ABSTRACT
Extracellular RNA in circulation mediates intercellular communication in normal and pathological processes. One mode of circulating miRNA transport in bodily fluids is within 30–150 nm small extracellular vesicles (sEVs) or exosomes. Uptake of sEVs can regulate gene expression in recipient cells enabling circulating miRNAs to exert paracrine and systemic effects. Complex regional pain syndrome (CRPS) is a debilitating pain disorder characterized by chronic inflammation. Our previous investigations identified a significant decrease of hsa-miR-939 in whole blood from CRPS patients compared to control; we also observed that overexpression of miR-939 can negatively regulate several proinflammatory genes in vitro. Though downregulated in whole blood, miR-939 was significantly upregulated in sEVs isolated from patient serum. Here we investigated miR-939 packaging into sEVs in vitro under inflammation induced by monocyte chemoattractant protein-1 (MCP-1), a chemokine that is upregulated in CRPS patients. Stimulation of THP-1 monocytes by MCP-1 led to elevated levels of miR-939 in sEVs, which was abrogated using inhibitors of exosome secretion. miRNAs loaded into exosomes largely contain short miRNA sequence motifs called EXOmotifs. Mutation analysis of miR-939 showed that EXOmotif is one of the possible cellular mechanisms responsible for packaging miR-939 into sEVs. We confirmed gene expression changes in recipient cells following the uptake of sEVs enriched in miR-939 using RNA sequencing. Additionally, our data from primary immune cell-derived sEVs of CRPS patients and controls demonstrate that while the relative expression of miR-939 is higher in sEVs derived from B cells, T cells and NK cells relative to monocyte-derived sEVs in controls, only the B cell-derived sEVs showed a significantly higher level of miR-939 in CRPS patients. Differential miRNA sorting into exosomes and its functional impact on recipient cells may contribute to the underlying pathophysiology of CRPS.
Introduction
Circulating miRNA mediates intracellular communication by regulating gene expression in recipient cells under normal and pathological conditions. Biological functions of circulating miRNAs can thus be elucidated by determining their cellular source, factors determining their release, and gene expression changes upon miRNA uptake in recipient cells. In disease states, understanding the stress signal and its impact on miRNA release, whether it is an uncontrolled process or conversely, secreted in a regulated fashion as a compensatory mechanism, can help assess the role and biomarker utility of extracellular RNAs.
Table 1. Relative expression of miR-939 in the whole blood of CRPS patients (n = 37) and healthy controls (n = 18) [Citation26] and serum-derived sEVs (n = 6 for both CRPS patients and controls) [Citation28].
Cell-free miRNAs in circulation are found either enclosed within membranous vesicles such as exosomes, or in association with high-density lipoproteins and RNA-binding argonaute (Ago) proteins [Citation1–Citation4]. Exosomes are small 30 to 150 nm membrane vesicles found in various bodily fluids. Exosomes contain diverse classes of biomolecules including miRNAs, mRNAs, proteins, and lipids that are co-expressed, packaged, and secreted by cells into bodily fluids and this composition can differ under normal and disease states [Citation5–Citation10]. Exosomes are formed from endosomes, which in turn arise from an inward budding of the plasma membrane into the cytoplasm. Inward budding of the endosomal membrane gives rise to intraluminal vesicles called multivesicular bodies (MVB), which involves the use of endosomal sorting complex required for transport (ESCRT) proteins and other associated proteins like TSG101 and ALIX in the late endosomal pathway [Citation11]. MVBs can fuse with the plasma membrane thereby releasing the exosomes to the adjacent cells or into circulation. Exosome release represents a novel pathway that allows biomolecules to travel long distances and the uptake of this cargo can impact function in recipient cells [Citation12].
Studies have demonstrated that the RNA content of exosomes is distinct from that found in the cytoplasm of donor cells from which they were derived, suggesting active RNA sorting into these vesicles [Citation13–Citation16]. One suggested mechanism of exosomal miRNA sorting involves the association of miRNA with Argonaut 2 (Ago2) component of the RNA-inducing silencing complex (RISC) to form Ago2-miRNA complexes that are found both in the extracellular space as well as in secreted exosomes [Citation17]. In another example, exosome sorting and export of miR-193a in mouse colon cancer cells were regulated by major vault protein (MVP), where knocking out MVP resulted in accumulation of miR-193a within donor cells and subsequently downregulating its target mRNA, Caprin-1 [Citation18]. Additionally, RNA-binding proteins like hnRNPB2A1, YBX1 and SYNCRIP are known to bind specific sequences/motifs within the miRNA and aid the recruitment of miRNA into exosomes [Citation19–Citation21]. A recent RNA-sequencing study of macrophages and their secreted exosomes showed that the sorting of miRNAs into exosomes can be modulated by changing either cellular miRNA expression or their respective endogenous target mRNA levels in the donor cells [Citation22]. Thus, several molecular pathways have been described to impact miRNAs sorting in different cellular contexts [Citation19,Citation23,Citation24].
Complex regional pain syndrome (CRPS) is a debilitating pain disorder that can significantly impact the quality of life. The pathophysiology of CRPS varies and can change during the course of the illness contributing to different clinical symptoms. Neurogenic inflammation is an important feature of this disorder along with sensory, autonomic and trophic impairments [Citation25]. In our previous study exploring the utility of circulating miRNAs as biomarkers for patient stratification, we profiled miRNAs in whole blood from 41 CRPS patients and 20 controls. We identified differential expression of 18 miRNAs, of which miR-939 ranked first and showed a 4.3-fold downregulation in whole blood from CRPS patients [Citation26]. Our in vitro studies showed miR-939 can be an anti-inflammatory miRNA targeting several genes encoding various proinflammatory mediators [Citation27]. Interestingly, though they are downregulated in whole blood (relative to total RNA including RNA derived from all circulating immune cells) [Citation26], miR-939 is significantly upregulated in the sEV fraction from patient serum (normalized to sEV RNA)() [Citation28]. Thus, miR-939 packaging into exosomes could serve as a unique model to understand miRNA alterations in an inflammatory state and how exosome-mediated gene regulation can influence the pro and anti-inflammatory homeostasis.
Proinflammatory signals like cytokines can trigger the secretion of chemokines, which play an important role in selectively recruiting immune cells such as monocytes and neutrophils [Citation29]. Monocyte chemoattractant protein-1 (MCP-1), also known as C-C motif chemokine ligand 2 (CCL2) is a chemokine with elevated plasma levels in CRPS patients [Citation26,Citation30]. Here we investigated whether higher MCP-1 levels can influence miR-939 packaging into sEVs and if the uptake of sEVs overexpressing miR-939 can alter gene expression in recipient cells using human monocytic THP-1 cells.
Results
Isolation and characterization of THP-1 cell-derived sEVs
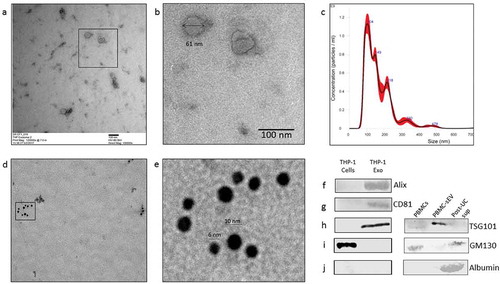
Small extracellular vesicles (sEVs) isolated from the human monocytic leukaemia THP-1 cells by differential ultracentrifugation were characterized for size and purity. These sEVs displayed a size below 100 nm under a transmission electron microscope (TEM) ()). Nanoparticle tracking analysis of THP-1 sEVs indicated particles with a mean diameter of 85.7 ± 0.9 nm and a concentration of 3.94 × 1011 ± 1.6 × 1010 particles/ml ()). Exosomes are characterized by the enrichment of endosome-derived membrane proteins such as tetraspanins (CD9, CD63, CD81), flotillins, Alix and Tsg101. The presence of such characteristic exosome proteins was also assessed using immunogold double labelling technique for CD81 and CD9 by TEM (,e)) and western blotting for Alix, CD81 and TSG101 ()). In addition, the sEV preparations isolated from both THP-1 cells and peripheral blood mononuclear cells (PBMCs) from CRPS patients were negative for the non-EV markers like GM130 ()) and serum contaminants like albumin which is eliminated into the supernatant post-ultracentrifugation (), post-UC sup). The metrics of sEV isolation, purity and characterization were submitted on EV-track [Citation31] and can be accessed using EV-TRACK ID: EV190028. The EV-metric score is 44%.
Figure 1. Characterization of THP-1 derived sEVs. (a). Transmission electron micrograph of THP-1 derived sEVs obtained from the culture supernatant of 60 million cells. (b). Magnified image of the selected region in 1a. (c). Nanoparticle tracking analysis of sEVs indicated particles with a mean diameter of 103.2 ± 4.9 nm and a particle concentration of 1.3 × 1011 ± 2.09 × 109 particles/ml. (d). Transmission electron micrograph of sEVs double-immunostained for CD81 and CD9 (conjugated to 6nm and 10 nm gold particles, respectively). (e). Magnified image of the selected region in 1d. Western blotting of sEV pellet was performed to confirm the presence of exosome marker protein, Alix (f), CD81 (g), and TSG101 (h). Absence of the routine contaminants of EV preparations like GM130 (Golgi marker) and serum albumin was also ascertained in the isolated sEVs from both THP-1 cell-derived sEVs and PBMC-derived sEVs (i, j, respectively).
Inflammation induces the cellular expression and packaging of miR-939 into sEVs
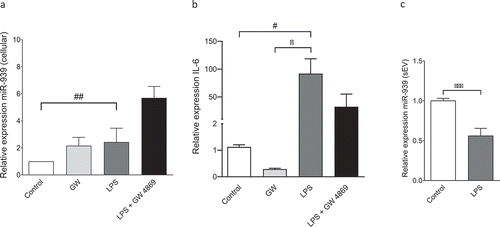
To assess if inflammation could modulate levels of the miR-939 in vitro, we stimulated THP-1 cells with bacterial lipopolysaccharide (LPS). THP-1 cells cultured in an exosome-depleted media overnight were stimulated with LPS (1 µg/ml) for 4 h. Endogenous levels of cellular miR-939 and target mRNAs were measured by qRT-PCR. A significant increase in cellular miR-939 was observed following LPS stimulation. Neutral sphingomyelinase 2 (nSMase2) is a critical enzyme in exosome biogenesis. GW4869, a synthetic nSMase2 inhibitor is used to inhibit exosome secretion [Citation32]. Cellular miR-939 were noticeably augmented in the presence of GW4869 both under naive and LPS stimulated conditions ()). Human IL-6 mRNA is a validated target of miR-939 and is downregulated by the overexpression of miR-939 [Citation27]. Hence, we quantified the levels of intracellular IL-6 mRNA following LPS stimulation of THP-1 cells. While LPS stimulation increased the cellular IL-6 mRNA levels, a marked decrease was observed in the presence of GW4869 ()). Although LPS treatment increased the overall cellular levels of miR-939, there was a moderate decrease in its incorporation into the sEVs ()). This could be attributed to the overexpressed cellular miR-939 being expended in counteracting the proinflammatory genes upon LPS stimulation. Hence, although LPS-mediated inflammatory stimulus plays a role in miR-939 expression, other mechanisms may govern its packaging and secretion within sEVs.
Figure 2. Regulation of miR-939 levels under inflammation. THP-1 cells were treated for 4 h with an inflammatory stimulus (LPS) following a two-hour pretreatment with an inhibitor for exosome biogenesis (GW4869). Cellular RNA was isolated and Taqman-based qRT-PCR used to determine the relative expression of miR-939 (a) and IL-6 (b) normalized to U6 snRNA and GAPDH, respectively. Significance was determined by one-way ANOVA with Sidak’s multiple comparison test *, # p < 0.05, ## p < 0.01. RNA was isolated from corresponding sEVs and Taqman-based qRT-PCR used to determine miR-939 expression normalized to miR-223. Significance was determined by Student unpaired t-test *** p < 0.001.
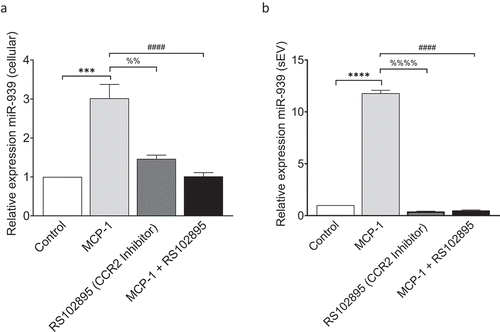
Several studies have reported an increase in plasma levels of MCP-1, also known as C-C motif chemokine ligand 2 (CCL2) in CRPS patients compared to healthy donors [Citation26,Citation30,Citation33]. Since we did not observe a significant change in miR-939 packaging into sEVs upon LPS stimulation, we further tested the hypothesis that inflammation induces packaging of miR-939, using MCP-1. We used MCP-1 to stimulate THP-1 cells in vitro and measured the levels of miR-939 in cells and their secreted sEVs. Addition of MCP-1 (10 ng/ml) increased both the cellular miR-939 and sEV miR-939 ()). CCL2 binds to its cognate receptor CCR2 present on the cell surface and exerts its function. RS 102895 hydrochloride is a small molecule antagonist of CCR2. It binds with high affinity to the β subunit of CCR2 and results in the inhibition of CCR2 signalling [Citation34]. Pretreatment with RS 102895 abrogated the increase of both cellular and sEV miR-939 observed after MCP-1 treatment ()). Based on this observation, we conclude that specific inflammatory stimulus induces the cellular expression of miR-939 and its concomitant packaging into sEVs.
Figure 3. MCP-1/CCL2 induces miR-939 upregulation and sEV packaging of miR-939. THP-1 cells were treated for 4 h with MCP-1 following a two-hour pretreatment with a CCR2 antagonist. RNA was isolated from the cells and corresponding sEVs secreted in the supernatant. Taqman-based qRT-PCR was used to determine the relative expression of miR-939 in cells (a) and sEVs (b) normalized to U6snRNA and miR-223, respectively. Significance was determined by one-way ANOVA with Tukey’s multiple comparisons test %% p < 0.01, *** p < 0.001, ****, ####, %%%% p < 0.0001.
Overexpression of miR-939 by transfection induces its packaging into sEVs
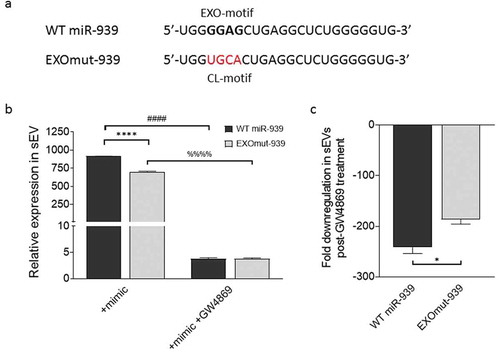
Short signature sequences called EXOmotifs are nucleotides within the 5ʹ end of miRNAs that can bind to RNA-binding proteins and thereby regulate their packaging into exosomes [Citation19]. Sequence analysis indicated that the 5ʹ end of miR-939 possesses an EXOmotif ()). Therefore, we hypothesized that overexpressing miR-939 in THP-1 cells could consequentially package miR-939 into the secreted sEVs. We transfected THP-1 cells with the miR-939 mimic to assess the packaging and secretion within sEVs after 24 h. Our results showed that transfection of miR-939 mimics into THP-1 cells induced the incorporation of miR-939 into the secreted sEVs ()).
Figure 4. Overexpression of miR-939 drives its packaging into sEVs via cellular machinery. miR-939 harbours the EXOmotif which aids in exosomal packaging of miRNAs (a). THP-1 cells were transfected with miR-939 mimics using lipofectamine and the sEVs were isolated from the supernatant after 24 h. Taqman-based qRT-PCR was used to determine the relative expression of sEV miR-939 normalized to miR-223 (b). Taqman analysis of recipient HUVEC cells incubated with THP-1 cell-derived sEVs overexpressing miR-939 for 24 h shows a decrease in miR-939 targets following uptake (c). Significance was determined by student t-test for miR-939 or two-way ANOVA with Tukey’s multiple comparisons test for the target mRNAs ** p < 0.01, ****/#### p < 0.0001.
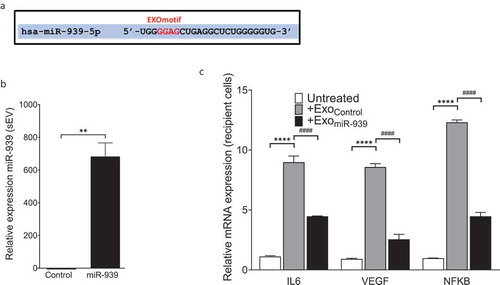
We further sought to address whether the sEVs overexpressing miR-939 are taken up by the recipient cells and if they could modulate the expression of the target mRNAs of miR-939. THP-1 cell-derived sEVs (from control or miR-939 transfection) were added to recipient HUVEC cells and incubated for 24 h, following which target gene expression was quantified in the recipient cells. The uptake of sEVs overexpressing miR-939 significantly reduced the mRNA expression of validated targets IL-6, VEGF and NFKB2 in HUVEC cells ()). This suggests that THP-1 derived sEVs are taken up by recipient HUVEC cells and can lead to a functional effect.
To further validate if the EXOmotif present on miR-939 is responsible for incorporating it into the sEVs, we mutated this particular sequence (GGAG) in the miR-939 mimic to a cytoplasm-localizing motif also known as CL motif ()). Lipofectamine-mediated transfection of THP-1 cells with either wild type (WT miR-939) or EXOmotif mutant (EXO-mut miR-939) mimics allowed their incorporation into the sEVs ()). Similarly, the addition of GW4869 abrogated the increase of both WT and EXOmut mir-939 mimics within sEVs. However, the extent of downregulation in the sEVs observed post-GW4869 treatment was greater for WT compared to the mutant mimic, thereby suggesting EXOmotif to be one of the possible cellular mechanisms responsible for packaging miR-939 within sEVs ()).
Figure 5. EXOmotifs determine the packaging of miR-939 into sEVs upon overexpression. (a). Presence of an EXOmotif towards the 5ʹ terminus of miR-939 could aid its exosomal recruitment via binding to RNA-binding proteins. This EXOmotif was mutated to a cytoplasmic localizing sequence (CL motif) in the EXOmut-939 mimic. (a). THP-1 cells were transfected with miR-939 or EXOmut-939 mimics using lipofectamine RNAiMAX and the sEVs isolated from the supernatant after 24 h. Taqman-based qRT-PCR was used to determine the relative expression of sEV mimics normalized to miR-223 (b). Significance was determined by two-way ANOVA with Sidak’s multiple comparison test ****, #### p < 0.0001. Fold downregulation of the miR-939 and EXOmut-939 mimics secreted from the transfected cells following inhibition of exosome secretion was also determined (c) and significance was determined by two-tailed unpaired Student t-test *p < 0.05.
Increasing the efficiency of sEV miR-939 packaging via electroporation
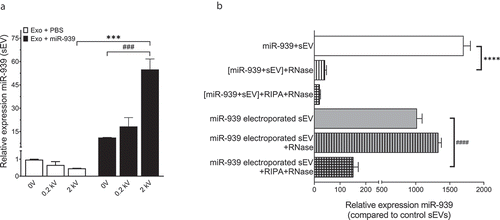
Although there was a reduction of proinflammatory gene expression in recipient cells following uptake of ExomiR-939 in comparison to Exomock, endogenous proinflammatory gene expression compared to untreated control was always higher upon the addition of sEVs derived from lipofectamine-treated cells ()). Hence, to avoid the detrimental inflammatory effect of transfection on sEV content, we sought to package naïve THP-1 cell-derived sEVs directly with miR-939 using several modifications to a siRNA packaging protocol reported for EVs [Citation8]. Ten micrograms of THP-1 cell-derived sEVs were resuspended in an electroporation buffer and electroporated with 30 picomoles of miR-939 mimic at different electric field strengths. Electroporation of naïve THP-1 cell-derived sEVs could efficiently package the miR-939 mimics into the sEVs ()). While mere incubation of the sEVs with miRNA mimic or electroporation at 200 V (used in eukaryotic cell electroporation) resulted in considerable packaging of miRNAs, the efficiency of packaging was significantly improved at a higher voltage (2 kV, used in prokaryotic cell transformation). Additionally, to assess whether miR-939 is packaged within the sEV or present on the surface of the sEVs upon electroporation, we performed an RNase protection assay of the vesicles. In brief, sEVs either passively incubated with miR-939 or electroporated at 2 kV were repurified by ultracentrifugation and treated with RNase in presence or absence of detergents. RNase treatment of sEVs passively incubated with miR-939 drastically reduced its relative expression indicating that miR-939 is almost entirely surface-bound on the sEV ()). In contrast, the miR-939 in the electroporated sEVs were resistant to RNAse activity, implying that it is protected within the vesicle. However, when the sEV membrane was disrupted by detergents and subjected to RNase, there was a considerable reduction in miR-939 expression.
Figure 6. Electroporation efficiently packages miRNA within THP-1 cell-derived sEVs. (a). Ten micrograms of THP-1 cell-derived sEVs were electroporated with 30 picomoles of miR-939 at different voltages and the amount of miR-939 incorporated within sEVs was estimated by Taqman-based qRT-PCR. (b). The sEVs subjected to passive incubation or 2 kV electroporation with miR-939 were subsequently treated with a combination of RNase A and detergents to evaluate the surface versus internal incorporation of miRNA. miR-939 within electroporated sEVs were resistant to RNase A digestion, while miR-939 incubated passively with sEVs were significantly degraded. Taqman-based qRT-PCR was used to determine the miR-939 content (normalized to miR-223) and significance was determined by one-way ANOVA with Sidak’s multiple comparison test ***, ### p < 0.001, ****, #### p < 0.0001.
Uptake of miR-939 overexpressing sEVs by recipient cells
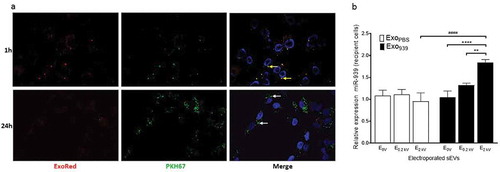
To determine if the sEVs obtained via electroporation were intact and capable of exerting a functional effect, it is essential to first confirm sEV uptake. Hence, miR-939 enriched sEVs double-labelled with Exo-red (RNA binding red fluorescent dye) and PKH67 (membrane labelling green fluorescent dye) were incubated with recipient HUVEC cells and cells were observed by confocal microscopy at different time points. RNA colocalizes with sEV membrane at 1 h (yellow speckles) indicating intact sEVs bound to the cell surface. However, at 24 h post-incubation absence of red punctae indicate the possible release of RNA molecules inside the donor cells upon fusion of sEVs. The sEV membrane was still visible at 24 h and appeared green further confirming the dissipation of sEV RNA inside recipient cells ()). We also confirmed that recipient THP-1 cells incubated with miR-939 overexpressing sEVs for 24 h showed an increased cellular miR-939 content when compared to control-electroporated sEVs by qPCR ()). Additionally, miR-939 levels in recipient cells increased significantly only with electroporated sEVs compared to non-electroporated sEVs (within the Exo939 group). This corroborates with the RNase protection experiment, wherein passive incubation of sEVs with miR-939 although show an increase in miR-939 levels, are susceptible to nucleases in the medium before they are taken up by recipient cells. In contrast, electroporation of the sEVs effectively packages the miRNA within the vesicle for functional cargo delivery.
Figure 7. Cellular uptake of miR-939 electroporated sEVs. sEVs electroporated with miR-939 were double-labelled with Exo-red (RNA binding red fluorescent dye) and PKH67 (membrane labelling green fluorescent dye). Labelled sEVs were incubated with recipient HUVEC cells and the nucleus was live-stained with Hoechst 33342. Cells were observed by CLSM under an Olympus FV1000 microscope (63 X magnification) after 1 h and 24 h of incubation with sEVs (b). RNA colocalizes with sEV membrane at 1 h (solid yellow arrows), however at 24 h post-incubation, much of the RNA has been delivered into the cell, and the sEV membrane is probably incorporated into the plasma membrane (solid white arrows). Taqman-based qRT-PCR was used to determine the cellular miR-939 content (normalized to U6snRNA) in recipient cells following uptake (c). Significance was determined by two-way ANOVA with tukey’s multiple comparisons test ** p < 0.01, ****, #### p < 0.0001.
Functional effects of sEV uptake in recipient cells
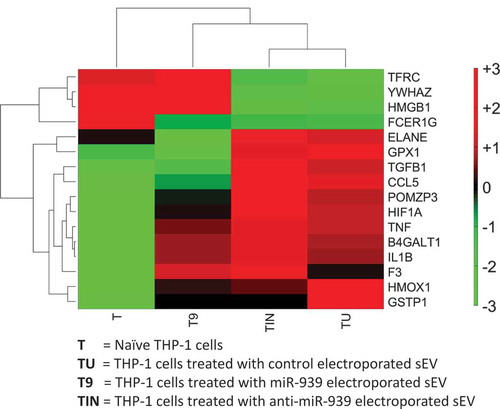
The functional effects of an sEV cargo can be determined by manipulating the source and hence its content. We sought to obtain sEVs derived from THP-1 cells overexpressing and conversely knocked out for miR-939. In humans, miR-939 is located on the long arm of chromosome 8, at the exon-intron junction of the cleavage and polyadenylation specificity factor subunit 1 (CPSF1). CPSF1 is part of the large protein complex essential for processing mRNA precursors. Hence, we did not pursue a gene knockout for miR-939 due to the anticipated perturbation of this essential gene and deleterious consequences on cell survival. As an alternative, we incubated recipient THP-1 cells with sEVs electroporated either with miR-939 or anti-miR-939 for 24 h and performed a global gene expression analysis using RNA-sequencing. We observed that several inflammatory genes are differentially expressed in THP-1 cells upon incubation with control sEVs as we had previously observed (See ), with most of these genes upregulated indicating a heightened inflammatory response (). Anti-miR-939 treated sEVs display a similar transcriptional profile as control sEVs. Incubation with miR-939 electroporated sEVs reverses, albeit not completely, the inflammatory state compared to control sEVs, bringing the transcriptional profile of inflammatory genes closer to naïve state. Complete data is shown in Supplementary Table 1. This supports a protective role of miR-939 in attenuating inflammation.
Figure 8. Clustering of inflammatory gene expression levels in THP-1 recipient cells incubated with different types of sEVs. A clustergram of the expression levels for differentially regulated inflammatory genes. Each column represents a particular type of sEV incubated with recipient THP-1 cells. T: no sEVs, TU: untreated sEVs, T9: miR-939 treated sEVs, TIN: anti-miR-939 treated sEVs. The heat map represents normalized gene expression values, with the colour indicating multiples of the standard deviation above or below the average value for that row in the heat map. Red, high; black, average; green, low.
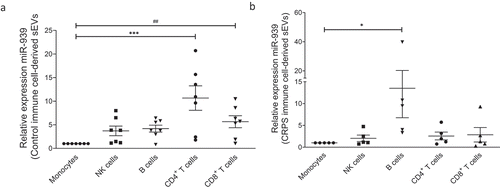
Ex vivo miR-939 expression in immune cell-derived sEVs
To determine if miR-939 alterations can be attributed to specific immune cells, and whether the packaging of miR-939 into sEVs differ in CRPS and control, we isolated CD4 + T cells, CD8 + T cells, monocytes, NK cells and B cells from CRPS patients and control donors. The cells were incubated in exosome-depleted media for 24 h, and the relative expression of miR-939 was determined in the corresponding sEVs that were secreted.
Monocyte-derived sEVs demonstrated a consistently low expression of miR-939 in both controls and CRPS patients and were hence used as a normalizer. The sEVs secreted by B cells, T cells and NK cells demonstrated a higher expression of miR-939 (fold change>2) compared to monocyte-derived sEVs in controls ()). In contrast, the ability of immune cells to secrete miR-939 within sEVs diminished in CRPS patients (fold change<2); only B cell-derived sEVs showed a significantly higher level of miR-939 among all the immune cell-derived sEVs ()).
Figure 9. Ex vivo expression of miR-939 in control and CRPS patient immune cell-derived sEVs. Freshly isolated PBMCs from healthy donors (n = 7) or CRPS patients (n = 6) were separated using magnetic bead-conjugated antibodies (MACS) to immune cell sub-populations, namely monocytes, NK cells, B cells, CD4 and CD8 positive T cell populations. The immune cells were incubated separately in exosome-depleted medium and the sEVs were isolated from the supernatant after 24 h. Taqman-based qRT-PCR was used to determine the relative expression of sEV miR-939 normalized to miR-223 in controls (a) and CRPS patients (b). Significance was determined by one-way ANOVA with Dunnett’s test *, # p < 0.05, *** p < 0.001.
Discussion
Exosomes while previously were deemed to be extracellular debris are now appreciated as vesicles aiding intercellular communication. Because of their pivotal roles in the initiation, propagation and regulation of a spectrum of inflammatory disorders, there is an increased interest in exploring the utility of EVs as therapeutic vehicles and as targets for the treatment and prevention of inflammatory diseases [Citation35]. A recent study showed that miRNA availability for exosome secretion is controlled, at least in part, by the cellular levels of their targeted transcripts [Citation22]. Thus, changes in the transcriptome in response to cellular activation may modulate miRNA sorting to the exosomes, and exosomal miRNA secretion is a mechanism to adjust miRNA:mRNA homeostasis. Here, we report the cellular overexpression and packaging of specific miRNA into exosomes under inflammatory stimuli.
In 2018, the International Society for Extracellular Vesicles (ISEV) has updated the previously proposed Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines for the field [Citation36]. It outlines the protocols and steps to follow to document specific EV-associated functional activities. We have adhered to MISEV guidelines and have performed extensive characterization of these vesicles with respect to quantitation, morphology, size and protein markers. The vesicles obtained by the isolation procedures in our study would be appropriately classified as small extracellular vesicles (sEVs) according to this classification.
Our studies showed that miR-939 is packaged into sEVs under inflammation and inhibiting exosome secretion leads to its retention within cells. This was specific for MCP-1 and not observed when donor cells were stimulated with LPS indicating that sEV composition is governed by the stimulus. Increase in miR-939 also resulted in the lowering of its target mRNA IL-6. We also observed that the sEV miR-939 was decreased upon LPS stimulation. This could be due to increased cellular IL-6 acting as a miRNA sponge and reducing the effective levels of miR-939 to be packaged. A recent study has shown that sorting of miRNAs into exosomes can be modulated by changing either cellular miRNA expression or their respective endogenous target mRNA levels in the donor cells [Citation22].
Proinflammatory gene expression was always higher in recipient cells following uptake of miR-939 enriched sEVs from lipofectamine-transfected donor cells. This was deemed to be a deleterious effect of lipofectamine-mediated transfection on sEV content, wherein other proinflammatory molecules may be packaged into sEVs in addition to miR-939. This prompted us to pursue electroporation of naïve sEVs instead. A recent study highlights the caution to be observed with transfection reagents, especially in RNA interference studies, where they are known to cause subtle changes in inflammatory gene expression [Citation37].
One of the proposed mechanisms for incorporation of specific miRNAs into the exosomes involves the EXOmotifs mostly located at the 3ʹ end of the miRNA, which is recognized by certain RNA-binding proteins that help in their recruitment [Citation19]. The presence of one such motif (GGAG) at the 5ʹ segment of miR-939 prompted us to postulate whether the cellular machinery could drive the packaging of miR-939 into sEVs under inflammatory stimuli. Mutations in this region towards a CL-motif (responsible for cytosolic compartmentalization) did not however completely abrogate their incorporation into sEVs, although it was noticeably reduced compared to the wild type miR-939. Hence, additional mechanisms may influence the sEV packaging of miR-939.
In order to efficiently package sEVs with miRNA, we used a high electric field strength, in the order of 2 kV to observe a discernible effect. This is due to the smaller surface area of the vesicle compared to a eukaryotic cell surface [Citation38]. Additionally, a high ionic strength and a high viscosity solvent like iodixanol was used in the electroporation buffer to attenuate aggregation and maximize colloidal stability, since the low zeta potentials of exosomal membranes (−30 mV) make them susceptible to aggregation, especially following electroporation [Citation39].
Exosome uptake studies were conducted using both HUVEC and THP-1 cells as recipient cells. HUVEC cells being adherent cells, were used primarily for confocal imaging to observe the uptake, while THP-1 cells were predominantly used to assess the functional effect of sEV uptake on inflammatory stimulus in recipient cells. Primarily, THP-1 being a monocytic cell line, was more suited to study the changes in gene expression of proinflammatory mRNAs. We also postulated optimal sEV uptake via endocytosis in an identical recipient cell type (since donor cells were THP-1) due to similar surface markers on sEVs and recipient cells. However, recent evidence in epithelial cell lines shows that recipient cell type is of a greater deterministic value than donor exosomes, which is more non-selective [Citation40]. Global RNA-sequencing studies of recipient cells upon uptake of sEVs overexpressing miR-939 reversed inflammation supporting a protective anti-inflammatory role for miR-939. Though the transcriptional changes moved closer to a naive state, this reversal was not complete which could be due to the fine tuner function of miRNAs in regulating gene expression.
We next investigated the packaging of miR-939 into sEVs using immune cells from CRPS patients and control donors. Our studies suggest differences in sEV miR-939 released by B cells in CRPS. Studies in well-characterized tibial fracture/cast model of CRPS in mice showed that depletion of CD20 + B cells resulted in attenuation of allodynia, postural unweighting, and vascular changes [Citation41]. The authors attributed the CRPS-like changes in the murine model to the antibodies produced by the B cells and suggested that therapies directed at reducing B-cell activity may be beneficial in treating CRPS patients [Citation41]. Additional studies are needed to verify if other miRNAs altered in CRPS stem from specific immune cells.
Future studies will focus further on investigating aberrations in exosomal uptake in patients of multiple inflammatory disorders. This will be pivotal in understanding if impaired exosome uptake and thus signalling is an underlying disease mechanism. Pharmacological interventions capable of selectively inducing the production of exosomes that are protective represent a novel strategy to be explored. Conversely, blocking the uptake of specific exosomes that may be pathogenic might be beneficial. Exosome biology will undoubtedly open novel avenues in understanding normal physiology, disease mechanisms, and therapeutic intervention strategies for several inflammatory disorders and pain.
Methods
Study approval
All subjects were enrolled after giving informed consent as approved by the Drexel University College of Medicine Institutional Review Board. All experimental protocols were approved by Drexel University College of Medicine Institutional Review Board. The methods were carried out in accordance with the approved guidelines.
Cell culture and maintenance
HEK293 cells (American Type Culture Collection [ATCC]) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. Human monocytic leukaemia THP-1 cells (TIB202, ATCC) were maintained in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin. Human umbilical vein endothelial cells (HUVECs) were maintained in endothelial growth medium kit (EGM2) from Lonza (Allendale, NJ).
Overexpression of miR-939 in THP-1 cells
Transfections were performed following the manufacturer’s protocol for RNAiMax transfection reagent (Life Technologies) using either miR-939-5p mimic (MC 12517), custom synthesized EXOmut-miR-939 mimic (Assay ID CTMFW3V, Thermofisher Scientific), or negative control mimic (4464058) (Life Technologies) with the following modifications. For each well of a 6-well plate, 7.5 µl RNAiMax reagent was diluted in 150 µl of serum-free media, and 30 pmol of miR-939 or control mimic was diluted in 150 µl of serum-free media individually. The dilutions were combined and incubated at room temperature for 15 min. This transfection complex (300 µl) was added to 0.5 × 106 cells/well in 1.7 ml serum containing media in 6-well plates and incubated for 6 h at 37°C, after which the media was replaced with complete RPMI and incubated for 24 h. Cells were collected by centrifugation at 135 × g for 5 min at 4°C. The cell pellet was washed with 1× phosphate-buffered saline and resuspended in RNA lysis buffer (mirVana kit; Life Technologies) containing 0.5 U/µl RNAsin Plus (Promega; Madison, WI).
Isolation of sEVs
THP-1 cells (60 million) or immune cells collected from CRPS patients and healthy controls were incubated for 24 h at 37°C in RPMI without FBS or containing 2% exosome-depleted FBS. Cells were collected by centrifugation at 135 × g for 5 min at 4°C and the supernatant was stored at −20°C. The sEVs were isolated by differential ultracentrifugation as mentioned previously. Briefly, cells were pelleted at 200 g for 5 min and supernatant was centrifuged at 2000 x g for 15 min to remove dead cells and at 12,000 × g for 45 min to remove cell debris. The supernatant was then filtered through a 0.2-micron filter and centrifuged at 120,000 × g to obtain an sEV pellet. The pellet was washed with 25 ml of 1X PBS (without calcium and magnesium ions, with 1 U/µl RNAse inhibitor). The pellet was resuspended in 100 µl of the same buffer and stored at −80°C until further use.
Characterization of sEVs
sEVs were characterized based on their morphology, vesicle concentration, size distribution and surface markers. EVs in PBS were analysed for size distribution and concentration by nanoparticle tracking analysis (NTA) using the NanoSight NS300 according to the manufacturer’s protocol (Malvern Instruments, MA, USA). Samples were diluted to ∼107–109 particles/ml and continuously injected with a syringe pump and three videos (30 s each) were captured for particle analysis. Nanoparticle tracking analysis was performed using NTA 3.2 software.
Ten microliters of PBS resuspended sEVs were coated on Ni-formvar grids (300 meshes) and incubated for 20 min at RT. The grids were washed on 50 µl drops of 0.1 M Sorensen’s phosphate buffer (pH 7.2) for 5 s each for a total of five times. The grids were blot dried perpendicularly on whatman #1 filter paper. Negative staining and embedding were performed by incubating the grids on 0.5% Uranyl acetate (in a 0.1% Methylcellulose) for 10 min at 4°C. The excess solution was blotted on a whatman paper, air-dried and imaged in a JEOL Transmission Electron Microscope (JEM 1230). Alternatively, the sEVs were crosslinked with 1% glutaraldehyde, coated on Cu-formvar grids (300 meshes) and immunolabelled for the exosome markers using rabbit anti-CD81 (SAB3500454, Sigma Aldrich, MO, USA) and goat anti-CD9 (ab92726, Abcam, MA, USA), and probed with 6 nm gold-conjugated goat anti-rabbit (#25104,Electron microscopy sciences, PA, USA) and 10 nm gold-conjugated donkey anti-goat secondary antibodies (#25805, Electron microscopy sciences, PA, USA), followed by negative staining and embedding.
The presence of exosome-specific protein markers and absence of non-EV markers on the isolated vesicles were determined by western blot. Briefly, protein concentration was estimated using a DC Protein assay (Bio-Rad Laboratories, CA, USA). Cellular extracts or 5 µg of sEVs were resolved on a reducing 12% SDS-PAGE, transferred to PVDF membrane and blocked in Odyssey Blocking buffer (927–50100, LI-COR Biosciences) for 2 h. The membrane was incubated in primary antibody cocktail (diluted in blocking buffer) overnight at 4°C, washed thrice in TBST, 10 min each, and incubated in secondary antibody cocktail for 45 min at RT, washed thrice in TBST for 10 min each, and imaged on an Odyssey Fc imaging system. The antibodies used were rabbit anti-CD81 antibody (HPA007234, Sigma Aldrich), 1:3000; mouse anti-Alix antibody (53540, Santa Cruz Biotechnology), 1:200; rabbit anti-GM130 antibody (12480, Cell Signaling Technology), 1:1000; rabbit anti-albumin antibody (4929S, Cell Signaling Technology), 1:1000; mouse anti-TSG101 antibody (sc-136111, Santa Cruz Biotechnology), 1:250; goat anti-rabbit 680RD IgG (925–68071, LI-COR Biosciences), 1:10000 and donkey anti-mouse 800CW IgG (926–32212, LI-COR Biosciences) 1:10000. We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190028) [Citation31].
cDNA synthesis and quantitation for mRNAs and miRNAs
RNA was isolated using the miRVana kit (Life technologies). The Maxima cDNA synthesis kit (Thermofisher Scientific) was used to generate cDNA and 2 µl cDNA was used for Taqman based quantitative real-time mRNA analysis containing 10 µl Taqman Fast Universal polymerase chain reaction (PCR) master mix (2×) no AmpErase UNG (Life Technologies), 1 µl Taqman primer probe (20×), in up to 20 µl nuclease-free water. GAPDH was used as the normalizer and one-way ANOVA was used to perform statistical analysis. Assay IDs were as follows: Hs01113624_g1 [TNFα], Hs00900055_m1 [VEGFA], Hs00985639_m1 [IL-6], Hs01028901_g1 [NFκB2] (Applied Biosystems, Carlsbad, CA). High capacity cDNA reverse transcription kit was used to generate cDNA for miRNAs. One microliter cDNA was used for Taqman based quantitative real-time mRNA analysis containing 5 µl Taqman Universal PCR master mix (2×) no AmpErase UNG (Life Technologies), 0.5 µl TaqMan microRNA assay (Assay ID 002182, Applied Biosystems), in up to 10 µl nuclease-free water. U6 snRNA (Assay ID 001973) for cellular or miR-223 (Assay ID 000526) for sEVs was used for normalization.
Inhibitor treatment of THP-1 cells
THP-1 cells were pretreated with DMSO, 5 µM RS102895 (MCP1 inhibitor) or 5 nM GW4869 (nSMase2 inhibitor) for 2 h followed by treatment with 50 nM MCP-1 or 100 ng/ml LPS for 4 h. The cells were centrifuged at 200 x g for 5 min at 4°C and the supernatant processed for isolation of sEVs and subsequent isolation of sEV RNA and compared to the cellular RNA isolated from the cell pellet.
Packaging of miR-939 in sEVs by electroporation
sEVs were isolated from 300 ml of THP-1 cell culture conditioned medium and packaged with miR-939 mimics with modifications to a siRNA packaging protocol for EVs [Citation8]. Briefly, 30 picomoles of miR-939 mimic was added to 200 ng of sEVs in 400 µl electroporation buffer (21% Optiprep, 25 mM KCl, 100 mM potassium phosphate, pH 7.2) and transferred to a 4 mm electroporation cuvette (Eppendorf, Hauppauge, NY). The sample was electroporated with three pulses at 0–2000 V in an E2510 electroporator (Eppendorf, Hauppauge, NY) and stored for 5 min at 4°C. Electroporated sEVs were diluted in 3 ml of 1X PBS and isolated by differential ultracentrifugation as mentioned earlier. Unincorporated miRNAs were removed by a PBS wash and the sEV pellet was resuspended in 30 µl of 1X PBS to be used for uptake studies or microscopy.
Evaluation of miR-packaging within intact sEVs
RNAse treatment was performed to determine if the miRNA was surface bound or packaged within sEVs. In brief, 10 µg sEVs were either passively incubated or electroporated at 2 kV with 30 picomoles of miR-939 mimic followed by ultracentrifugation to remove any unbound extravesicular RNA. The sEVs were resuspended in PBS and incubated with 20 µg/µl RNase (Purelink RNase A, Life technologies) for 20 min at 37°C. Additionally, miRNA cargo specificity was ascertained by detergent treatment to disrupt vesicle membrane integrity, using RIPA buffer for 20 min followed by RNase treatment as described above. Following incubation with RNase A, lysis buffer was added to inhibit the reaction and RNA was isolated. Taqman miRNA assay was performed to determine the relative expression of miR-939 in comparison to miR-223 used as a control.
RNA sequencing of recipient cells following sEV uptake
THP-1 cells (1 × 10^6 cells) were incubated with either control, miR-939 or anti-miR-939 electroporated sEVs for 24 h. Total cellular RNA from recipient THP-1 cells was purified using the miRVana total RNA isolation kit. RNA-seq sequencing libraries were prepared using a NuGEN Ovation RNA-Seq System (NuGEN Technologies). Libraries were multiplexed and sequenced on an Illumina HiSeq 2000 platform to obtain >20 million 50 bp paired-end reads per sample.
Low-quality RNAseq reads with more than 50% bases of quality score 15 were filtered out and the reads were mapped using HISAT [Citation42] and gene expression was quantified using RSEM [Citation43]. Differential expression was calculated using PossionDis [Citation44]. A false discovery rate (FDR) corrected p-value of 0.01 was used to determine significantly differentially expressed genes. Inflammatory response genes were identified using gene ontology (GO:0006954). Relative gene expression values (FPKM, Fragments Per Kilobase of transcript per Million mapped reads) and differential expression analysis results are available in the supplementary data.
Isolation of immune cell-derived sEVs
Blood (6–10 ml) withdrawn from the cubital vein of 10 healthy individuals and CRPS patients were used to isolate peripheral blood mononuclear cells (PBMCs) using Ficollpaque premium (GE healthcare) density gradient separation according to the manufacturer’s protocol. Immune cell populations (CD4 + T cells, CD8 + T cells, monocytes, NK cells and B cells) were separated using magnetic bead-based MACS® technology (Miltenyi Biotec) according to manufacturer’s protocol. An average of 2–3 million cells of each cell type was isolated by sequential separation of monocytes, B cells, NK cells, CD4 + T cells and CD8 + T cells. The fractions were analysed by flow cytometry (LSR Fortessa, BD Biosciences) to determine the purity of each cell type. The cells were individually incubated in exosome-depleted RPMI for 24 h and the supernatant was used to isolate the secreted sEVs.
Statistical analysis
Data are presented as mean ± the standard error of the mean from three or more independent experiments. Treatment effects were analysed with a one-way analysis of variance (ANOVA). Error probabilities with p < .05 were considered statistically significant.
Supplemental Material
Download Zip (769 KB)Disclosure statement
The authors report no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121(25):4977–14.
- Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33(2):186–192.
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233.
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518.
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659.
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593.
- Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44(1):11–15.
- El Andaloussi S, Lakhal S, Mäger I, et al. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65(3):391–397.
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920.
- El Andaloussi S, Mäger I, Breakefield XO, et al. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357.
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5(9). doi:10.1101/cshperspect.a016766.
- Kalra H, Gilbert TL, Overly CC. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450.
- Baglio SR, Rooijers K, Koppers-Lalic D, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6(1):127.
- Guduric-Fuchs J, O’Connor A, Camp B, et al. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13(1):357.
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282.
- Buermans HPJ, Buermans HPJ, Waasdorp M, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–9285.
- McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15(5):978–987.
- Teng Y, Feygenson M, Page K, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448.
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980.
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276.
- Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799–808.
- Squadrito ML, Baer C, Burdet F, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8(5):1432–1446.
- Janas T, Janas MM, Sapoń K, et al. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589(13):1391–1398.
- Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24.
- Birklein F, Schlereth T. Complex regional pain syndrome-significant progress in understanding. Pain. 2015;156(Suppl 1):S94–s103.
- Orlova IA, Alexander GM, Qureshi RA, et al. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9(1):195.
- McDonald MK, Ramanathan S, Touati A, et al. Regulation of proinflammatory genes by the circulating microRNA hsa-miR-939. Sci Rep. 2016;6:30976.
- McDonald MK, Tian Y, Qureshi RA, et al. Functional significance of macrophage-derived exosomes in inflammation and pain. PAIN®. 2014;155(8):1527–1539.
- Deshmane SL, Kremlev S, Amini S, et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326.
- Parkitny L, McAuley JH, Di Pietro F, et al. Inflammation in complex regional pain syndrome: a systematic review and meta-analysis. Neurology. 2013;80(1):106–117.
- Consortium E-T, Mestdagh P, Agostinis P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228–232.
- Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452.
- Alexander GM, Peterlin BL, Perreault MJ, et al. Changes in plasma cytokines and their soluble receptors in complex regional pain syndrome. J Pain. 2012;13(1):10–20.
- Mirzadegan T, Diehl F, Ebi B, et al. Identification of the binding site for a novel class of CCR2b chemokine receptor antagonists: binding to a common chemokine receptor motif within the helical bundle. J Biol Chem. 2000;275(33):25562–25571.
- Buzas EI, György B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10(6):356–364.
- Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750.
- Yang H-Y, Vonk LA, Licht R, et al. Cell type and transfection reagent-dependent effects on viability, cell content, cell cycle and inflammation of RNAi in human primary mesenchymal cells. Eur J Pharm Sci. 2014;53(p):35–44.
- Weaver JC. Electroporation: a general phenomenon for manipulating cells and tissues. J Cell Biochem. 1993;51(4):426–435.
- Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Anal Biochem. 2014;448:41–49.
- Horibe S, Rahimi K, Köbel M, et al. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018;18(1):47.
- Li WW, Guo T-Z, Shi X, et al. Autoimmunity contributes to nociceptive sensitization in a mouse model of complex regional pain syndrome. Pain. 2014;155(11):2377–2389.
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357.
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323.
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995.
