ABSTRACT
Bacteria release nano-sized extracellular vesicles (EVs) into the extracellular milieu. Bacterial EVs contain molecular cargo originating from the parent bacterium and have important roles in bacterial survival and pathogenesis. Using 8-plex iTRAQ approaches, we profiled the EV proteome of two Escherichia coli strains, uropathogenic (UPEC) 536 and probiotic Nissle 1917. For these strains, we compared the proteome of crude input EVs prepared by ultracentrifugation alone with EVs purified by either density gradient centrifugation (DGC) or size exclusion chromatography (SEC). We further compared the proteome of EVs from bacterial cultures that were grown in iron-restricted (R) and iron-supplemented (RF) conditions. Overall, outer membrane components were highly enriched, and bacterial inner membrane components were significantly depleted in both UPEC and Nissle EVs, in keeping with an outer membrane origin. In addition, we found enrichment of ribosome-related Gene Ontology terms in UPEC EVs and proteins involved in glycolytic processes and ligase activity in Nissle EVs. We have identified that three proteins (RbsB of UPEC in R; YoeA of UPEC in RF; BamA of Nissle in R) were consistently enriched in the DGC- and SEC-purified EV samples in comparison to their crude input EV, whereas conversely the 60 kDa chaperonin GroEL was enriched in the crude input EVs for both UPEC and Nissle in R condition. Such proteins may have utility as technical markers for assessing the purity of E. coli EV preparations. Several proteins were changed in their abundance depending on the iron availability in the media. Data are available via ProteomeXchange with identifier PXD011345. In summary, we have undertaken a comprehensive characterization of the protein content of E. coli EVs and found evidence of specific EV cargos for physiological activity and conserved protein cargo that may find utility as markers in the future.
Abbreviation: DGC: density gradient centrifugation; DTT: 1,4-dithiothreitol; EV: extracellular vesicles; FDR: false discovery rate; GO: Gene Ontology; R: iron-restricted; RF: iron-supplemented; iTRAQ: isobaric tags for relative and absolute quantitation; OMV: outer membrane vesicle; SWATH-MS: sequential window acquisition of all theoretical mass spectra; SEC: size exclusion chromatography.
Introduction
Bacteria, Archaea and Eukaryota all release nano-sized extracellular vesicles (EVs) into the extracellular environment. EVs of bacteria, also commonly called “membrane vesicles”, are 20–250 nm in size and comprise specific subsets of molecular cargo including protein, DNA, RNA, lipids and various metabolites packaged from the parent bacterium [Citation1,Citation2]. These bacterial EVs are released constantly during the cell growth but are known to be produced in a more controlled and regulated manner under certain environmental conditions [Citation3]. As an example, iron restriction, which reflects the bacterial environment at the start of an infection [Citation4], is known to increase the production of bacterial EVs in some species [Citation5,Citation6]. Bacterial EVs play a crucial role in both bacteria-to-bacteria and bacteria-to-host interactions [Citation7], and they are involved in bacterial survival, biofilm formation, horizontal gene transfer, stress response, nutrient acquisition, toxin delivery, antibiotic resistance, immunomodulation and pathogenesis [Citation2,Citation8–Citation10].
EVs are released from various pathogenic and non-pathogenic bacterial species, for which the cargo of virulence factors, and their functional roles have been summarized in a recent review [Citation11]. EVs of Gram-negative bacteria are frequently referred to as outer membrane vesicles (OMVs) because they are mainly released by blebbing from the outer membrane of the cell envelope [Citation12]. However, an increasing number of studies have shown heterogeneity in bacterial EVs and proposed the existence of multiple mechanisms of EV biogenesis [Citation2,Citation6,Citation13–Citation16]. In the bacteria research field, it is common to study “crude input” EVs isolated by sequential filtration and ultracentrifugation. Occasionally, these crude input samples are further purified by density gradient centrifugation (DGC) to remove protein aggregates and cellular structures like flagellae. The extra DGC step provides more defined, cleaner EV samples but is laborious. For bacterial EVs, the DGC protocol has not been standardized, and there are no established “minimal experimental requirements”. The implication is that some reported findings could be partly due to purification artefacts, instead of the true biogenesis differences. We believe that understanding the impact of the purification method of bacterial EV subpopulations is now required for better interpretation of EV biology and EV contributions to pathogenesis.
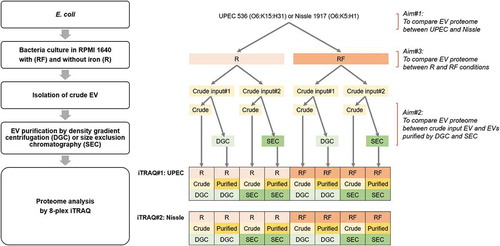
In a recent study, we reported the molecular heterogeneity of EVs from different bacterial species and the effect of culture under iron restriction on the molecular diversity [Citation17]. The study assessed the relative utility of DGC and size exclusion chromatography (SEC) to purify the “crude input” EVs harvested by ultracentrifugation of the cell-free growth medium. We investigated two Gram-negative Escherichia coli (E. coli) strains – uropathogenic E. coli (UPEC) 536 and probiotic Nissle 1917 – and the acid-fast bacterium Mycobacterium smegmatis; and reported diversity in EV subpopulations by various means including particle counts, protein profiles on PAGE, and total yields of protein, RNA and endotoxin [Citation17]. In this present study, we extend this bacterial EV purification and characterization by using quantitative proteomics analysis with isobaric tags for relative and absolute quantitation (iTRAQ) mass spectrometry approaches. The iTRAQ method uses covalent labelling of N-terminus and side chain amines of peptides with isobaric tags. Each sample is labelled with one of eight isobaric tags (all have an identical overall mass), and samples are then pooled and analyzed together by tandem mass spectrometry (MS/MS). During peptide fragmentation, a tag-specific reporter ion is released in proportion to the amount of peptide in each sample. Analysis of the fragmentation data enables to identify the labelled peptides and hence the corresponding proteins, and relatively quantify proteins between different-labelled samples [Citation18]. This method allows for the simultaneous identification and quantification of proteins from up to eight samples in a single run, reducing inter-sample variations, time and cost. Using this iTRAQ approach, here we aimed to: (1) identify the difference in the EV proteome of two E. coli strains (UPEC 536 and Nissle 1917), which may provide us an insight on EV proteins involved in pathogenicity; (2) compare the protein content between crude input EVs and EVs purified by DGC or SEC, which will show the benefit of DGC or SEC purification on bacterial EV proteome and help us to identify technical markers of EV; and (3) compare proteome between EVs isolated from bacteria cultured in iron-supplemented and -restricted conditions, which will help us to determine the effect of iron and to identify potential EV markers for infection. A flow diagram of the research strategy is depicted in .
Materials and methods
Bacterial growth conditions
UPEC 536 (O6:K15:H31) [Citation19] and Nissle 1917 (O6:K5:H1) [Citation20] were grown to exponential phase for 5 hr at 37°C with shaking at 200 rpm in 20 ml of RPMI 1640 (Thermo Fisher Scientific) supplemented with 10 µM FeCl3 (RF) or without (R). The cultures were then diluted 100-fold in 2 L of RF or R and grown to stationary phase for 16 hr at 37°C [Citation21].
Crude input EV preparation
Bacterial cells were removed from the culture broths by centrifugation twice at 7,000 × g for 10 min at 4°C followed by filtration through 0.22 μm PES syringe filter (Merck Millipore), and then concentrated using 100 kDa Vivaflow 200 (Sartorius AG). Crude input EV was prepared from the cleared supernatants by ultracentrifugation in polycarbonate tube (29 x 104 mm; Beckman Coulter) at 75,000 × g for 2.5 hr at 4°C in Avanti J-30I centrifuge with JA-30.50 Ti rotor (Beckman Coulter). Resulting EV pellets were resuspended in 1 ml of PBS (Sigma-Aldrich), filtered through 0.22 μm filter and further concentrated using 100 kDa Vivaspin 500 columns (Sartorius AG) before storage at −80°C [Citation21]. These crude input EV preparations were used for further purification by either DGC or SEC as per [Citation17]. Details are shown below and Supplementary Data 1.
EV fractionation by DGC
A 60% OptiPrep stock (Sigma-Aldrich) was diluted with appropriate amounts of PBS to create different density solutions. Forty-five percent OptiPrep was created by adding 1.5 ml of 60% OptiPrep stock to 0.5 ml of crude input EV in PBS. A density gradient was prepared by layering six successive OptiPrep solutions (2 ml of 45% OptiPrep/Sample at the bottom onto which 2 ml each of 40%, 35%, 30%, 25% and 20% OptiPrep were sequentially layered) in a 15 ml Ultra-Clear tube (16 × 95 mm; Beckman Coulter). DGC was performed by centrifugation at 100,000 × g for 16 hr at 4°C in Avanti J-30I centrifuge with JS-24.15 rotor (Beckman Coulter). Resulting fractions were removed from the top according to their visual banding patterns. Each fraction was diluted to 50 ml in PBS to remove OptiPrep, and concentrated to ~500–1500 µl using 100 kDa Vivaspin 20 (Sartorius AG) before storage at −80°C.
EV fractionation by SEC
SEC was carried out using qEV original columns (IZON) according to the manufacturer’s instructions. The column was washed with 10 ml of PBS, and 0.5 ml of input EV sample with known protein concentration was applied to the column. Six ml of PBS was added to the column, and thirteen or fourteen 0.5 ml fractions were collected manually. The column was regenerated by washing with 20 ml of PBS, then with 10 ml of 0.5 M NaOH, followed by 3–5 column volumes of PBS and stored with 20% ethanol in water. Eluted fractions were stored at −80°C.
Protein assay, NTA and TEM
The protein concentrations of crude input and each fraction of DGC and SEC purification were measured by BCA protein assay (Thermo Scientific) according to the manufacturer’s instruction. The protein assay results enabled us to select the EV-rich fractions, and these were pooled as indicated in the Supplementary Data 1 to generate “purified” EV samples.
Particle count and size of EV samples were measured by nanoparticle tracking analysis (NTA) using a Nanosight NS300 system (Malvern Instruments Ltd). The system was calibrated and cleaned between samples with ultrapure MilliQ water. Each sample was diluted 1:200 to 1:2000 in DPBS (Gibco) to reach a concentration within the recommended measurement range (1–10 × 108 particles/ml), loaded onto 1 ml syringes and infused into the NS300 system by a syringe pump (Harvard Apparatus, Cat# 98–4730). Each measurement was recorded in a set of three videos 30 s each with 5 s delay between recordings under the same setting (Capture – screen gain 1.0, camera level 10; Process – screen gain 10.0, detection threshold 10), and the data were analyzed using NTA software version 3.0. Each sample was measured in triplicate, and the results were averaged.
Negative stain was used to prepare EV samples for transmission electron microscopy (TEM). Firstly, EV samples (8 μl of crude or 40 μl of purified) were diluted to 2 ml in UltraPure distilled water (Invitrogen) and concentrated to ~50 μl using 100 kDa Vivaspin 500 columns (Sartorius AG) to exchange buffer. Twenty μl of resulting EV samples were adsorbed onto Formvar-coated copper grids for 2 min before blotting off the excess with filter paper. Each grid was transferred to a 20 μl drop of 2% (w/v) aqueous uranyl acetate for 2 min. The excess was blotted off and the grids were air-dried. All grids were viewed in a Technai G2 Spirit TWIN transmission electron microscope (FEI, Hillsboro, OR, USA) at 120 kV accelerating voltage, and images were captured using a Morada digital camera (OSIS GmbH, Munster, Germany).
Peptide preparation
Proteins were extracted from DGC- and SEC-purified pooled EV samples and their matching crude input EV samples, using the methanol-chloroform precipitation/lipid removal method described by Wessel et al. [Citation22]. Briefly, samples (50 µg of protein for each UPEC sample; 42.5 µg for each Nissle sample) were first concentrated using a Savant SpeedVac concentrator (Thermo Fisher Scientific), and volumes balanced to 100 µl with addition of 50 mM ammonium bicarbonate as necessary. Protein precipitation and lipid removal were performed using four volumes (400 µl) of methanol, one volume (100 µl) of chloroform, and three volumes (300 µl) of water. Samples were mixed by vortexing after each solvent addition. Phase separation was achieved by centrifugation at 12,800 × g for 2 min. The upper phase was discarded, and proteins present at the interface were recovered by adding four volumes (400 µl) of methanol followed by centrifugation at 12,800 × g for 2 min. Protein pellets were dried under vacuum using a SpeedVac concentrator before being resolubilized in 50 µl lysis buffer containing 7 M urea, 2 M thiourea, 10 mM 1,4-dithiothreitol (DTT), 50 mM ammonium bicarbonate. Disulfide bonds were reduced by incubation in lysis buffer at 56°C for 45 min. Cysteines were alkylated by addition of iodoacetamide to a final concentration of 50 mM, followed by incubation at room temperature for 30 min in the dark. Excess iodoacetamide was quenched by the addition of DTT to 20 mM. Samples were diluted 10-fold with 50 mM ammonium bicarbonate and digested with trypsin (Promega, Madison, WI, USA) at 37°C for 16 hr (enzyme:substrate ratio of 1:25). Digested peptides were acidified with formic acid, and desalted on Oasis HLB Vac cartridges (1 cc, 10 mg sorbent, 30 µm; Waters Corp., Milford, MA, USA). Desalted peptides were eluted with 300 µl of 50% acetonitrile, 0.1% formic acid, and dried using SpeedVac concentrator.
Peptide labelling with iTRAQ reagents
Peptide samples were resuspended in 500 mM triethylammonium bicarbonate and labelled with iTRAQ 8-plex reagents (AB Sciex, Framingham, MA, USA) according to the manufacturer’s supplied protocol. Labelling reactions were allowed to proceed for 2 hr before samples were concentrated and peptides resuspended in 0.1% formic acid at a concentration equivalent to 1 µg/µl starting material. For each sample pool, equal amounts of labelled peptide were pooled and desalted on Oasis HLB 1 cc Vac cartridges as described above. All eight UPEC-derived EV samples were pooled together to be analyzed simultaneously in a single run (named as iTRAQ#1) and all the other eight Nissle EV samples were pooled together similarly for the second run (iTRAQ#2).
Mass spectrometry and protein identification and quantitation
Data were acquired on an AB Sciex TripleTOF 6600 Quadrupole-Time-of-Flight mass spectrometer with an Eksigent ekspert nano HPLC system (AB Sciex). Solvents were 0.1% formic acid in water (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B). Samples were injected onto a 0.3 × 10 mm trap column packed with Reprosil C18 media (Dr Maisch, Ammerbuch, Germany) and desalted at 2 µl/min with 2% acetonitrile in 0.1% formic acid before being separated on a 0.075 × 200 mm PicoFrit column (New Objective, Woburn, MA, USA) packed in-house with Reprosil C18 media. For elution of peptides, the following solvent B gradient was formed at 250 ml/min: 0 min 1%; 2 min 1%; 105 min 35.2%; 110 min 98%; 115 min 98%; 116 min 1%; 120 min 1%. The mass spectrometer was programmed to perform cycles of one survey scan with a duration of 250 ms (m/z 350–1600), followed by 40 ms MS/MS scans on the 40 most abundant multiply-charged peptides (m/z 80–1600). The mass spectrometer and HPLC system were under the control of the Analyst TF 1.7 software package (AB Sciex).
Proteins were identified and quantified using ProteinPilot software (v5.0.0.0, 4769; AB SCIEX) that utilizes the Paragon algorithm [Citation23] and integrated false discovery rate (FDR) analysis [Citation24]. Spectral data files were searched against the UniProt E. coli proteome database (19,517 sequence entries, downloaded on 17 August 2015 from www.uniprot.org) with following analysis parameters: Sample type – iTRAQ 8-plex (Peptide Labeled); Cysteine alkylation – iodoacetamide; Digestion – trypsin; Instrument – TripleTOF 6600; Search effort – thorough; FDR analysis – yes. Automatic bias correction was applied to normalize the variability due to unequal mixing of different-labelled peptide samples. Proteins were grouped by the Pro Group algorithm of ProteinPilot to minimize redundancy. ProteinPilot Protein Summary output lists all the proteins found from each iTRAQ run, ratio between samples and additional information related to the protein (e.g. accession number, protein sequence, details from UniProt database), and can be found in the Supplementary Data 2 and 3 for iTRAQ#1 (UPEC) and iTRAQ#2 (Nissle) experiments, respectively, and relevant Peptide Summary showing the peptide details are also provided as the Supplementary Data 4. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [Citation25] partner repository with the dataset identifier PXD011345. Proteins with Unused ProtScore >2 (99% confidence; global FDR all <1%) were reported as “identified proteins”. Venn diagram was created using the UniProt names of identified proteins acquired from the ProteinPilot Protein Summaries. iTRAQ ratio and p value were available for a subset of identified proteins, and such proteins were reported as “quantified proteins”. Quantified proteins with iTRAQ ratio of <0.83 (under-represented) or >1.2 (over-represented), and p value <0.05 were considered to be differentially abundant.
Functional analysis was performed on the identified proteins using Gene Ontology (GO) (http://geneontology.org/) [Citation26] with the following selections: Analysis type – PANTHER Overrepresentation Test (release 13/4/2017); Annotation – GO database, Released 2017-08-14; Analyzed list – UniProt names of identified proteins from iTRAQ experiments; Reference list – Escherichia coli (Database – PortEco; Annotated gene products – 3545; Annotations – 20,323; submission date – 24/5/2017); Bonferroni correction – TRUE. Selected key categories with Bonferroni-corrected p value <0.05 are presented in .
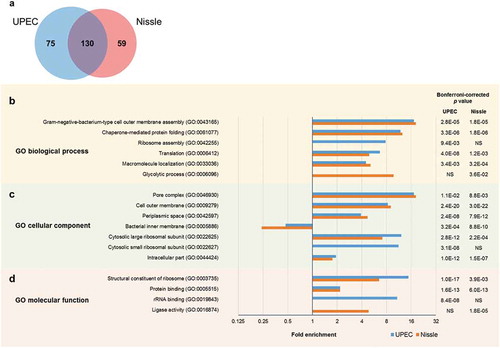
Figure 2. Comparison of EV protein profile between UPEC and Nissle by (a) Venn diagram, and the Gene Ontology (GO) enrichment analysis by (b) biological process, (c) cellular component and (d) molecular function. For the GO analysis, categories with Bonferroni-corrected p value <0.05 were selected. Fold enrichment greater than 1 indicates the over-represented category and less than 1 indicates the under-represented category.
Results
Isolation and characterization of EV samples
Protein yield of crude input samples isolated from 2 L bacteria culture ranged 938–1,980 μg (Supplementary Data 5), and protein recovery following DGC or SEC purification ranged 41–74% of the relative crude input samples, except the SEC-purified EVs of Nissle in RF media (only 15%; Supplementary Data 1). NTA results (Supplementary Data 5) revealed that the mean size of UPEC EVs (range 118–156 nm) was smaller than the Nissle EVs by ~40 nm (range 149–189 nm), EVs of R condition were generally ~20 nm larger than that of RF condition, and the particle count per μg (range 3.2–20 × 109) in each sample was variable. EVs in each sample were visualized and validated using TEM (Supplementary Figure 1).
Comparison of overall EV protein profile between UPEC and Nissle
First, a combination of all eight UPEC-derived EV samples were analyzed simultaneously in the iTRAQ#1 experiment, and all eight Nissle samples in the iTRAQ#2 experiment () to identify the overall EV proteomes for each strain, irrespective of growth or purification conditions. The number of peptides and proteins identified in the two iTRAQ experiments are summarized in with a total of 205 and 189 proteins identified in UPEC and Nissle EV samples, respectively. Of these, 130 proteins were common to both UPEC and Nissle EVs, 75 were unique to UPEC EV and further 59 in Nissle EV only (Supplementary Data 6; Venn diagram in )). Outer membrane proteins like ferrienterobactin receptor FepA, outer membrane protein OmpA, and penicillin-binding protein activator LpoA were found to be abundant (based on “N”, the rank of specified protein relative to all other proteins in the list of detected proteins) and commonly present in both UPEC and Nissle EVs. Hemolysin was N = 1 and had the highest “Unused ProtScore” (the amount of total unique peptide evidence related to a given protein), and the highest “Peptides (95%)” score (the number of distinct peptides having ≥95% confidence), and was identified only in UPEC EV, confirming that this well-known virulence factor [Citation27] is highly abundant in EVs of the pathogenic strain. GO functional enrichment analysis (full results in Supplementary Data 7) highlighted significant enrichments (Bonferroni-corrected p values <0.05) – some common between the strains and some unique to a strain due to their protein variability (). In both UPEC and Nissle EVs, cell outer membrane components (GO:0009279) were highly enriched, but bacterial inner membrane components (GO:0005886) were significantly depleted ()), supporting the outer membrane origin of EVs [Citation14]. In contrast, ribosome-related GO terms such as ribosome assembly (GO:0042255), cytosolic small ribosomal subunit (GO:0022627) and rRNA binding (GO:0019843) were uniquely enriched in UPEC EV, whereas proteins involved in glycolytic process (GO:0006096) and ligase activity (GO:0016874) were enriched in Nissle EV, indicating some differences between the strains ().
Table 1. Samples used in two 8-plex iTRAQ experiments.
Table 2. Summary of spectra, peptides and proteins identified and quantified in two 8-plex iTRAQ experiments.
Effect of purification method on the proteome of UPEC EVs produced under different iron availability conditions
The effect of the purification method on the UPEC EV proteome was studied in the iTRAQ#1 experiment, and the results are summarized in Supplementary Figure 2 (data available in Supplementary Data 2). We first compared the protein profile between four iron-restricted UPEC EV samples (R DGC-crude input, R SEC-crude input, R DGC-purified, R SEC-purified) using “R DGC-crude input” as a denominator (Supplementary Figure 2(a)) and using “R SEC-crude input” as a denominator (Supplementary Figure 2(b)), to dissect the effect of purification methods in R condition. We then repeated the comparison for the RF condition (Supplementary Figure 2(c,d)). Noting that two separate preparations of crude input EVs were used for the DGC and SEC, we observed greater similarity in the UPEC proteome between crude input EVs and their resultant purified EVs (e.g. R DGC-crude input and R DGC-purified) than between two crude input EVs (e.g. R DGC- and R SEC-crude inputs) for both R and RF conditions, indicating relatively high biological variability between input EV preparations (Supplementary Figure 2(a-d)). In addition, the two crude input EV samples prepared from UPEC bacteria grown under iron restriction (R DGC- and R SEC-crude inputs) (Supplementary Figure 2(a,b)) exhibited more variability in protein profile than those grown in the presence of iron (RF DGC- and RF SEC-crude inputs) (Supplementary Figure 2(c,d)). The overall difference in proteome between crude input and purified EV samples was minor in comparison to the difference between crude input UPEC EVs prepared from iron-supplemented and -restricted conditions.
Overall, different sets of proteins were enriched or depleted with DGC and SEC purifications (Supplementary Figure 2(a-d)). We identified individual proteins that were consistently under- or over-represented in both DGC- and SEC-purified UPEC EV samples relative to their crude input EV preparations, suggesting that the extra purification does deplete/enrich for protein cargo. Ribose import binding protein RbsB in R media and uncharacterized protein YoeA in RF media were found enriched in EVs after both purification methods (; Supplementary Figure 2(a-d)). In contrast, 4 proteins including hemolysin, 60 kDa chaperonin GroEL, flagellin and iron uptake system component EfeO were found more in the crude input UPEC EVs of R media than the respective DGC- or SEC-purified EVs (; Supplementary Figure 2(a,b)), suggesting that some may in part be contaminating vesicle-free proteins.
Table 3. Key findings from iTRAQ#1 experiment on the UPEC EV proteins that compared three EV preparation methods (crude input for DGC or SEC purifications, DGC-purified and SEC-purified) in R (without iron) and RF (with iron) growth conditions. Comparison A) indicates a protein enriched by both DGC and SEC purification methods in iron-restricted condition. Comparison B) indicates proteins depleted by both DGC and SEC purification methods in iron-restricted condition. Comparison C) indicates a protein enriched by both DGC and SEC purification methods in iron-supplemented condition. Comparison D) shows a protein depleted by both DGC and SEC purification methods in iron-supplemented condition. Comparison E) indicates proteins enriched in iron-supplemented condition. Comparison F) indicates proteins enriched in iron-restricted condition. N is the rank of the specified protein relative to all other detected proteins (sorted by Unused ProtScore). The details shown below are based on the winner protein selected by the ProteinPilot software for the specific N.
Iron is a key micronutrient for bacteria, and its availability in the host environment is restricted during bacterial infection [Citation4]. To study the proteome of EVs in an infection scenario, we isolated EVs from bacteria grown in the iron-restricted (R) media and compared their proteome with that grown in iron-supplemented (RF) media (i.e. comparisons between R and RF for each DGC-crude input, SEC-crude input, DGC-purified, SEC-purified) (Supplementary Figure 2(e)). As there were inherent differences in protein abundance between samples prepared by different purification techniques, only the proteins altered in at least 3 out of 4 samples were considered important in this comparison. Three proteins categorized as outer membrane components (outer membrane proteins OmpF and Slp, and a putative acyl-CoA thioester hydrolase YbhC), and a cytosolic protein (formate acetyltransferase 1, PflB) were detected more in the RF condition (with iron) (). Conversely, five proteins of various cellular component origins (colicin I receptor CirA, putative lipoprotein AcfD homolog, uncharacterized protein YncE, hemolysin HlyA, iron uptake system component EfeO) were found enriched in the samples of R condition (without iron) (), suggesting that iron restriction affects the EV packaging process. We note that the last two proteins (HlyA and EfeO) were detected more (~1.5-fold) in the crude input EVs than purified EVs, as previously mentioned.
Effect of purification method on the proteome of Nissle EVs produced under different iron availability conditions
The effect of purification method and iron availability on the Nissle EV proteome was studied in the iTRAQ#2 experiment, and the results were summarized in Supplementary Figure 3 (data available in Supplementary Data 3) in a similar manner to Supplementary Figure 2 for UPEC. In comparison to the UPEC, the biological variation from crude input EV preparations was less prominent in the Nissle strain (Supplementary Figure 3(a-d)). In the Nissle iTRAQ experiment, the outer membrane protein assembly factor BamA was found more enriched in DGC- and SEC-purified EVs than the respective crude input EV samples in R media (; Supplementary Figure 3(a,b)). In contrast, 60 kDa chaperonin GroEL and D-galactonate dehydratase family member RspA were over-represented in the crude input EVs of R media than the DGC- or SEC-purified EVs (; Supplementary Figure 3(a,b)). For the RF condition, we did not detect any proteins that were consistently under- or over-represented with the DGC- and SEC-purification in comparison to the crude input EV samples.
Table 4. Key findings from iTRAQ#2 experiment on the Nissle EV proteins that compared 3 EV preparation methods (crude input for DGC or SEC purifications, DGC-purified and SEC-purified) in R (without iron) and RF (with iron) growth conditions. Comparison A) indicates a protein enriched by both DGC and SEC purification methods in iron-restricted condition. Comparison B) indicates proteins depleted by both DGC and SEC purification methods in iron-restricted condition. Comparison C) indicates a protein enriched by both DGC and SEC purification methods in iron-supplemented condition. Comparison D) shows a protein depleted by both DGC and SEC purification methods in iron-supplemented condition. Comparison E) indicates proteins enriched in iron-supplemented condition. Comparison F) indicates proteins enriched in iron-restricted condition. N is the rank of the specified protein relative to all other detected proteins (sorted by Unused ProtScore). The details shown below are based on the winner protein selected by the ProteinPilot software for the specific N.
Seven proteins were enriched in EVs from RF cultures, when compared to EVs from R cultures in 3 out of 4 samples: type-1 fimbrial protein Fim1C, DNA protection during starvation protein Dps, flagellar hook-associated protein 3 FlgL, penicillin-binding protein activator LpoA, serine protease Sat autotransporter, peptidoglycan-associated lipoprotein Pal, and long-chain fatty acid transport protein FadL. LpoA, Sat, Pal, and FadL are categorized as outer membrane components (; Supplementary Figure 3(e)). Moreover, another three outer membrane proteins were enriched in RF condition than R for all four samples tested: OmpC, endolytic peptidoglycan transglycosylase RlpA and putative outer membrane porin protein NmpC (; Supplementary Figure 3(e)). This indicates that 8 out of 11 proteins enriched in the Nissle RF EVs were from the outer membrane origin. In contrast, eight proteins were enriched in the R condition than RF: elongation factor Tu 1 TufA, periplasmic dipeptide transport protein DppA, uncharacterized protein YncE, iron uptake system component EfeO, FKBP-type peptidyl-prolyl cis-trans isomerase FkpA, uncharacterized lipoprotein YbaY, thiol peroxidase Tpx and outer membrane lipoprotein SlyB (; Supplementary Figure 3(e)). Of the latter eight proteins, SlyB is the only protein localized at the outer membrane.
Common findings from UPEC and Nissle EV iTRAQ experiments
We finally compared the results from both iTRAQ experiments to identify any common changes to both UPEC and Nissle EVs when iron availability in culture and subsequent purification methods were modified. In summary, the 60 kDa chaperonin GroEL was found over-represented in the crude input EVs than the DGC- and SEC-purified EVs for both UPEC and Nissle in R media ( and ), suggesting a potential utility as a technical marker for purification. We also found two proteins (iron uptake system component EfeO and uncharacterized protein YncE) that were enriched in the R condition for both UPEC and Nissle ( and ), suggesting a potential role for these EV proteins in iron acquisition. Further, our results on both strains showed that EVs released in the RF condition were more enriched with proteins of outer membrane origin, whereas EVs released in the R condition contain proteins of various cellular origins, suggesting different EV biogenesis mechanisms may be employed depending on the availability of iron.
Discussion
Here we determined the UPEC and Nissle EV proteomes to investigate the effect of DGC and SEC purification and changes in iron availability during culture using an iTRAQ approach. The majority of the proteins identified in this study were able to be confirmed in previous studies on the bacterial EV proteome [Citation28]. However, there has not been a study to date that has compared the EV proteome of UPEC and Nissle between different EV purification methods, grown in both iron-restricted and -supplemented media.
Difference in EV between UPEC and Nissle
UPEC 536 is a pathogenic strain that is a causative agent of urinary tract infections [Citation29], whereas the Nissle 1917 is probiotic, non-invasive, non-pathogenic and known to have a beneficial effect on many intestinal diseases [Citation30]. In view of the total EV proteomes, 130 proteins identified in this study were shared by UPEC and Nissle, and the rest were unique (75 and 59 proteins; 37% and 31% of total protein identified, respectively). This reflects a considerable difference in the EV protein content in two different strains of the same E. coli species. It was interesting to see that proteins involved in the glycolytic process and ligase activity were found enriched in Nissle EV, but ribosomal proteins and ribosome assembly enriched in UPEC EV. Recent studies have shown that ribosomal proteins in both bacteria [Citation31] and human [Citation32] have extra-ribosomal functions, acting as regulatory proteins or cellular components. Further, many ribosomal proteins (including S4 and S7 detected in UPEC here) are known to be universal and present in both prokaryotes and eukaryotes [Citation31], meaning that they could potentially have some roles in disease involvement. Contrary to our proteome results, the genomes of UPEC and Nissle have high homology. BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of UPEC genome sequence (Accession#CP000247) to Nissle (Accession#CP000247) resulted in 93% query coverage and 99% identical sequences. Comparing those strain-specific EV proteins identified in this study and the genome distribution between two strains revealed that some were also unique to a single strain at the genome level. For example, Nissle lacks the hemolysin gene hlyA [Citation33], whereas hemolysin protein is highly abundant in the EVs of pathogenic UPEC. In the case of carbamoyl-phosphate synthase large chain however, both UPEC and Nissle carry the same gene, but the protein was only detected in UPEC EV. This protein could either be expressed in Nissle bacteria but not in their EV or present in their EV in undetectable amount.
Potential ubiquitous markers
Among the 130 proteins that were common to both UPEC and Nissle EVs, a few proteins of outer membrane origin including FepA, LpoA and OmpA were found abundantly present in both species. Moreover, OmpA has been previously used to quantify bacterial EVs [Citation34,Citation35]. Such outer membrane proteins may be able to serve as ubiquitous protein markers of E. coli EVs in the way that Alix and CD63 are used as common markers for eukaryotic exosomes [Citation36,Citation37]. Further studies will be needed to validate this finding.
Potential markers for purification
Bacterial EV heterogeneity and multiple EV biogenesis mechanisms proposed by recent studies [Citation2,Citation6,Citation13] led us to study the effect of purification on the EV proteome. “Crude input” samples were prepared by a common method that involves sequential filtration and ultracentrifugation. DGC and SEC, two methods frequently used for eukaryotic EV research [Citation38], were used to further purify the bacterial EVs here. By comparing EV proteome prepared by different purification methods, we found a few potential technical methods and purification markers. Three proteins (RbsB of UPEC EV in R; YoeA of UPEC EV in RF; BamA of Nissle EV in R) were consistently enriched in the DGC- and SEC-purified EV samples in comparison to their input EVs. RbsB is a part of the ABC transporter complex RbsABC and involved in chemotaxis. RbsB has been previously identified in EVs of Salmonella enterica serovar Typhimurium, which is a major cause of enteric diseases [Citation39], but has not been previously detected in UPEC EVs. YoeA (UniProt#P76356) is an uncharacterized outer membrane protein and has a TonB-dependent Receptor Plug Domain (pfam07715; amino acid residues 54–159 of 167; https://www.ncbi.nlm.nih.gov/Structure/cdd/), suggesting a biological role as a receptor. BamA is a part of the outer membrane protein assembly complex, previously identified in EVs of two E. coli strains DH5α [Citation40] and Nissle [Citation41]. RbsB, YoeA and BamA enriched in purified EVs may represent true EV-associated proteins, and may be useful protein markers for assessing purity after EV purification or for antibody-assisted EV purification.
Proteins depleted in purified EVs
The crude input sample contains proteins that are depleted by the purification process. Are these contaminant proteins present in “extra-vesicle” aggregates that co-pellet with EVs? If this is the case these proteins can be used as contamination markers. Alternatively, these proteins may be present in the EVs and in protein aggregates, and so be less useful as purity markers. Hemolysin, flagellin and GroEL are examples that were detected at higher levels in the crude input EVs than the purified EVs from UPEC grown under iron-restriction (refer to data).
Hemolysin is a secreted virulence factor associated with upper urinary tract infection [Citation27]. Our results show HlyA is present in high abundance in EVs from UPEC, with most found in EVs from cultures grown in iron-restricted conditions, supporting a role in the early events of infection [Citation42]. A previous study has demonstrated that cytotoxically active hemolysin is tightly associated with DGC-purified E. coli EVs [Citation43]. One explanation of our findings is that we have detected the small fraction of hemolysin that is tightly associated with purified EVs, while the majority of soluble hemolysin is removed by DGC or SEC.
Flagellin is frequently identified in gram-negative bacterial EVs [Citation28]. Flagellin is the major subunit of flagellae, multi-subunit structures that are usually considered an unwanted contaminant in EV preparation, especially as they could exert an immunomodulatory effect and interfere with subsequent analyses [Citation16]. Proteomics results () and TEM images (Supplementary Figure 1) in our study suggest that additional purification step by SEC helps to reduce this filament-like contaminating structural protein more effectively than by DGC.
The 60 kDa chaperonin GroEL, which is required for proper protein folding, was the only protein over-represented in the crude input EVs from both UPEC and Nissle cultured in R. GroEL is a major heat shock protein of E. coli, but abundant and essential even when the cells are not stressed [Citation44]. GroEL could be in crude input EVs because it is complexed with misfolded or damaged proteins that are being excreted. Or it could be a contaminant of preparations from extra-vesicle aggregates or from lysed cells that stick to the outside of EV. If it is confirmed to be a contaminating waste protein, it could be used as a technical marker for purification. However, this protein has been previously detected in many EV studies on Gram-negative bacteria [Citation28] including E. coli strains such as non-pathogenic DH5α [Citation40], Nissle [Citation41], MG1655 [Citation45]; enterohemorrhagic O157 [Citation46] and extraintestinal pathogenic O25b:H4-ST131 [Citation47]. Most of these studies used DGC for purification, although none compared its protein amount before and after the DGC. Further, a recently released commercial kit for bacterial EV isolation (System Biosciences, Cat#EXOBAC100A-1) uses GroEL as an EV marker. Further tests to validate the presence of GroEL as a true-EV associated protein (e.g. using antibody-mediated microscopy techniques to localize GroEL in crude and purified EVs) now seem warranted.
Benefit from DGC or SEC purification
We initially hypothesized that a DGC- or SEC-purification step applied to the crude input EVs would result in a clear depletion of contaminating proteins from parent bacterial cells. In reality, however, the biological variability from input EV preparations was large, whereas the difference between input, DGC- and SEC-purified EVs was comparatively small. Overall, our results showed that the additional purification of crude input EV preparations by DGC or SEC only helps to remove a small number of contaminating proteins under the defined growth media conditions we used, although such purification step was shown to be beneficial for other contents of bacterial EV (e.g. RNA, endotoxin) in our previous study [Citation17]. In addition, different sets of proteins were enriched or depleted with DGC and SEC purification method, so it is difficult to say which method is superior.
Effect of iron on EV proteome
Iron is an important micronutrient for bacteria, and its availability is often restricted in the host environment at the beginning of an infection and further reduced when the host’s innate immune responses become active. This iron restriction is a stressful environment for the bacteria and inhibits in their growth, but can increase the production of EVs in Mycobacterium tuberculosis [Citation5], Haemophilus influenzae, Vibrio cholerae and E. coli [Citation6], and influence their content in Helicobacter pylori [Citation48]. Roier et al. recently proposed a novel EV biogenesis mechanism based on phospholipid accumulation in the outer leaflet of the outer membrane, which is regulated by iron availability [Citation15].
It has been previously shown that iron restriction causes alteration in the cellular proteome of bacteria [Citation49–Citation52], where the proteins that were altered in abundance were often those involved in iron acquisition, homeostasis and transport [Citation49,Citation50,Citation52]. Our study detected several proteins whose abundance in EVs was dependent on the iron availability in the bacterial growth media. An interesting observation here was that the EVs released in iron-supplemented condition were enriched with proteins of outer membrane origin, whereas the EVs from iron-restricted condition had no particular enrichment of outer membrane proteins. It suggests that there could be a selective packaging or an alternative mechanism of EV formation when the iron is depleted.
In our study, OmpF in UPEC and three proteins (RlpA, NmpC, OmpC) in Nissle were more abundant in the iron-supplemented condition in all four EV samples (DGC-crude input, SEC-crude input, DGC-purified, SEC-purified) and all localized at the outer membrane. Such proteins may represent markers for iron availability. Conversely, YncE, which is an uncharacterized, predicted periplasmic protein with DNA-binding activity [Citation53], was specifically enriched in EVs of R condition for both UPEC and Nissle. A few reports have identified YncE in the UPEC EVs. Wurpel et al. detected YncE in the crude EV proteomes of a large collection of UPEC [Citation54] and in the EDTA heat-induced UPEC EV after growth in human urine [Citation55]. Moriel et al. [Citation56] showed the presence of YncE in crude EV proteome of UPEC EC958 strain and its regulation by Fur, confirming a finding by McHugh et al. [Citation57]. In addition, they identified YncE as a novel protective vaccine candidate, by demonstrating that it is immunogenic, highly prevalent, highly conserved across E. coli, soluble, stable and expressed during infection [Citation56]. Considering the infection scenario, YncE significantly enriched in the iron-restricted condition in our study would potentially be an EV marker for iron restriction or early infection, and a valuable target for vaccine and drug development. Other than the YncE, proteins involved in iron uptake (EfeO in UPEC and Nissle; CirA in UPEC) were found more abundant in iron-restricted condition, suggestive of other potential markers for infection. A previous study suggested that M. tuberculosis EVs can capture iron and deliver it back to the bacteria, indicating a crucial role of EVs in iron acquisition [Citation5]. E. coli may function similarly, and the latter two proteins may be involved in this process. But EfeO is a periplasmic protein [Citation58] and was more abundant in the crude input EVs than purified EVs in this study, so its suitability as a useful vesicular marker is questionable.
Challenges with EV heterogeneity
EVs released by bacteria are composed of heterogeneous EV populations of different size, content and structure [Citation46,Citation59], owing to multiple mechanisms of biogenesis [Citation2,Citation6,Citation13]. Such heterogeneity is also present in eukaryotic EVs [Citation60], which are better characterized than those from bacteria. Overall, however, a clear discrimination between different types of vesicles is still not well established for both prokaryotes and eukaryotes. This highlights the need for using purified EV subpopulations to better understand their roles in physiology and pathogenesis. The issue of bacterial EV heterogeneity is further complicated by the impact of the environment. In the case of iron restriction, it exerts a dramatic effect on the E. coli cells [Citation61] and can alter the amount of EV production [Citation5,Citation6], EV content (lipid [Citation5] and protein [Citation47]) and the DGC banding pattern [Citation17] of the EVs. In addition, here we detected substantial changes in EV proteome in response to iron restriction. Thus, the heterogeneity of bacterial EVs presents a challenge that requires further in-depth study. Moreover, the consideration of results from studies that used different EV purification techniques or different conditions to grow bacteria requires care as the results from one particular condition should not be used to infer in vivo outcomes.
Limitation of this study
The iTRAQ approach [Citation18] allows proteins to be analyzed simultaneously in up to eight different samples, eliminates the inter-sample variations of protein identification and quantification from the data-dependent acquisition, and reduces the analysis time and experimental cost. iTRAQ has helped to discover disease biomarkers and therapeutic targets [Citation62]. Using the iTRAQ approach, we have compared the EV proteome from different purification methods and from EVs from cells cultured in iron-supplemented and -restricted conditions for UPEC (iTRAQ#1) and Nissle (iTRAQ#2). In doing so, we were able to identify a few potential markers for EV purification and for growth under iron restriction. We note however that there are limitations to this study. Firstly, we analyzed eight different samples in each run to compare three purification methods and two iron conditions. The use of more replicates and fractionated iTRAQ would have provided more robust, deeper proteome data. Secondly, quantification for the label-based iTRAQ approach relies on how well the chemical tags are incorporated into the peptides. In this respect, more recently developed SWATH-MS (Sequential Window Acquisition of all THeoretical Mass Spectra) uses label-free, data-independent analysis for quantification, so could be used as an alternative to iTRAQ [Citation63]. Thirdly, some proteins might not have been fully solubilized after methanol-chloroform precipitation method, and thus not be detected. But our initial sample inputs were of variable volume and concentration, and this methanol-chloroform precipitation method produced a pellet for each sample and allowed us to digest an equal volume of protein lysates with trypsin. In a test study (Supplementary Data 8), we detected a great improvement in the number of protein identifications from a pool of four E. coli EV samples with the methanol-chloroform precipitation (214 proteins with Unused ProtScore >2) in comparison to the one prepared without the precipitation step (only 17 with Unused ProtScore >2); hence, we have decided to add this precipitation step. Fourthly, proteins satisfying both 1.2-fold cutoff and p < 0.05 were considered differentially abundant proteins here. Higher fold-change or lower p value cutoff would give results with higher confidence, but less number of altered proteins would be detected in return. And the 1.2-fold cutoff is frequently used in iTRAQ data analysis [Citation64–Citation66]. Thus, we believed that less conservative 1.2-fold cutoff is reasonable to use in this exploratory study to find a few candidate proteins. Lastly, lack of completeness and uniformity in annotations in different strains of E. coli limit the proper profiling of EV proteome in both UPEC 536 and Nissle. Supported by the high genome similarities between different E. coli strains (discussed above), here we used all E. coli entries present in the database to identify proteins using ProteinPilot and to perform GO functional analysis on the UPEC 536 and Nissle proteins.
Conclusion
We have provided a global proteome of EVs isolated from two E. coli strains – the pathogen UPEC 536 and the probiotic Nissle 1917 grown in in vivo relevant conditions. EV proteins that were common and unique to each strain were identified. We identified proteins that were enriched in DGC- or SEC-purified EVs, representing potential technical method or purification markers. Furthermore, we identified EV proteins affected by the simple addition of iron to the growth media where the identification of such proteins may provide a rationale to investigate the mechanisms that control EV packaging in the process of a bacterial infection.
Data deposition
ProteomeXchange (ID#PXD011345)
Supplemental Material
Download Zip (26.6 MB)Acknowledgments
The authors would like to thank Ratish Kurian from the Biomedical Imaging Research Unit, University of Auckland for his assistance with TEM analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 2015;15(6):375–16.
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from gram-negative bacteria. Microbiol. 2014;160(Pt 10):2109–2121.
- Orench-Rivera N, Kuehn MJ. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol. 2016;18(11):1525–1536.
- Dauros-Singorenko P, Swift S. The transition from iron starvation to iron sufficiency as an important step in the progression of infection. Sci Prog. 2014;97(Pt 4):371–382.
- Prados-Rosales R, Weinrick BC, Pique DG, et al. Role for mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol. 2014;196(6):1250–1256.
- Roier S, Zingl FG, Cakar F, et al. A novel mechanism for the biogenesis of outer membrane vesicles in gram-negative bacteria. Nat Commun. 2016;7:10515.
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19(22):2645–2655.
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184.
- Haurat MF, Elhenawy W, Feldman MF. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol Chem. 2015;396(2):95–109.
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74(1):81–94.
- Jan AT. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front Microbiol. 2017;8:1053.
- Hoekstra D, van der Laan JW, de Leij L, et al. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta. 1976;455(3):889–899.
- Perez-Cruz C, Delgado L, Lopez-Iglesias C, et al. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One. 2015;10(1):e0116896.
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13(10):605–619.
- Roier S, Zingl FG, Cakar F, et al. Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microb Cell. 2016;3(6):257–259.
- Klimentova J, Stulik J. Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol Res. 2015;170:1–9.
- Dauros Singorenko P, Chang V, Whitcombe A, et al. Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. J Extracell Vesicles. 2017;6(1):1324731.
- Wiese S, Reidegeld KA, Meyer HE, et al. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340–350.
- Knapp S, Hacker J, Jarchau T, et al. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J Bacteriol. 1986;168(1):22–30.
- Nissle A. Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Med Klin. 1918;2:29–33.
- Blenkiron C, Simonov D, Muthukaruppan A, et al. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. Plos One. 2016;11(8):e0160440.
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–143.
- Shilov IV, Seymour SL, Patel AA, et al. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6(9):1638–1655.
- Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7(9):3661–3667.
- Vizcaino JA, Csordas A, del-Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456.
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29.
- Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4(1):80–128.
- Lee J, Kim OY, Gho YS. Proteomic profiling of Gram-negative bacterial outer membrane vesicles: current perspectives. Proteomics Clin Appl. 2016;10(9–10):897–909.
- Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11–19.
- Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microb Ecol Health Dis. 2009;21(3–4):122–158.
- Aseev LV, Boni IV. Extraribosomal functions of bacterial ribosomal proteins. Mol Biol. 2011;45(5):739–750.
- Wang W, Nag S, Zhang X, et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35(2):225–285.
- Grozdanov L, Raasch C, Schulze J, et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186(16):5432–5441.
- Chutkan H, Macdonald I, Manning A, et al. Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol Biol. 2013;966:259–272.
- McBroom AJ, Johnson AP, Vemulapalli S, et al. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188(15):5385–5392.
- Bobrie A, Colombo M, Krumeich S, et al. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1:18397.
- Clark DJ, Fondrie WE, Liao Z, et al. Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster analysis. Anal Chem. 2015;87(20):10462–10469.
- Witwer KW, Buzas EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360.
- Bai J, Kim SI, Ryu S, et al. Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar Typhimurium. Infect Immun. 2014;82(10):4001–4010.
- Lee EY, Bang JY, Park GW, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7(17):3143–3153.
- Aguilera L, Toloza L, Gimenez R, et al. Proteomic analysis of outer membrane vesicles from the probiotic strain Escherichia coli Nissle 1917. Proteomics. 2014;14(2–3):222–229.
- Ristow LC, Welch RA. Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger? Biochim Biophys Acta. 2016;1858(3):538–545.
- Balsalobre C, Silvan JM, Berglund S, et al. Release of the type I secreted alpha-haemolysin via outer membrane vesicles from Escherichia coli. Mol Microbiol. 2006;59(1):99–112.
- Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171(3):1379–1385.
- Kulkarni HM, Nagaraj R, Jagannadham MV. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol Res. 2015;181:1–7.
- Bielaszewska M, Ruter C, Bauwens A, et al. Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog. 2017;13(2):e1006159.
- Chan KW, Shone C, Hesp JR. Antibiotics and iron-limiting conditions and their effect on the production and composition of outer membrane vesicles secreted from clinical isolates of extraintestinal pathogenic E. coli. Proteomics Clin Appl. 2017;11(1–2). DOI:10.1002/prca.201600091
- Keenan JI, Allardyce RA. Iron influences the expression of helicobacter pylori outer membrane vesicle-associated virulence factors. Eur J Gastroenterol Hepatol. 2000;12(12):1267–1273.
- Folsom JP, Parker AE, Carlson RP. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J Bacteriol. 2014;196(15):2748–2761.
- Lim CK, Hassan KA, Tetu SG, et al. The effect of iron limitation on the transcriptome and proteome of pseudomonas fluorescens Pf-5. PLoS One. 2012;7(6):e39139.
- Lima DC, Duarte FT, Medeiros VK, et al. The influence of iron on the proteomic profile of chromobacterium violaceum. BMC Microbiol. 2014;14:267.
- Pieper R, Fisher CR, Suh MJ, et al. Analysis of the proteome of intracellular Shigella flexneri reveals pathways important for intracellular growth. Infect Immun. 2013;81(12):4635–4648.
- Kagawa W, Sagawa T, Niki H, et al. Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 12):1045–1053.
- Wurpel DJ, Moriel DG, Totsika M, et al. Comparative analysis of the uropathogenic Escherichia coli surface proteome by tandem mass-spectrometry of artificially induced outer membrane vesicles. J Proteomics. 2015;115:93–106.
- Wurpel DJ, Totsika M, Allsopp LP, et al. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J Proteomics. 2016;131:177–189.
- Moriel DG, Tan L, Goh KG, et al. A novel protective vaccine antigen from the core Escherichia coli genome. mSphere. 2016;1(6):pii: e00326–16.
- McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, et al. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278(32):29478–29486.
- Sturm A, Schierhorn A, Lindenstrauss U, et al. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J Biol Chem. 2006;281(20):13972–13978.
- Elmi A, Watson E, Sandu P, et al. Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect Immun. 2012;80(12):4089–4098.
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383.
- Rowe MC, Withers HL, Swift S. Uropathogenic Escherichia coli forms biofilm aggregates under iron restriction that disperse upon the supply of iron. FEMS Microbiol Lett. 2010;307(1):102–109.
- Fuller HR, Morris GE. Quantitative proteomics using iTRAQ labeling and mass spectrometry. In: Leung HE, editor. Integrative proteomics; 2012. 347–362. London (England): InTechOpen.
- Bourassa S, Fournier F, Nehme B, et al. Evaluation of iTRAQ and SWATH-MS for the quantification of proteins associated with insulin resistance in human duodenal biopsy samples. PLoS One. 2015;10(5):e0125934.
- Yang W, Ding D, Zhang C, et al. iTRAQ-based proteomic profiling of Vibrio parahaemolyticus under various culture conditions. Proteome Sci. 2015;13:19.
- Liang V, Ullrich M, Lam H, et al. Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell Mol Life Sci. 2014;71(17):3339–3361.
- Rhein V, Song X, Wiesner A, et al. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proc Natl Acad Sci U S A. 2009;106(47):20057–20062.