to compare the presence or absence of these proteins between biological replicates and cell lines. The statistical environment R (version 3.4.1) (R-Core Team, 2017) was used to perform the PCA, with the functions princomp() and fviz_pca_ind() from factoextra package to plot the results.
Ontology enrichment analysis
Proteomics data were assessed to identify enriched gene ontologies using the ToppFun application within the ToppGene suite [Citation16]. Where proteomic-based gene annotation did not match HUGO gene symbols, ToppFun performed automatic re-annotation. All p-values were obtained using the probability density function and were Bonferroni-corrected. Further analysis was performed using default settings. All Venn diagrams were prepared using the Venn diagram tool from the Bioinformatics Institute Ghent (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Results
In this study, we chose to examine proteomic profiles of EVs captured from the cell-culture supernatant of three breast cell lines that span the spectrum of breast cancer progression: i.e. MCF-10A (normal immortalized breast epithelial cells) [Citation17] MCF-7 (epithelial-like breast cancer cells) [Citation18] and MDA-MB-231 (mesenchymal-like metastatic breast cancer cells) [Citation19]. In order to compare the degree of protein overlap between extraction methods, we considered two biological replicates for each cell line analysed in technical triplicate by nanoLC-MS/MS.
EV protein isolation and Western blot analyses
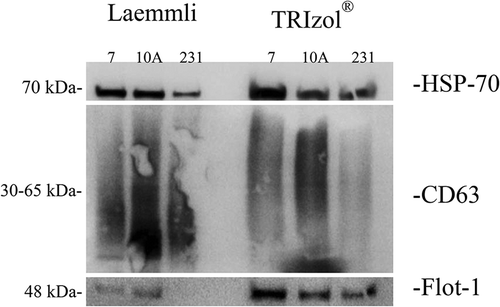
Total Vn96-isolated EV protein extracted by the Laemmli and TRIzol© methods was examined using Coomassie blue staining (Supplementary Figure #1). We next performed Western blot analyses to determine if the Vn96-isolated material contained canonical protein markers of EVs. In each cell line, from both the Laemmli and TRIzol©-extracted material, we probed for the presence of HSP (HSC) 70, CD63, CD81, CD9 and Flotillin-1 in accordance with the guidelines published by the International Society of Extracellular Vesicles [PMC4275645]. While CD81 and CD9 were not detected in any sample (data not shown), we did observe the presence of the other markers in EVs isolated from each of the cell lines (). Indeed, both protein-extraction methods also detected each of the markers, with the exception of Flotillin-1 from MDA-MB-231 cell EVs extracted with the Laemmli protocol. These data therefore demonstrate that, as previously reported, Vn96 successfully captured EV material, and EV proteins can be extracted by both Laemmli and TRIzol©-based methods.
Figure 2. Verification of EV isolation. Laemmli and TRIzol©-extracted samples were interrogated by Western blot for the expression of the canonical EV markers HSP-70, CD63, Flotillin-1, CD81 and CD9. MCF-7 (7), MCF-10A (10A) and MDA-MB-231 (231) cell EVs were examined. HSP-70 (top), CD63 (middle) and Flotillin-1 (bottom) were detected in the isolated EVs.
Proteomic analysis of human breast cell-derived EVs
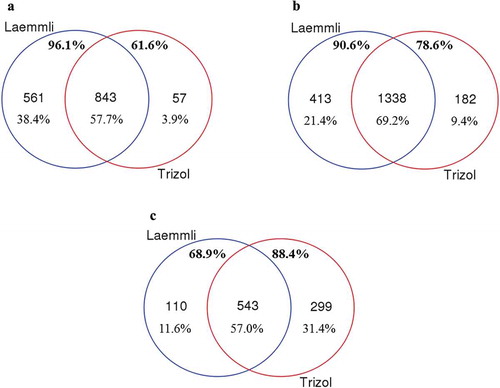
The use of Laemmli buffer to extract proteins carried by EVs captured from two MCF-10A cell culture media biological replicates produced totals of 1661 and 1616 proteins when run as technical triplicate nanoLC-MS/MS sample injections. Of these, 1404 proteins were found to be common to both biological replicates. Slightly fewer overall proteins were observed using TRIzol© for extraction, with 1356 and 1364 in each biological replicate respectively, and a total of 900 found in common. In all, 843 EV proteins were found to be common to both extraction methods for EVs isolated from the culture supernatant of MCF-10A cells ()). Of the total unique proteins detected in both biological replicates from each extraction method (1461), 96.1% were identified within the Laemmli-extracted EVs, and 61.6% were identified within the TRIzol©-extracted EVs from this cell line.
Figure 3. Comparison of the Laemmli and TRIzol© protein-extraction methods for the extraction of protein cargo from (A) MCF-10A, (B) MCF-7 and (C) MDA-MB-231 EVs. The unique proteins (number and percentage) from Laemmli are shown in blue, while TRIzol©-extracted proteins are shown in red. The common proteome is shown in the central region of the Venn diagram. Percentages below these numbers indicate the percentage of total proteins identified in each region. The upper percentages in bold show the fraction of proteins isolated from each methodology.
We compared the proteins identified in EVs from MCF-10A cells to the list of the top 25 proteins listed by Exocarta, as shown in [Citation20,Citation21], and found that 17 proteins identified in both Laemmli and TRIzol© extraction methods matched this list, with one additional protein detected uniquely in the Laemmli extraction. Of the top 200 EV proteins listed in EVpedia [Citation22], we found that 106 of these are common to both extraction methods, with 15 matches unique to Laemmli extraction and three protein matches unique to TRIzol© extraction (Supplemental File #1).
Table 1. List of top 25 proteins from the Exocarta web site that were identified in EVs captured from cell-culture-conditioned media of MCF-10A, MCF-7 and MDA-MB-231 cells.
As with MCF-10A EVs, we next examined the commonality of proteins extracted from MCF-7 EVs using both Laemmli and TRIzol© extraction. The use of Laemmli buffer to extract proteins from EVs captured from MCF-7 cell culture media produced totals of 1933 and 2290 proteins per biological replicate, respectively. Of these, 1751 proteins were found to be common to both biological replicates. TRIzol© extraction identified 1700 and 2145 proteins, with a total of 1520 found in common between the two biological replicates. In all, 1338 EV proteins were found to be common to both extraction methods for EVs isolated from the culture supernatant of MCF-7 cells ()). From these EVs, we identified a total of 1933 unique proteins from both extraction methods; 90.6% of these proteins were detected in the Laemmli-extracted samples, and 78.6% were identified in the TRIzol©-extracted samples.
Three publications have reported specifically on proteins identified in MCF-7 EVs [Citation23–Citation25]. Additionally, based on the results from 59 cancer cell lines, Hurwitz [Citation23] compiled a list of 213 proteins with unique gene symbols found to be common to multiple cancer cell lines, including MCF-7. We identified a total of 129 of the 213 proteins in our dataset of common proteins for both extraction methods, with 11 protein matches unique to Laemmli extraction and three protein matches unique to TRIzol© extraction (Supplemental File#1). Staubach [Citation24] reported 28 proteins from EVs of MCF-7 cells, of which we observed 16 using both extraction methods and just one unique match for Laemmli extraction (Supplemental File#1). Finally, Kruger [Citation25] tabulated a total of 59 EV proteins from MCF-7 cells, of which we identified 26 using both protein-extraction methods and one exclusively with TRIzol© (Supplemental File #1). Comparison of our MCF-7 dataset with the top 200 proteins listed by EVpedia shows that we identified 121 matching proteins, where four are unique to Laemmli extraction, and one is exclusive to TRIzol© extraction (Supplemental File #1). Finally, of the top 25 EV proteins listed in Exocarta, 16 proteins were found in both extraction methods, whereas two were unique to Laemmli extraction ().
Lastly, we performed a comparison of Laemmli and TRIzol© extractions of proteins for MDA-MB-231 EVs. In the same manner as described above, we identified totals of 750 and 1062 proteins from two biological replicates, with a common total of 653 using Laemmli extraction. For EVs from this cell line, more proteins were identified using TRIzol© for extraction, with 966 and 1356 from the two biological replicates and a total of 842 found in common. In all, 543 EV proteins were found to be common to both extraction methods for EVs isolated from the culture supernatant of MDA-MB-231 cells ()). A total of 68.9% of the identified proteins (952) were identified in the Laemmli extraction, whereas 88.4% were identified via TRIzol© extraction.
Hurwitz [Citation23], Kruger [Citation25] and Palazzolo [Citation2] have each published lists of proteins identified from EVs produced by MDA-MB-231 cells. Compared with the 213 EV proteins listed by Hurwitz [Citation23], we identify a total of 105 proteins in common with 72 matches for both extraction methods, seven matches unique to Laemmli extraction and 26 matches exclusively with TRIzol© extraction (Supplemental File#1). Palazzolo et al. [Citation2] provided a list of 22 proteins that appear to be enriched in ‘exosomes-like’ vesicle fractions from cell-culture supernatant of MDA-MB-231 cells. Here, we have identified nine of these proteins using both extraction methods, one that is unique to Laemmli extraction and two protein matches that are exclusive to TRIzol© extraction (Supplemental File#1). Of the 113 proteins identified by Palazzolo using MALDI-TOF mass spectrometric fingerprinting, N-terminal sequencing, Western blotting and/or LC-MS/MS [Citation2], we found 44 of these proteins in both extraction methods (Supplemental File #1). We also found five of these proteins exclusively with TRIzol© extraction and seven exclusively using Laemmli extraction (Supplemental File #1). Comparisons with the EV proteins identified by Kruger [Citation25] from MDA-MB-231 EVs showed that we identified 28 of 88 reported proteins (Supplemental File#1). From the top 25 proteins listed in Exocarta, 16 proteins were identified using both extraction methods, while a single hit was exclusive to Laemmli extraction (). Of the top 200 proteins from EVpedia, 79 proteins were found in both extraction methods, eight were unique to Laemmli extraction, and 21 were identified only using TRIzol© extraction (Supplemental File#1).
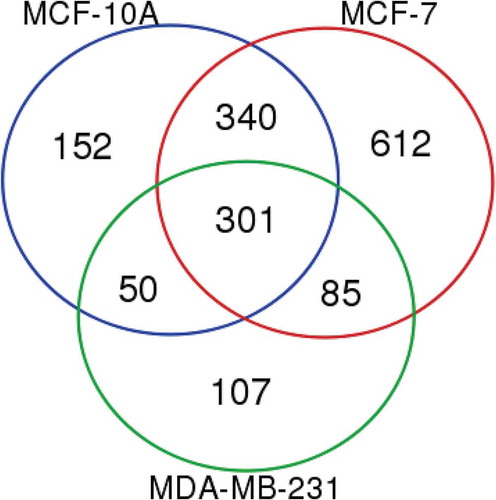
We then compared the lists of common proteins isolated using both Laemmli and TRIzol© extraction from MCF-10A, MCF-7 and MDA-MB-231 EVs (Supplemental File #2). Overall, we found 301 proteins common to EVs from all three cell lines and both extraction methods (). The largest number of proteins was identified as common to both MCF-10A and MCF-7 with a further 340. Only 50 and 85 proteins were identified as common between MCF-10A/MDA-MB-231 and MCF-7/MDA-MB-231, respectively. Finally, there were 152, 612 and 107 unique proteins for each of MCF-10A, MCF-7 and MDA-MB-231 EVs, respectively.
Figure 4. Proteomic analysis of MCF-10A, MCF-7 and MDA-MB-231 EVs revealing sets of common, overlapping and unique proteins from different EV groups. Proteins included in the analysis of each cell type EVs were present using both Laemmli and TRIzol© extraction reagents in both biological replicates.
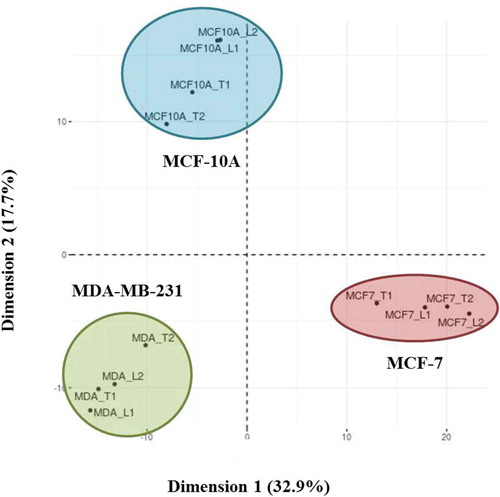
PCA
A PCA was performed on the duplicated biological replicates of each cell line to determine if the Laemmli and TRIzol©-based EV protein-extraction methods produced similar (and cell line-characteristic) data sets (). The total list of proteins identified by either Laemmli or TRIzol© within the biological replicates was compiled. We then compared the presence or absence of these proteins from each extraction within each cell line. We found that regardless of cell line, both Laemmli and TRIzol©extracted proteins clustered together by cell line. These data demonstrate that irrespective of the protein-extraction method chosen, similar protein data were captured from the EVs, and the biological distinctiveness between cell lines, but not between methods, was predominant.
Figure 5. Principal component analysis (PCA) of Laemmli and TRIzol©-extracted EV proteins from MCF-10A, MCF-7 and MDA-MB-231 cells. The two Laemmli (L) and TRIzol© (T) biological replicates selected from each cell line that were used for all analyses were compared. The total list of extracted proteins within each biological replicate of the extraction method was compiled from the technical replicates.
Gene ontology analysis
We also performed an ontology analysis on the proteins extracted from the EVs captured from these three cell lines. Using ToppFun [Citation16], the list of proteins common to TRIzol© and Laemmli extraction methods for EVs captured from each cell line was analysed for enriched gene ontology (GO) terms. The associated ontologies were the total of the biological process (BP), molecular function (MF) and cellular component (CC) categories (Supplemental File #3).
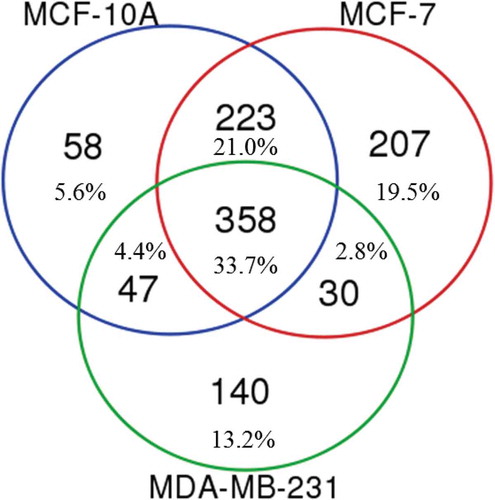
In total, we found that 348 GO terms were common to proteins extracted from EVs of the three cell lines using both extraction reagents (). There were 58 unique ontologies to MCF-10A EV proteins, a further 205 associated with MCF-7 EVs and 138 from MDA-MB-231 EV proteins. There was considerable ontological commonality between the MCF-10A and MCF-7 EV proteins, with 222 common GO terms. MCF-7 and MDA-MB-231 EV proteins shared only 32 ontologies, whereas MCF-10A and MDA-MB-231 EV proteins had a further 48 common ontologies. These data suggest that the intrinsic biological distinctiveness of the cell types is reflected in the data that can be captured using either Laemmli- or TRIzol©-extraction of proteins from the EVs produced by these cells.
Figure 6. Unique and overlapping gene ontologies (GOs) from MCF-10A, MCF-7 and MDA-MB-231 EVs. The GO terms were identified using ToppFun from the lists of proteins identified using both Laemmli and TRIzol© extracted EV proteins.
Finally, we examined the lists of MCF-10A, MCF-7 and MDA-MB-231 unique EV proteins to determine if there was any cell-type specific information captured. We first examined the BPs represented in each protein list, reasoning that these processes represent the unique physiological activities from these cell lines. MCF-10A EV proteins showed significant associations with cell-substrate interactions and various metabolic processes (Supplemental File #3). Considerably more BPs were associated with MCF-7 EV proteins. Many of these were also associated with metabolism-related functions; however, there were also associations with cytosolic vesicle trafficking, as well as nucleotide repair, synthesis and mitotic functions. Proteins extracted from MDA-MB-231 EVs were strongly associated with the regulation of cell movement, chemotaxis and the negative regulation of cell adhesion, which are all indicative of metastatic disease.
We further examined if the proteins extracted using the TRIzol© method for each cell line have specific pathway associations, or have disease-related annotations. We therefore analysed the pathway and disease associations of the unique MCF-10A, MCF-7 and MDA-MB-231 EV proteins (Supplemental File #3) that were common to both extraction reagents/protocols. MCF-10A EV proteins were associated with TCA cycle and translation pathways. The disease associations unique to MCF-10A proteins primarily represented disorders of connective tissue. In comparison, MCF-7 EV proteins were associated with fewer unique pathways, but these included epithelial cell signalling (E-cadherin junction signalling) and the regulation of mRNA processing. Moreover, although only a few disease associations were captured for MCF-7 EV proteins, these associations were uniformly associated with cancer and included general tumour progression with specific associations with ovarian and colon tumours. Finally, MDA-MB-231 EV proteins were associated with multiple extracellular matrix regulating pathways, including ECM regulators, those moderating elastin and collagen formation and ECM remodelling processes. Unlike MCF-10A and MCF-7 EV proteins, MDA-MB-231 EV proteins were associated with a large number of disease conditions. A comprehensive list of calculated p-values for each association and cell line can be found in Supplemental File #3. A substantial number of these were related to different cancer types including glioma, osteosarcoma, pancreatic tumours, ovarian cancers and mammary tumours. The MDA-MB-231 EV proteins were uniquely associated with stage IV breast cancer (the most aggressive, latest stage malignancy). Other captured disease associations included endometriosis, cystic fibrosis and ECM/connective tissue disorders such as Pterygium.
Discussion
Nanoparticle tracking analysis (NTA) and electron microscopy (EM) have been performed on Vn96 mediated extracellular vesicle isolations of MCF-10A and MCF-7 cell culture media previously [Citation7,Citation26]. Both studies found particle size distributions consistent with small EVs. Herein, we did not analyse or attempt to count EVs from Vn96 isolations using NTA or EM because the EVs tend to aggregate, and the use of reagents to disperse them can negatively impact downstream gel fractionation and protein profiling.
In this study, we sought to determine whether the TRIzol© reagent is suitable for the extraction and analysis of biologically relevant proteins from Vn96-captured EVs. We therefore compared a standard Laemmli-mediated protein extraction with TRIzol©-mediated protein extraction for EVs captured from the MCF-10A, MCF-7 and MDA-MB-231 breast cell lines, and found that 57.7% (843), 69.2% (1338) and 57.0% (543), respectively, of the identified EV proteins were common to both extraction methods. We typically observe 65–75% overlap of protein identifications over triplicate technical replicate nanoLC-MS/MS analyses using conventional Laemmli extraction, which is comparable with other published values [Citation27]. In consideration of this technical variability of the nanoLC-MS/MS protein identification workflow, these data suggest that TRIzol© extraction of proteins from EVs yields results that are comparable with those for Laemmli extraction.
The cell lines used in this study represent healthy immortalized breast tissue (MCF-10A), minimally metastatic, hormone receptor-positive breast cancer (MCF-7) and a highly aggressive, metastatic, triple negative breast cancer (MDA-MB-231). We compared our datasets with published lists of EV proteins that have been identified as both common and cell-type specific. For these analyses, we used two databases that provide lists of common EV proteins, Exocarta (exocarta.org) and EVpedia (student4.postech.ac.kr/evpedia2_xe/xe/), as well as publications that report specifically on the proteins present in MCF-7 [Citation23–Citation25] and MDA-MB-231 EVs [Citation2,Citation23,Citation25]. These comparisons demonstrate that using our Vn96/LC-MS/MS workflow we have identified EV proteins that are congruent to those reported in other studies, but we also determined via nanoLC-MS/MS that our EV isolation and protein-extraction platform identifies unique sets of EV proteins.
Our proteomic analysis of these three breast cancer cell lines confirms that the proteins extracted from EVs using TRIzol© can be used to identify biologically relevant data. We found that MCF-10A EVs carried 843 common proteins, MCF-7 EVs have 1338 common proteins, and MDA-MB-231 EVs have 543 common proteins identified by the Laemmli and TRIzol© extraction methods. A total of 301 proteins were identified as common to all three groups of EVs, suggesting that they may constitute a common EV proteome from these breast cancer cell lines. In future studies, these data may be useful in identifying the underlying components that are essential for, or universally incorporated by, EVs from various cell types. Furthermore, all three cell lines studied originated from human breast tissue. Despite the inherent underlying biological similarity of the three cell lines examined, we identified a number of cell-line-specific protein cargoes captured in the EVs. These data suggest that functionally or process-relevant information is captured in these EV proteomes. This clearly demonstrates that the use of Laemmli or TRIzol© EV-protein-extraction methods is amenable for proteomics-based discovery research. Therefore, not only is TRIzol©-based protein extraction capable of preserving and isolating biological distinctions between similar cell backgrounds, but unlike Laemmli-based protein isolation, only TRIzol© can facilitate multi-parametric analysis through simultaneous extraction of nucleic acid in the TRIzol© aqueous phase.
Our analysis of EV proteins extracted using the TRIzol© method shows that, despite modest differences in the proteins that are extracted compared with Laemmli extraction, we are able to obtain information that is representative of the parental cell from which the EVs are derived, despite using cell lines of a common tissue type. Our analysis also demonstrates that these EV proteins demonstrate clear physiological differences based on their cells of origin and may be useful for assessing the biological activity of their parental cells. The benefit of using the TRIzol© reagent is that both protein and nucleic acids can be isolated from the same limited sample. While the use of this TRIzol©-based proteomics method does not depend on any specific EV-isolation technology, it is suitable for affinity isolation methods. The benefit of using Vn96 as a fast and highly efficient affinity reagent for the capture of EVs further enhances the proteomic analysis of EVs from breast cancer cells. Between protein and RNA, we found that the amount of RNA available from isolated EVs is generally more limiting for downstream analysis than the amount of protein. Overall, our results suggest that Vn96-mediated EV capture combined with TRIzol© extraction provides a powerful methodology for multi-parametric analysis of EVs. Future studies may therefore be able to adapt and further develop this methodology not only for fundamental research activities but also for clinical research settings.
supp_data.zip
Download Zip (3.8 MB)Acknowledgements
The authors would like to acknowledge the Atlantic Canada Opportunities Agency for financial assistance through the Atlantic Innovation Fund.
Disclosure statement
The Vn96 EV extraction reagent and protocol were developed by the Atlantic Cancer Research Institute and commercialized in partnership with New England Peptide (Gardner, MA).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Yáñez-Mó M, Siljander PR.-M, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:1–12.
- Palazzolo G, Albanese NN, Cara GD, et al. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012;32(3):847–860.
- Akers JC, Gonda D, Kim R, et al. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113:1–11.
- Revenfeld ALS, Baek R, Nielsen MH, et al. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Therapeut. 2014;36(6):830–846.
- Ung TH, Madsen HJ, Hellwinkel JE, et al. Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci. 2014;105(11):1384–1392.
- Li M, Zeringer E, Barta T, et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci. 2014;369:1652–1660.
- Ghosh A, Davey M, Chute IC, et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLOS One. 2014;9(10):1–12.
- Knol JC, deReus I, Schelfhorst T, et al. Peptide-mediated ‘miniprep’ isolation of extracellular vesicles is suitable for high-throughput proteomics. EuPA Open Proteom. 2016;11:11–15.
- Ayre DC, Chute IC, Joy AP, et al. CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo. Sci Rep. 2017;7. DOI:10.1038/s41598-017-08094-8.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):6.
- Rio DC, Ares Jr M, Hannon GJ, et al. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010.
- Osteikoetxea X, Sodar B, Nemeth A, et al. Differential detergent sensitivity of extracellular vesicle subpopulations. Org Biomol Chem. 2015;13(38):9775–9782.
- Hummon AB, Lim SR, Difilippantonio MJ, et al. Isolation and solubilization of proteins after TRIzolR extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42(4):5.
- Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Nat Acad Sci. 2016;113(8):E968–E977.
- Tabb DL. The SEQUEST family tree. J Am Soc Mass Spectrom. 2015;26(11):6.
- Chen J, Bardes EE, Aronow BJ, et al. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nuclei Acids Res. 2009;37:7.
- Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086.
- Soule HD, Vazquez J, Long A, et al. A human cell line from a pleural infusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51(5):1409–1416.
- Cailleau R, Olive M, Cruciger QVJ. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14(11):911–915.
- Keerthikumar S, Chisanga D, Ariyaratne D, et al. Exocarta: A web-based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688–692.
- Mathivanan S, Simpson RJ. Exocarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4.
- Choi D-S, Kim D-K, Kim Y-K, et al. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2014;1–17. DOI:10.1002/mas.21420
- Hurwitz SN, Rider MA, Bundy JL, et al. DGM. Proteomic profiling of NCI-60 extracellular vesicles unconvers common protein cargo and cancer type-specific biomarkers. Oncotarget. 2016;7(52):86999–87015.
- Staubach S, Razawi H, Hanisch F-G. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics. 2009;9:2820–2835.
- Kruger S, Elmageed ZYA, Hawke DH, et al. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer. 2014;14(44):1–10.
- Griffiths SG, Cormier MT, Clayton A, et al. Differential proteome analysis of extracellular vesicles from breast cancer cell lines by chaperone affinity enrichment. Proteomes. 2017;5(25):1–16.
- Tabb DL, Vega-Montoto L, Rudnick PA, et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9(2):761–794.