ABSTRACT
Extracellular vesicles (EVs) are membrane-enclosed particles that play an important role in cancer progression and have emerged as a promising source of circulating biomarkers. Protein S-acylation, frequently called palmitoylation, has been proposed as a post-translational mechanism that modulates the dynamics of EV biogenesis and protein cargo sorting. However, technical challenges have limited large-scale profiling of the whole palmitoyl-proteins of EVs. We successfully employed a novel approach that combines low-background acyl-biotinyl exchange (LB-ABE) with label-free proteomics to analyse the palmitoyl-proteome of large EVs (L-EVs) and small EVs (S-EVs) from prostate cancer cells. Here we report the first palmitoyl-protein signature of EVs, and demonstrate that L- and S-EVs harbour proteins associated with distinct biological processes and subcellular origin. We identified STEAP1, STEAP2, and ABCC4 as prostate cancer-specific palmitoyl-proteins abundant in both EV populations. Importantly, localization of the above proteins in EVs was reduced upon inhibition of palmitoylation in the producing cells. Our results suggest that this post-translational modification may play a role in the sorting of the EV-bound secretome and possibly enable selective detection of disease biomarkers.
Introduction
Protein S-acylation is a reversible post-translational modification (PTM) where long-chain fatty acids are covalently attached to cysteine residues via labile thioester bonds [Citation1]. Since the vast majority of S-acylated proteins are modified with palmitate [Citation2], S-acylation is frequently called palmitoylation, and hereafter referred to as palmitoylation. Palmitoylation increases protein hydrophobicity, transiently targeting cytosolic proteins to cell membranes. Among all lipid modifications, palmitoylation is the most pervasive, affecting about 20% of the proteome [Citation3,Citation4]. Additionally, palmitoylation tethers proteins into membrane microdomains such as lipid rafts, thus modulating protein activity, stability, and multiprotein complex formation [Citation5]. Palmitoylation is frequently altered in various diseases, including cancer [Citation6,Citation7]. However, the biological role of this PTM is complex and multifaceted and has not been fully investigated due to technical challenges associated with the low abundance of palmitoyl-proteins in general, the difficulty of enriching palmitoyl-proteins with high specificity, and the high hydrophobicity of intact palmitoyl-peptides.
Extracellular vesicles (EVs) are lipid-enclosed particles that play an important role in cancer progression and have emerged as a promising source of circulating biomarkers. Various classes of EVs that differ in size, cargo, biogenesis and function have been reported in the last decade [Citation8–Citation12]. However, markers that clearly define these classes across cells of diverse origin are currently lacking [Citation13]. To address this challenge the International Society for Extracellular Vesicles has proposed that EV should be classified into two major populations of vesicles based on their size: small EVs (S‐EVs) and large EVs (L‐EVs). S-EVs usually include vesicles in the 40–150 nm size range while L-EVs include vesicles in the 200 nm–10 μm size range [Citation13]. S-EVs are represented both by exosomes, which originate from endocytic machinery and by small ectosomes, which originate from membrane blebs pinching off the plasma membrane [Citation14,Citation15]. However, endosomal origin of S-EVs is difficult to prove and current isolation methods do not separate endosome‐derived S‐EVs from non‐endosomal S‐EVs [Citation15–Citation20]. In contrast, L-EVs frequently form at the plasma membrane by direct shedding of membrane blebs from the cells, and are represented by microvesicles, large oncosomes, apoptotic bodies and other types of vesicles, mostly in the micrometre size range [Citation21]. Several pathways involved in regulating the biogenesis of these two EV classes have been proposed [Citation14], but a lot is still unknown.
Due to the high membrane to cargo ratio in EVs, palmitoylation may play a role in targeting proteins to EVs. However, a whole palmitoyl-proteomics analysis of EVs has not been performed and palmitate residues are frequently lost during conventional proteomics thus hampering the study of this PTM. We employed a novel method that uses a metabolic labelling-independent, cysteine centric approach, namely low-background acyl-biotinyl exchange (LB-ABE) to identify specific palmitoyl-proteomic signatures in highly purified EVs isolated by density gradient [Citation8–Citation10,Citation22] by comparing the palmitoyl-proteome of L- and S-EVs versus their parental cells. We also investigated whether these signatures reflect a specific subcellular origin and/or biological function and sought to identify prostate cancer specific palmitoyl-proteins that are highly abundant in EVs.
Methods
Cell culture
The PC3 cell line was obtained from the American Type Culture Collection (ATCC). The DU145DIAPH3-KD cell line, stably transfected with DIAPH3 shRNA, was generated in our laboratories [Citation23]. PC3 and DU145DIAPH3-KD cell lines were cultured in DMEM (Invitrogen). All cells were supplemented with 10% foetal bovine serum (Denville Scientific), 2 mM L-glutamine (Invitrogen) and 1% PenStrep (Invitrogen). DU145DIAPH3-KD cells were additionally selected with 2 µg/mL puromycin as described [Citation23]. All cells were grown at 37°C and 5% CO2. Cell viability of the EV-producer cells was tested with the 0.4% Trypan Blue (Sigma) exclusion method. All cell lines were routinely tested for mycoplasma contamination by using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza). Finally, in order to collect EVs from cells in which palmitoylation was inhibited, PC3 cells were treated for 24 hours with 2-bromohexadecanoic acid (Sigma), also known as 2-bromopalmitate (0.5, 1, 5, 10, 20, 50 and 100 µM), in serum-starvation.
Isolation of extracellular vesicles (EVs)
The isolation of EVs was conducted as previously described with minor modifications [Citation9,Citation10,Citation22]. Cells were grown on 18 × 150 cm2-cell culture plates (Corning) until 90% confluence, washed in PBS and serum-starved for 24 hours before the collection of conditioned cell media. The conditioned media was cleared by differential centrifugation of floating cells at 300 g, of cell debris at 2,800 g for 10 min, and spun in an ultracentrifuge at 10,000 g for 30 min (4°C, k-factor 2547.2) for the collection of L-EVs. The supernatant was then spun at 100,000 g for 60 min (4°C, k-factor 254.7) for the collection of S-EVs. Both 10,000 g and 100,000 g pellets were then subjected to Optiprep™ (Sigma) density gradient purification. Fresh pelleted EVs were resuspended in 0.2 µm-filtered PBS and deposited at the bottom of an ultracentrifuge tube. Next, 30% (4.3 mL, 1.20 g/mL), 25% (3 mL, 1.15 g/mL), 15% (2.5 mL, 1.10 g/mL), and 5% (6 mL, 1.08 g/mL) iodixanol solutions were sequentially layered at decreasing density to form a discontinuous gradient. Separation was performed by ultracentrifugation at 100,000 g for 3 h 50 min (4°C, k-factor 254.7) and EV-enriched fractions collected either at 1.10–1.15 g/mL for L-EVs or 1.10 g/mL for S-EVs [Citation10]. Purified EVs were then washed in PBS (100,000 g, 60 min, 4°C) and resuspended in the appropriate buffer. All ultracentrifugation spins were performed in a SW28 swinging rotor (Beckman Coulter). We have submitted all relevant data from our experiments to the EV-TRACK knowledgebase [Citation24] (EV-TRACK ID: EV190069).
Whole cell and membrane protein lysates from EV-producer cells
Whole cell lysate (WCL) and membrane preparations (M) were obtained upon 24-hour serum starvation and collection of conditioned cell media. Cell monolayers were scraped and washed in chilled PBS (×3). For WCL, cells were directly lysed in DTT-free 4% SDS/Tris-HCl lysis buffer [Citation25]. For M preparations, cells were gently scraped, washed in PBS (×3) and resuspended in filtered PBS containing 1% protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail, Roche). Cell suspensions were immediately subjected to 20 cycles of sonication (5 sec) in ice to induce cell disruption. Membrane suspensions were then cleared of intact cells at 500 g, pelleted at 16,000 g (20 min, 4°C), washed in PBS (×3) and resuspended in 4% SDS/Tris-HCl lysis buffer. All protein lysates were stored at −80°C until use.
LB-ABE enrichment of palmitoyl-proteins
Protein concentration was determined with the Pierce 660 nm protein assay (Pierce). 300 µg of WCL, M and L-EVs (×3); and 250 µg of S-EVs (×2) were subjected to LB-ABE coupled to label-free mass spectrometry as described [Citation26]. Briefly, proteins were reduced with 50 mM tris(2-carboxyethyl)phosphine (TCEP), sequentially alkylated with 50 mM N-ethylmaleimide (NEM) and 25 mM 2,2ʹ-dithiodipyridine (DTDP), and biotinylated with 1 mM biotin-HPDP in the presence or absence of 2 M neutral hydroxylamine (Hyd). Palmitoyl-proteins were enriched by streptavidin affinity purification, eluted by 50 mM TCEP, and precipitated by methanol/chloroform.
In order to evaluate the specificity of the LB-ABE method, 1 mg of WCL and M protein lysates were subjected to LB-ABE. 2 µg of the recovered palmitoylated fractions were resolved on a SDS-acrylamide gel alongside with 2 µg of the whole protein and non-palmitoylated fractions. Finally, silver staining of the gel was performed with the Silver Stain Kit (Pierce) following manufacturer’s recommendations. Alternatively, for the validation of select palmitoyl-proteins in WCL, M, L-EVs and S-EVs, 300 µg of total protein per group were processed as above with minor modifications. Briefly, samples were split into two equal halves in order to constitute the experimental (Hyd+) and negative control (Hyd-) groups prior to LB-ABE chemistry [Citation26]. Experimental groups were subjected to 2 M Hyd treatment as above, whereas control groups were incubated with Tris/HCl buffer in order to evaluate the unspecific recovery of non-palmitoylated proteins. Upon recovery of palmitoylated proteins, experimental and control samples were re-dissolved in loading buffer and 10% (v/v) of total recovered proteins was loaded onto SDS-PAGE gels for immunoblotting analysis.
LC-MS/MS analysis and data processing
Enriched palmitoyl-proteins were digested with MS-grade trypsin (Promega) by filter-aided sample preparation (FASP) as described previously [Citation26,Citation27]. Tryptic peptides were then recovered, dried down in a SpeedVac concentrator (Thermo Scientific), and re-dissolved in 0.2% formic acid (Sigma) up to a concentration of 0.15 µg/mL. Label-free proteomic analysis was performed using an EASY-nLC 1000 connected to an LTQ Orbitrap Elite hybrid mass spectrometer essentially as we previously described [Citation28,Citation29]. Briefly, 7 μL of peptide solution was loaded onto a 2-cm trap column (75 μm × 2 cm, C18) and separated on a 50-cm EASY-Spray analytical column (PepMap RSLC C18, 2 μm, 100 Å, 50 μm × 15 cm) heated to 55°C, using a 2 h-gradient consisting of 2–40% B in 150 min, 40–100% B in 20 min, and 100% B in 10 min at the flow rate of 150 nL/min. Separated peptides were ionized with an EASY-Spray ion source. Mass spectra were acquired in a data-dependent manner, with automatic switching between MS and MS/MS scans. In MS scans, the lock mass at m/z 445.120025 was applied to provide real-time internal mass calibration. The full MS scan (400–1600 m/z) was performed in 240,000 resolution at m/z of 400 Th, with an ion packet setting of 1 × 106 for automatic gain control and a maximum injection time of 500 ms. Up to 20 most intense peptide ions with charge state of ≥2 were automatically selected for MS/MS fragmentation by rapid collision-induced dissociation (rCID), using 7,500 resolution, 1 × 104 automatic gain control, 50 ms maximum injection time, 10 ms activation time, and 35% normalized collision energy. To minimize redundant spectral acquisition, dynamic exclusion was enabled with a repeat count of 1, an exclusion during of 30 s, and a repeat duration of 60 s.
The acquired MS data were searched against the Uniprot_Human database (released on 01/22/2016, containing 20,985 protein sequences) using the Andromeda [Citation30] algorithm in theMaxQuant (v1.5.5.1) [Citation31] environment. The searching parameters were set as follows: trypsin/P as the protease; oxidation (M), acetyl (protein N-term), NEM (C) and carbamidomethyl (C) as variable modifications; up to two missed cleavages; minimal peptide length as 7; mass tolerance for MS1 was 4.5 ppm for main search and for MS2 was 0.5 Da; identification of second peptides enabled; label free quantification (LFQ) enabled, and match-between-runs within 2 min were enabled. A stringent false discovery rate (FDR) <0.01 was used to filter PSM, peptide, and protein identifications.
Identification of high-confidence and high-abundance proteins
To retain high-confidence proteins from the raw LFQ data, we applied criteria that proteins should be detected with at least 2 peptides and in at least 2 replicates, and low abundant proteins less than 5% of absolute protein abundance for each identification were discarded for subsequent analyses. Absolute protein abundance for each identification was defined as the median value of replicates. We assumed that the total protein amount and composition of protein species are different between WCL, M, L-EVs and S-EVs. To compare the protein abundance between them, a max-min normalization method was used to scale the protein expression values between 0 to 1. Protein expression value 1 was given to the most abundant protein within each sample and protein value 0 was given to the least abundant protein identified. To determine the cut-off value of the protein expression difference, we computed the protein expression differences of randomly permuted samples, fitted a Gaussian distribution to the random protein expression difference, and then calculated the 99th percentile corresponding to α = 0.01 (protein expression difference cut-off = 0.1). Finally, in order to detect significantly abundant proteins within each sample, the rank product algorithm was applied to the normalized protein abundance for each identification [Citation32]. In order to identify high-abundance proteins in EVs, WCL or M, we performed integrated hypothesis test. Briefly, Student’s t-test and log2-median ratio test were performed. We estimated empirical null distributions of T values and log2-median ratio value by randomly permuting all samples 1,000 times and calculating t-test and log2-median ratio test p values. We then integrated these two p value into an overall p value using Stouffer’s method [Citation33]. FDR was corrected by Storey’s method [Citation34]. We then selected proteins with a FDR <0.05 and normalized expression difference ≥0.1.
Functional annotation of the palmitoyl-proteome
The percentage of putative high-confidence (identified by two independent methods in palmitoyl-proteomes or experimentally validated) human palmitoyl-proteins in cells was directly retrieved from the SwissPalm database (v2) (www.swisspalm.org) [Citation4]. For EVs, the percentage of putative palmitoyl-proteins was estimated by direct comparison to the number of human proteins retrieved from the ExoCarta database (www.ExoCarta.org) [Citation35]. To assess the biological relevance of the putative palmitoyl-proteins in the proteome of prostate cancer EVs, we selected for a subset of proteins uniquely found or differentially expressed (FDR<0.05, Fold change ≥ ±1.5) in the proteome of L-EVs and S-EVs from a previously published study [Citation10]. Characterization of the highly and differentially expressed proteins was performed by the Ingenuity Pathway Analysis (IPA, QIAGEN) [Citation36] and DAVID [Citation37] tools. Differentially expressed proteins among groups were selected based on an averaged relative expression difference >0.1 and a p value<0.05 by one-sample t-test. Transcriptional information was recovered from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) [Citation38] databases.
Immunoblotting analysis
Immunoblotting analysis was performed as described [Citation26]. Primary antibodies used were HSPA5 (#3177, 1:1,000 dilution), GAPDH (#3683, 1:10,000 dilution), H3.1 (#9717, 1:10,000 dilution) and pan-SRC (#2108, 1:10,000 dilution) from Cell Signaling. Antibodies for TSG101 (sc-7964, 1:1,000 dilution), CD9 (sc-13118, 1:1,000 dilution), STEAP1 (sc-271872, 1:1,000 dilution), and Cav-1 (sc-894, 1:10,000 dilution) were obtained from Santa Cruz. Antibodies to KRT18 (ab93741, 1:10,000 dilution) and CD81 (ab79559, 1:10,000 dilution) were obtained from Abcam and ABCC4 (#GTX15602, 1:10,000 dilution) from GenTex. Antibody to STEAP2 (#PA5-25495, 1:1,000 dilution) was obtained from Thermo Scientific. Densitometric quantification of films was performed with the ImageJ software (v1.52a).
Tunable Resistive Pulse Sensing (TRPS) measurements
Concentration and particle size distribution of EVs were carried out in a qNano device (iZON Science, New Zealand). Freshly isolated EVs were diluted 1:40 in 0.2-µm filtered PBS and analysed either with a NP2,000-nm nanopore (resolution window 0.9–5.7 µm) for L-EVs or in a NP250-nm nanopore for S-EVs (resolution window 110–630 nm). Membranes were stretched at 47 mm and voltage set either at 0.04 V for L-EVs or 0.5 V for S-EVs in order to achieve a stable current baseline of about 120 nA. Particle size and concentrations were calibrated using Izon calibration particles (1:100 diluted TPK200 for S-EVs, and 1:1,000 diluted CPC2000 for L-EVs) and a minimum of 500 events were registered for each sample with a positive pressure of 5 mbar. Particle quantitation was performed in EVs obtained from ∼3.0×108 cells and resuspended in 200 µL of filtered PBS.
Flow cytometry analysis of prostate cancer cells and L-EVs
L-EVs were isolated as described above. Cells were fixed in 75% EtOH for 30 min at 4°C. L-EVs were fixed in 4% PFA for 10 min at room temperature. Cells and L-EVs were incubated for 30 min or 1 h, respectively, with one of the following primary antibodies: STEAP1 (H00026872-D01P, Abnova) at 1:50 dilution, STEAP2 (PA5-25495, Invitrogen) at 1:10 dilution, or ABCC4 (GTX15602, GeneTex) at 1:20 dilution. Excess antibody was washed off with PBS followed by incubation for 30 min with phycoerythrin-conjugated (111–116-144, Jackson ImmunoResearch; for STEAP1 and STEAP2) or FITC-conjugated (31629, ThermoFisher Scientific; for ABCC4) secondary antibody at 1:250 dilution. Both cell and L-EV samples were analysed using the LSR-II flow cytometer (Becton Dickinson) with settings optimized for the detection of cells or particles larger than 1 µm, respectively. Data were analysed using FlowJo software (Treestar).
Statistical analysis
Plots represent the mean and standard deviation of at least three independent replicates. Experimental groups were compared using Student’s t-test (unpaired, two-tails) and statistical significance established for a p value < 0.05.
Results
Low-background acyl-biotinyl exchange (LB-ABE) enables highly selective isolation of palmitoyl-proteins from whole cell lysates and membrane preparations
Because palmitoyl-proteins are anchored to cellular membranes [Citation1] and EVs are enriched in membrane components, we hypothesized that palmitoyl-proteins would be enriched in EVs. Using an in silico approach, we intersected the compendium of palmitoylated proteins SwissPalm [Citation39], with the ExoCarta database [Citation35] that contain proteins identified in EVs. As expected, we found a three-fold higher percentage of putative palmitoylated proteins in EVs versus cells (13.2% vs. 4.3%) (Suppl. Figure 1A). Further in silico analysis of the whole proteomes of prostate cancer cell-derived L- and S-EVs [Citation10], obtained by gradient centrifugation, showed that ~20% of the proteins identified in both EV fractions are putative palmitoylated proteins (Suppl. Figure 1B). Additionally, when we examined the proteins that are differentially expressed between the two EV fractions, this percentage increased to 43% in L-EVs and 32% in S-EVs (Suppl. Figure 1B). These results suggest that palmitoylated proteins are enriched in EVs in comparison to cells and might be differentially distributed in different EV populations.
Quantitative analysis of protein palmitoylation has been impaired by the lack of methodological approaches allowing efficient separation of palmitoylated proteins from non-palmitoylated proteins. We recently developed an improved acyl-biotinyl exchange (ABE) method, termed low-background ABE (LB-ABE) [Citation26], which consists of the blockage of non-palmitoylated cysteine residues by N-ethylmaleimide (NEM) and further by 2,2ʹ-dithiodipyridine (DTDP), followed by converting palmitoylated cysteine residues into biotinylated cysteines and specific purification of palmitoyl-proteins by streptavidin ()). This approach largely eliminates the co-isolation of non-palmitoylated proteins, thus enabling a comprehensive and specific palmitoyl-proteomic analysis [Citation26].
Figure 1. Selective enrichment of putative palmitoylated proteins in the proteome of EVs. (a) Schematic representation of the low-background acyl-biotinyl exchange (LB-ABE) method employed for selective enrichment of palmitoylated proteins. Free cysteines are sequentially blocked by NEM and DTDP incubations. Acyl-biotinyl exchange (ABE) allows specific labelling of palmitoyl-proteins and purification of the whole palmitoyl-proteome by biotin-streptavidin interaction. Non-palmitoylated proteins can be recovered in the flow-through upon specific capture of palmitoyl-proteins with streptavidin-functionalized beads. (b) Silver-stained PAGE gel of PC3 WCL and M protein lysates with versus without LB-ABE enrichment of palmitoyl-proteins. (c) Immunoblotting of the indicated proteins enriched or excluded from the palmitoyl-proteome of WCL and M in PC3 upon LB-ABE.
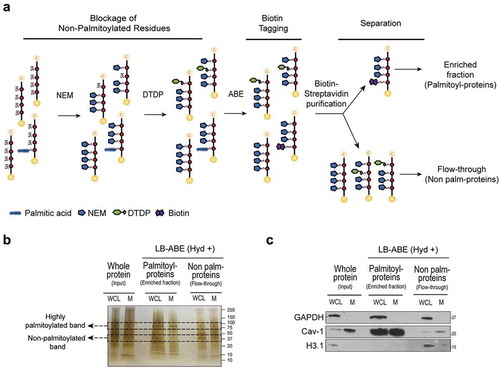
We tested the LB-ABE method in membrane preparations (M) from PC3 prostate cancer cells before applying it to EVs because M preparations are easier to generate and give a higher protein yield. Silver staining of M processed through LB-ABE enrichment showed distinct protein patterns in the fractions enriched with or depleted of palmitoyl-proteins ()). This enrichment was further validated by immunoblotting for proteins known to exhibit variable palmitoylation status. Caveolin-1 (Cav-1) was highly enriched in the palmitoyl-protein fraction (lanes 3–4, )) and almost completely absent from the palmitoyl-depleted fraction (lanes 5–6, )). This remarkable enrichment suggests that Cav-1 is predominantly present as a palmitoylated protein, in agreement with a recent report [Citation26]. In contrast, the nuclear histone H3 (H3.1), which is predicted to be a non-palmitoylated protein, was excluded from the palmitoyl-protein enriched fractions ()). GAPDH was detected in similar proportions in its palmitoylated and non-palmitoylated form. The localization of Cav-1 in M, and H3.1 and GAPDH in whole cell lysate (WCL) (lanes 1–2, )) confirmed the quality of the subcellular fractionation. The successful enrichment of palmitoylated proteins by LB-ABE in WCL and M encouraged us to apply this approach to studying large scale protein palmitoylation in EVs.
Comprehensive palmitoyl-proteomic analysis identifies differentially abundant palmitoyl-proteins in WCL, M, and EVs
In order to identify EV-specific palmitoyl-protein signatures, we employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to analyse candidate palmitoylated proteins isolated by LB-ABE in L-EVs and S-EVs, and performed a comparative analysis with the palmitoyl-proteins identified in WCL and M from PC3 cells. L-EVs and S-EVs were isolated from cell culture media by differential ultracentrifugation followed by density gradient purification (Suppl. Figure 1C), in line with the most recent MISEV2018 guidelines [Citation13] and previous studies that separated large oncosomes from exosomes [Citation9,Citation10,Citation22,Citation40]. Over 99% of the cells from which EVs were isolated were viable (Suppl. Figure 1D), limiting the possibility of contamination from apoptotic vesicles.
Tunable Resistive Pulse Sensing (TRPS) identified 1.76 × 109 particles/mL of 1.5–5 µm diameter, with a modal size of 1.77 µm in L-EV samples ()), and 2.43 × 1011 particles/mL of 90–600 nm diameter, with a modal size of 131 nm, in S-EV samples, in line with previous reports [Citation9]. Despite the higher number of EVs in the S-EV fraction, L-EVs contained significantly more protein than S-EVs derived from the same number of producing cells ()), in agreement with our previous observations [Citation10]. Finally, immunoblotting of proteins enriched in L- and S-EVs confirmed the nature of the EV preparations. L-EVs showed the enrichment of HSPA5 and KRT18 at 1.10–1.15 g/mL, which are enriched in large oncosomes [Citation9,Citation10,Citation22,Citation41], while S-EVs showed the enrichment of CD81 and TSG101 at 1.10 g/mL, that are typically enriched in exosomes ()) [Citation8–Citation10,Citation13,Citation22,Citation41].
Figure 2. Large-scale MS analysis identifies the palmitoyl-protein signature of prostate cancer cells and EVs. (a) Quantification and particle size distribution of PC3EVs by TRPS. Histogram plots depicted with a bin width of 100 and 10 nm, respectively. (b) Yield of purified PC3 EV-protein after differential ultracentrifugation and density-gradient purification of conditioned media. (c) Immunoblotting of select proteins enriched either in L-EVs (HSPA5 and KRT18) or S-EVs (TSG101 and CD81). (d) Venn diagram showing the number of palmitoylated proteins identified in PC3 cells by LB-ABE in comparison to LNCaP cells [Citation26] and the number of known human palmitoyl-proteins compiled in the SwissPalm database [Citation4]. (e) Venn diagram showing the number of unique and common palmitoylated proteins in WCL, M, L-EVs and S-EVs. (f) Unsupervised heat map and dendrogram of the normalized relative abundance of the common palmitoyl-proteins. Clustering analysis identifies a group of palmitoylated proteins highly abundant across all subcellular compartments and a group of proteins with varied abundance. (g) Left, dot plot shows the relative abundance of the indicated palmitoyl-proteins in WCL, M, L-EVs and S-EVs by MS analysis and max(1)-min(0) normalization. Middle, immunoblotting against the indicated palmitoyl-proteins from WCL, M, L-EVs and S-EVs in presence (Hyd+) or absence of hydroxylamine (Hyd-) confirms specific enrichment of palmitoyl-proteins by LB-ABE. Right, immunoblotting of WCL, M, L-EV and S-EV lysates taken prior the enrichment of palmitoylated proteins in order to confirm the expression and distribution of the indicated proteins.
![Figure 2. Large-scale MS analysis identifies the palmitoyl-protein signature of prostate cancer cells and EVs. (a) Quantification and particle size distribution of PC3EVs by TRPS. Histogram plots depicted with a bin width of 100 and 10 nm, respectively. (b) Yield of purified PC3 EV-protein after differential ultracentrifugation and density-gradient purification of conditioned media. (c) Immunoblotting of select proteins enriched either in L-EVs (HSPA5 and KRT18) or S-EVs (TSG101 and CD81). (d) Venn diagram showing the number of palmitoylated proteins identified in PC3 cells by LB-ABE in comparison to LNCaP cells [Citation26] and the number of known human palmitoyl-proteins compiled in the SwissPalm database [Citation4]. (e) Venn diagram showing the number of unique and common palmitoylated proteins in WCL, M, L-EVs and S-EVs. (f) Unsupervised heat map and dendrogram of the normalized relative abundance of the common palmitoyl-proteins. Clustering analysis identifies a group of palmitoylated proteins highly abundant across all subcellular compartments and a group of proteins with varied abundance. (g) Left, dot plot shows the relative abundance of the indicated palmitoyl-proteins in WCL, M, L-EVs and S-EVs by MS analysis and max(1)-min(0) normalization. Middle, immunoblotting against the indicated palmitoyl-proteins from WCL, M, L-EVs and S-EVs in presence (Hyd+) or absence of hydroxylamine (Hyd-) confirms specific enrichment of palmitoyl-proteins by LB-ABE. Right, immunoblotting of WCL, M, L-EV and S-EV lysates taken prior the enrichment of palmitoylated proteins in order to confirm the expression and distribution of the indicated proteins.](/cms/asset/01057105-6883-402b-97b9-96dba6604f62/zjev_a_1764192_f0002_oc.jpg)
Next we compared the palmitoyl-proteins identified in WCL, M, L-EVs and S-EVs. A total of 2,408 palmitoyl-proteins were identified with a false discovery rate (FDR) of <0.01. We then selected 2,133 high-confidence proteins detected in two or more replicates and no less than 5% of absolute protein abundance (Suppl. Table 1). Of these, 1,803 proteins have been reported as palmitoyl-proteins in the SwissPalm database [Citation4] and/or were identified in our recent deep palmitoyl-proteomic profiling of prostate cancer cells [Citation26] ()). Of note, we also identified 330 proteins that have not yet been reported as palmitoyl-proteins. Interestingly, 41% of them were identified in EVs. These results confirm the enrichment of palmitoyl-proteins in cell and EV preparations with the LB-ABE approach.
In order to perform quantitative analysis, we selected for high confidence palmitoyl-proteins resulting in a total of 1,875, 1,843, 1,355 and 603 palmitoyl-proteins in WCL, M, L-EVs and S-EVs, respectively () and Suppl. Table 1). Our stringent quality criteria resulted in a smaller number of proteins identified in S-EVs due to lower initial protein input. However, similar correlation coefficients among the technical replicates for S-EVs and L-EVs suggest a similar reproducibility for the enrichment of palmitoyl-proteins in both EV populations (Suppl. Figure 1E). Clustering of the palmitoyl-proteins detected in all 4 fractions (WCL, M, L-EVs and S-EVs) ()) identified 141 proteins that were consistently highly abundant across all fractions and 388 proteins with varied abundance ()). Among the proteins with varied abundance, 31 were more abundant in L-EVs and 98 in S-EVs (Suppl. Table 1), suggesting that S-EVs harbour a more discrete palmitoyl-protein signature than L-EVs. Functional enrichment analysis of the 141 highly abundant proteins identified in all four fractions including EVs showed significant association with major biological processes relevant to cancer (e.g., adhesion and cellular movement) (Suppl. Figure 2A). Importantly, vesicle-mediated transport was also identified as one of the most prominent functions of the highy abundant palmitoylated proteins identified (Suppl. Figure 2A). These data suggest that palmitoylation is functionally significant in the biology of cancer and EVs.
Western blotting confirmed expression of palmitoyl-proteins in the fractions enriched for palmitoylated proteins (Hyd+) but not in the control groups (Hyd-), confirming specific recovery following LB-ABE ()). Src-family tyrosine kinases and Cav-1, which are known to be highly palmitoylated [Citation42–Citation44], were identified with high confidence in all fractions ()). HSPA5 was enriched in L-EVs as total protein [Citation9,Citation10,Citation41] but we found it to be equally represented in L- and S-EVs as a palmitoylated protein. CD9 is enriched in S-EVs both as a total and as palmitoyl-protein. We also noticed that the palmitoylated form of CD81 was significantly enriched in L-EVs (Suppl. Table 1), despite its “canonical” description as an S-EV marker [Citation8–Citation10,Citation41], based on total protein analysis. This result suggests that the palmitoyl status of certain proteins can determine their sorting to different EV fractions.
L- and S-EVs exhibit distinct profiles that distinguish them from their parental cells
We and others have previously demonstrated that EVs contain not only proteins that are highly expressed in the originating cells, but they are also enriched in proteins of low abundance in the cell [Citation45]. Moreover, discrete EV populations contain a set of distinct proteins, suggesting cargo selection [Citation8,Citation10]. In order to determine whether this cargo selection is evident in the palmitoyl-proteome, we compared L- and S-EVs to the originating cells. Because palmitoylation is a membrane-anchoring modification and EVs are membrane encapsulated particles, we compared the palmitoyl-proteins identified in EVs and M fractions. The majority (roughly 90%) of them was identified in both compartments (Suppl. Figure 2B), in line with the rich membrane composition of EVs. However, when we looked at the relative abundance of these proteins in EVs versus M, we found a weak correlation (r = 0.444 in L-EVs/M; r = 0.317 in S-EVs/M). Conversely, the correlation between M and WCL was higher (r = 0.782 in WCL/M) ()), suggesting selective enrichment of palmitoylated proteins in EVs.
Figure 3. L- and S-EVs exhibit distinct palmitoyl-protein profiles that distinguish them from their parental cells. (a) Regression analysis of the relative abundance of the palmitoyl-proteins identified in WCL, M, L-EVs and S-EVs. Spearman’s coefficient (r) demonstrates a low correlation between EVs and M when compared to WCL and M. (b) The biological functions overrepresented in L- and S-EVs in comparison to M identified by functional enrichment analysis of the palmitoyl-proteome differentially expressed in EVs using DAVID software [Citation37]. (c) Volcano plots showing differential protein expression between L- and S-EVs compared to M. X and Y axes represent the normalized expression difference and -log10(FDR), respectively. Red and blue dots correspond to palmitoyl-proteins significantly enriched either in L- or S-EVs, respectively, compared to those enriched in the M (green dots). The blue and red boxes highlight functionally relevant proteins to the biological processes differentially represented in L-EVs and S-EVs shown in panel B. The green boxes highlight the top 5 proteins enriched in M.
![Figure 3. L- and S-EVs exhibit distinct palmitoyl-protein profiles that distinguish them from their parental cells. (a) Regression analysis of the relative abundance of the palmitoyl-proteins identified in WCL, M, L-EVs and S-EVs. Spearman’s coefficient (r) demonstrates a low correlation between EVs and M when compared to WCL and M. (b) The biological functions overrepresented in L- and S-EVs in comparison to M identified by functional enrichment analysis of the palmitoyl-proteome differentially expressed in EVs using DAVID software [Citation37]. (c) Volcano plots showing differential protein expression between L- and S-EVs compared to M. X and Y axes represent the normalized expression difference and -log10(FDR), respectively. Red and blue dots correspond to palmitoyl-proteins significantly enriched either in L- or S-EVs, respectively, compared to those enriched in the M (green dots). The blue and red boxes highlight functionally relevant proteins to the biological processes differentially represented in L-EVs and S-EVs shown in panel B. The green boxes highlight the top 5 proteins enriched in M.](/cms/asset/33a1abd0-c756-47de-ac51-dabe4e8353f1/zjev_a_1764192_f0003_oc.jpg)
Functional enrichment analysis revealed that the palmitoyl-proteins significantly enriched in both L- and S-EVs were involved in pyridine nucleotide metabolism and cell adhesion () and Suppl. Table 2), and included proteins known to be involved in modulating cell polarity, adhesion and migration [Citation46]. These comprised membrane-anchored integrins (ITGA2, ITGA6, ITGB4), claudins (CLDN1, CLDN11), and cytoskeletal proteins such as the Rho-associated moesin (MSN), vinculin (VCL), ezrin (EZR), ROCK2 and the Ras GTPase-activating-like protein IQGAP1 (Suppl. Table 2). Cell functions enriched in L-EVs versus M were associated with positive regulation of organelle and cell component organization, as well as actin cytoskeleton organization () and Suppl. Table 2). In contrast, cell functions enriched in S-EVs versus M were associated with cell and chemical homoeostasis () and Suppl. Table 2).
Detailed analysis of the palmitoylated proteins involved in these biological processes revealed that, while the differentially enriched proteins in L-EVs were mostly represented by cytoplasmic proteins involved in cell growth (GSN, FSCN1 and ATCR3) and signal transduction (ANXA1, ARF1, ROCK2 and CORO1B); the proteins enriched in S-EVs were mainly represented by two major groups of plasma membrane proteins ()). The first group contained two families of transmembrane palmitoyl-proteins with transporter activity: the P-type ATPase superfamily (ATP1A1, ATP1B1 and ATP2B1) and the cation transporters (SLC12A2, SLC39A6 and SLC46A1). The second group contained palmitoyl-proteins associated with signal transduction and cell communication (ANXA6, LPAR1, GNB1, LYN, NTSR1, OXTR and STX4) ()).
Palmitoyl-proteomic profiles of L- versus S-EVs reflect EV population-specific biological processes and subcellular origin
Ninety-seven palmitoyl-proteins were detected only in EVs, with 66 unique to L-EVs, 7 unique to S-EVs, and 24 common to both EV types ()). To further explore the difference between the palmitoyl-proteomes of L-EVs and S-EVs, we compared the proteins differentially enriched in each EV population (Suppl. Table 1). 581 proteins (24 + 12 + 529 + 16) were detected in both S-EV and L-EVs but not unique to them ()). Among these, 58 proteins were significantly enriched in L-EVs and 44 proteins were significantly enriched in S-EVs ()). Functional analysis showed that palmitoyl-proteins enriched in L-EVs were associated with protein localization, regulation of protein stability, regulation of cell component organization and cellular localization ()). This association was mainly represented by the Vacuolar Protein-sorting Associated Protein 35 (VPS35), which is involved in trafficking of proteins [Citation47], and by several members of the chaperone (HSP90AA1) and chaperonin-containing T-complex (CCT2, CCT3, CCT4, CCT7), which have been described to participate in the folding of nascent proteins [Citation48], as well as in vesicular transport [Citation49] ()). Functional analysis of the palmitoyl-proteins enriched in S-EVs showed enrichment for proteins associated with cell communication, signalling processes, proteolysis and regulation of phosphate metabolism ()), and included the metalloproteinase ADAM17, the death receptor FAS and the ubiquitin UBA52 ()). The palmitoylated form of CD9, a tetraspanin highly enriched in S-EVs, was identified as significantly enriched in S-EVs in comparison to L-EVs ()) – in agreement with the distribution observed for the non-palmitoylated form.
Figure 4. Palmitoyl-protein profiles of L- and S-EVs are associated with EV population-specific biological processes and subcellular origin. (a) Volcano plot showing differential protein expression between L-EVs and S-EVs. X and Y axes represent the normalized expression difference and -log10(FDR), respectively. The red and blue boxes highlight functionally relevant palmitoyl-proteins to the biological processes differentially represented in L- and S-EVs shown in panel B. (b) The biological functions overrepresented in L- and S-EVs identified by functional enrichment analysis of the palmitoyl-proteome differentially expressed in EVs using DAVID software. (c) Pie charts indicating the main subcellular localization of the palmitoylated proteins differentially enriched in L- and S-EVs, as defined by the Ingenuity Knowledge database.
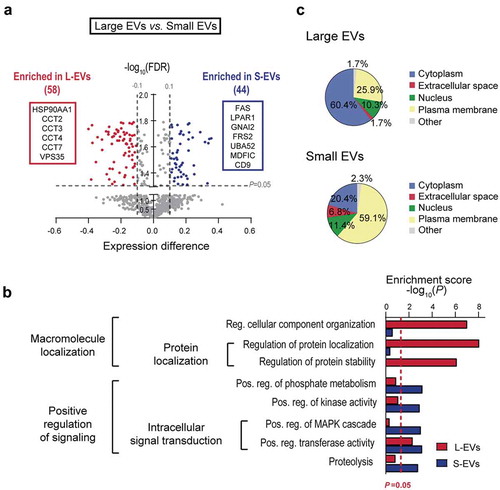
Next we examined subcellular distribution of the palmitoyl-proteins differentially enriched in L- and S-EVs. Palmitoyl-proteins enriched in L-EVs were primarily associated with cytoplasm (60.4%), while those enriched in S-EVs were mainly associated with the plasma membrane (59.1%) ()). Importantly, 34 of the 35 cytoplasmic proteins enriched in L-EVs have been previously identified as palmitoylated by independent approaches [Citation4] (Suppl. Table 3), suggesting that they are genuine palmitoyl-proteins. Interestingly, both EV populations contained a smaller portion of nuclear proteins, which were similarly represented in both EV types (10.3% in L-EVs, 11.4% in S-EVs). Altogether, these data suggest a different subcellular derivation and functional/biological profiles of the palmitoylated proteins in L- and S-EVs.
Prostate cancer-derived EVs contain cancer-specific palmitoylated proteins
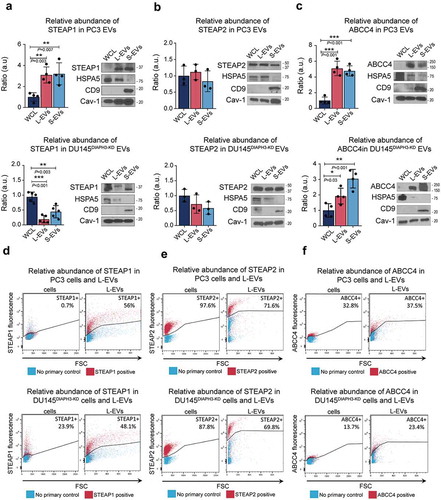
Altered palmitoyl transferase activity has been reported in cancer. In order to evaluate if the palmitoylation profile in EVs reflected cancer-associated functions, we selected for high-abundance palmitoylated proteins in EVs (Suppl. Table 1) and assessed their functional profile ()). We found a significant association with canonical cancer functions such as cell-to-cell signalling, adhesion, cell movement, and epithelial-to-mesenchymal transition (EMT). Using a panel of tissue-specific genes that combines the information from different databases [Citation50], we identified 92 prostate cancer-specific/enriched genes. The multidrug-resistance-associated protein 4 (ABCC4), and the six-transmembrane epithelial antigen of prostate 1 (STEAP1) and 2 (STEAP2) ()) were identified as abundant palmitoyl-proteins in EVs. Importantly, these 3 genes are encoded by RNA that is highly expressed in prostate cancer tissue in comparison to other types of cancer ()). Interestingly, even though these proteins were abundant in both EV types, palmitoylated STEAP1 was more abundant in S-EVs, while STEAP2 and ABCC4 were more abundant in L-EVs (Suppl. Figure 2C), confirming that EV heterogeneity is not limited to size but also affects cargo including protein post-translational modifications.
Figure 5. Prostate cancer-derived EVs are enriched in prostate cancer-enriched palmitoyl-proteins. (a) Cancer-associated functions of the palmitoyl-proteins abundant in L- and S-EVs identified using IPA. (b) Venn diagram showing the number of palmitoyl-proteins abundant (determined by the rank product algorithm) in L- and S-EVs which are specifically enriched in prostate cancer according to the TissGene GDB [Citation50]. (c) The gene expression of STEAP1, STEAP2 and ABCC4 determined as fragments per kilobase of transcript per million mapped reads (FPKM) for select carcinomas according to the cancer genome atlas (TCGA) database[Citation38].
![Figure 5. Prostate cancer-derived EVs are enriched in prostate cancer-enriched palmitoyl-proteins. (a) Cancer-associated functions of the palmitoyl-proteins abundant in L- and S-EVs identified using IPA. (b) Venn diagram showing the number of palmitoyl-proteins abundant (determined by the rank product algorithm) in L- and S-EVs which are specifically enriched in prostate cancer according to the TissGene GDB [Citation50]. (c) The gene expression of STEAP1, STEAP2 and ABCC4 determined as fragments per kilobase of transcript per million mapped reads (FPKM) for select carcinomas according to the cancer genome atlas (TCGA) database[Citation38].](/cms/asset/e31b2609-dead-40cd-bd17-3990c6102076/zjev_a_1764192_f0005_oc.jpg)
Immunoblotting confirmed strong expression of STEAP1 in both populations of EVs from highly metastatic PC3 cells in comparison with the cells themselves ()). This was not the case for purified EVs from DU145DIAPH3-KD cells (Suppl. Figure 2D-E), which is also highly metastatic, suggesting that STEAP1 expression in EVs is not indicative of disease aggressiveness. Conversely, STEAP2 was equally abundant in cells and both EVs from PC3 and DU145DIAPH3-KD cells ()). Finally, ABCC4 was highly enriched in EVs from PC3 and DU145DIAPH3-KD cells in contrast to the low expression detected in the parental cells ()). We also analysed these proteins at the single EV level using flow cytometry, and found that STEAP1 was highly expressed in both PC3 and DU145DIAPH3-KD L-EVs but virtually undetectable in PC3 cells ()). STEAP2 was highly abundant in both L-EVs and cells ()). ABCC4 was relatively enriched in L-EVs versus cells ()). Collectively, these results indicate that prostate cancer-specific proteins enriched in EVs are not necessarily abundant in cancer tissue and support the use of EV cargo as a source of clinically relevant circulating biomarkers.
Figure 6. Prostate cancer-specific proteins are enriched in EVs. (a–c) Representative immunoblot of STEAP1 (a), STEAP2 (b) and ABCC4 (c) in EVs and WCL from PC3 and DU145DIAPH3-KD cells along with the control proteins HSPA5 (L-EV enriched protein), CD9 (S-EV enriched protein) and Cav-1 (general EV protein). Bar plots represent the densitometric quantification across several blots. (d-f) FACS analysis ofSTEAP1 (d), STEAP2 (e) and ABCC4 (f) expression in cells and L-EVs from PC3 and DU145DIAPH3-KD cells.
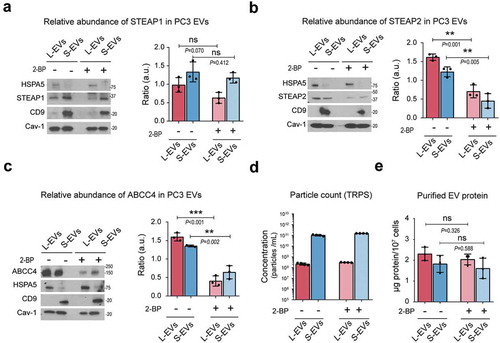
Finally, we investigated if palmitoylation had any influence on the localization of select proteins in EVs. To do so, we tritrated the concentration of 2-bromopalmitate (2-BP), a general inhibitor of palmitoylation, to a non-toxic concentration of 10 µM in PC3 cells (Suppl. Figure 3A) in order to avoid any alterations in the EV shedding () and Suppl. Figure B) and recovery of EV-protein ()). Importantly, a number of studies have demonstrated inhibition of palmitoylation by this compound at the indicated dose [Citation51].Treatment with 2-BP induced a decrease of STEAP1 ()), STEAP2 ()) and ABCC4 ()) in EVs, suggesting a role for protein palmitoylation in the trafficking and sorting of these proteins to EVs. Cav-1 levels in EVs did not change in response to 2-BP (Suppl. Figure 3C), suggesting that palmitoylation of Cav-1 is not a requisite for its EV localization. This is a novel observation but in line with previous studies that report that palmitoylation of Cav-1 is not necessary for its membrane localization [Citation52].
Figure 7. Inhibition of palmitoylation reduces the abundance of prostate cancer-specific palmitoyl-proteins in EVs. (a–c) Representative immunoblot of the abundance of STEAP1 (A), STEAP2 (B) and ABCC4 (C) along with the control proteins HSPA5 (L-EV enriched protein), CD9 (S-EV enriched protein) and Cav-1 (general EV protein) in PC3 L- and S-EVs after inhibition of palmitoylation with 10 µM 2-BP for 24 h. Bar plots represent the densitometric quantification across replicate blots. (d) TRPS quantification of PC3 L- and S-EVs after inhibition of palmitoylation. (e) Yield of purified EV-protein from PC3 cells treated with or without 2-BP.
Discussion
This is one of the most comprehensive palmitoyl-proteomic studies to date and the first study to profile the palmitoyl-proteome of EVs. It was prompted by the observation that EVs are enriched in putative palmitoylated proteins in comparison to the cells of origin. Interestingly, the palmitoylated form of some proteins typically enriched in specific EV populations did not replicate the enrichment observed for the native form of the protein. This suggests an important role of palmitoylation in the sorting of cargo to different EV populations that would be missed if only the native protein is investigated. Further studies on the palmitoyl-proteome of EVs may identify mechanisms that could be exploited to target specific proteins to, or to deplete specific proteins from EVs.
The selective packaging of palmitoyl-proteins in EVs is also supported by the weak correlation between the palmitoyl-proteins identified in EVs and cell membranes. These findings are in agreement with previous studies that demonstrated poor correlation between protein abundance in L- and S-EVs in comparison to their cells of origin [Citation45]. In that study, as well as in ours, the differences between EVs and cells were more pronounced in the S-EVs than L-EVs, in line with a different origin of these vesicles from distinct intracellular compartments [Citation14,Citation15]. Of note, our study points to the association to different subcellular compartments of the palmitoyl-proteins enriched in EVs. Palmitoyl-proteins enriched in L-EVs associated primarily with the cytoplasm, in line with previous reports that show the enrichment of cytoplasmic proteins in L-EVs [Citation10]. Conversely, the palmitoyl-proteins enriched in S-EVs were mostly associated with the plasma membrane. This likely reflects that L-EVs are often generated as a result of membrane blebs inflated by the cytoplasm whereas the enrichment of the plasma membrane proteins in S-EVs may be explained by their high membrane/cytoplasm ratio. Comparative studies of the palmitoyl and total proteome of EVs will be essential to clarify the role of protein palmitoylation in the shuttling of proteins across different subcellular compartments and EV populations.
Attempts to identify disease proteins biomarkers from biological fluids have been hampered by the presence of high abundant proteins which severely masks detection of low abundance but physiologically important proteins. Because most abundant proteins are not palmitoylated, the LB-ABE may allow their drastic depletion from biofluids. The ABE chemistry can be applied to any type of biological samples as it does not rely on metabolic labelling. In addition to S-palmitoylated proteins, LB-ABE may also enrich for other PTMs (e.g. myristate and stearate) that are linked to cysteine residues via thioester bonds [Citation53]. Nevertheless, palmitoylation represents the most frequent S-acylation [Citation2] and the LB-ABE method has proved to largely eliminate the co-isolation of non-S-acylated proteins [Citation26], thus representing a state-of-the-art discovery approach for large scale palmitoyl-protein profiling. The fact that 84% of the proteins identified have been previously identified as palmitoylated [Citation4,Citation26] strongly supports the recovery of S-acylated proteins with high specificity. Moreover, out of the 330 proteins identified as palmitoylated for the first time, 65% have cysteines predicted to be palmitoylated with high confidence by bioinformatic tools [Citation4], suggesting that they are genuine palmitoylated proteins. Further studies are required to validate the palmitoylation status of these candidate palmitoyl-proteins in cells and EVs. Taken together, these results support the use the LB-ABE for deep palmitoyl-proteomics with high specificity and sensitivity.
Given the altered palmitoylation in cancer and that palmitoylation targets proteins to EVs [Citation54], the LB-ABE may allow for the detection of prostate cancer-relevant proteins in EVs. We found three proteins that are abundant in EVs (STEAP1, STEAP2 and ABCC4) and whose expression has been reported to be specific to prostate cancer. While STEAP1 was enriched in S-EVs, STEAP2 was found to be equally abundant in S- and L-EVs. The observation that STEAP2 participates in the intracellular vesicular transport machinery and associates with endocytic and exocytic pathways [Citation55,Citation56] might explain its equal abundance in both L- and S-EVs. Of note, STEAP1 and ABCC4 were enriched in EVs in comparison to cells when we examined the total protein thus confirming the importance of interrogating the mechanisms of protein trafficking and cargo into EVs.
Interestingly, the loading of STEAP2 and ABCC4 into EVs was reduced by inhibition of palmitoylation, in line with a report showing that palmitoylation of TG-A transglutaminase is required for its secretion in exosomes [Citation57]. In contrast, 2-BP treatment did not alter the localization of other palmitoylated proteins such as Cav-1 in agreement with previous studies reporting that palmitoylation is not necessary for Cav-1 membrane localization [Citation52]. Of note, palmitoylation is essential for the protein localization into lipid rafts [Citation58], which participate in the biogenesis of EVs [Citation59–Citation62]. Therefore STEAP2 and ABCC4 may require palmitoylation for their loading into EVs via association with lipid rafts. In contrast, Cav-1 association to lipid rafts derives from its high affinity to cholesterol rather than its palmitoylation status [Citation63]. Further studies will elucidate the role of palmitoylation in directing proteins towards distinct EV populations. Even though the role of palmitoylation in the subcellular localization and trafficking of proteins has been widely reported [Citation1,Citation64], the role of palmitoylation in EV transport is a novel field of study.
In summary, this is the first large-scale analysis of the palmitoyl-proteomes of L- and S-EVs. We show that: (1) the LB-ABE method enables the isolation of palmitoyl-proteins from intracellular compartments, as well as L- and S-EVs; (2) L- and S-EVs exhibit EV population-specific palmitoyl-profiles that reflect EV biological processes and subcellular origin and distinguish them from their parental cells; (3) prostate cancer-derived EVs contain cancer-specific palmitoylated proteins; (4) palmitoylation may play a role in sorting and trafficking of proteins to EVs. Taken together, our results suggest that protein palmitoylation may be involved in the selective packaging of proteins to different EV populations and palmitoyl-proteomics may allow for better detection of disease biomarkers.
Geolocation
Cedars-Sinai Medical Center, Los Angeles, California.
Supplemental Material
Download Zip (1.9 MB)Acknowledgments
The authors are grateful to the Cedars-Sinai flow cytometry core for experiments on extracellular vesicles, and to Drs. Leonora Balaj, Valentina Minciacchi and Cristiana Spinelli for helpful discussions.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–16.
- Muszbek L, Laposata M. Covalent modification of platelet proteins by palmitate. Blood. 1989;74(4):1339–1347.
- Zhou B, An M, Freeman MR, et al. Technologies and challenges in proteomic analysis of protein S-Acylation. J Proteomics Bioinform. 2014;7(9):256–263.
- Blanc M, David FPA, Van Der Goot FG. SwissPalm 2: Protein S-Palmitoylation Database. Edited by ME Linder. Methods in Molecular Biology. Springer Nature. 2019; Chapter 16. p. 203–214.
- Yang W, Di Vizio D, Kirchner M, et al. Proteome scale characterization of human S-Acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9(1):54–70.
- Sanders SS, Martin DDO, Butland SL, et al. Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput Biol. 2015;11(8):e1004405.
- Ko P, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep. 2018;19(10):e46666.
- Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci. 2016;113(8):E968–E977.
- Vagner T, Spinelli C, Minciacchi VR, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7(1):1505403.
- Minciacchi VR, You S, Spinelli C, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6(13):11327–11341. .
- Willms E, Johansson HJ, Mäger I, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6(22519).
- Willms E, Cabañas C, Mäger I, et al. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. 2018;9(738).
- Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750.
- Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372.
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30(1):255–289.
- Li B, Antonyak MA, Zhang J, et al. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31(45):4740–4749.
- Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343.
- D’Souza-Schorey Crislyn C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26(12):1287–1299.
- Booth AM, Fang Y, Fallon JK, et al. Exosomes and HIV gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172(6):923–935.
- Antonyak MA, Li B, Boroughs LK, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A. 2011;108(42):17569.
- Zijlstra A, Di Vizio D. Size matters in nanoscale communication. Nat Cell Biol. 2018;20(3):225–232.
- Minciacchi VR, Spinelli C, Reis-Sobreiro M, et al. MYC mediates large oncosome-induced fibroblast reprogramming in prostate cancer. Cancer Res. 2017;77(9):2306–2317.
- Hager MH, Morley S, Bielenberg DR, et al. DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med. 2012;4(8):743–760. .
- Van Deun J, Mestdagh P, Agostinis P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228–232.
- Zhou B, Yan Y, Wang Y, et al. Quantitative proteomic analysis of prostate tissue specimens identifies deregulated protein complexes in primary prostate cancer. Clin Proteomics. 2019;16(15).
- Zhou B, Wang Y, Yan Y, et al. Low-background Acyl-biotinyl exchange largely eliminates the co-isolation of non-S-Acylated proteins and enables deep S-Acylproteomic analysis. Anal Chem. 2019;91(15):9858–9866.
- Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362.
- Morley S, You S, Pollan S, et al. Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci. Rep. 2015;5(1):12136.
- Han B, Zhou B, Qu Y, et al. FOXC1-induced non-canonical WNT5A-MMP7 signaling regulates invasiveness in triple-negative breast cancer. Oncogene. 2018;37(10):1399–1408.
- Cox J, Neuhauser N, Michalski A, et al. Andromeda: a peptide search engine integrated into the maxquant environment. J Proteome Res. 2011;10(4):1794–1805.
- Cox J, Mann M. maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372.
- Breitling R, Armengaud P, Amtmann A, et al. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92.
- Hwang D, Rust AG, Ramsey S, et al. A data integration methodology for systems biology. Proc Natl Acad Sci. 2005;102(48):17296–17301. .
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100(16):9440–9445.
- Keerthikumar S, Chisanga D, Ariyaratne D, et al. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688–692. .
- Krämer A, Green J, Pollard J, et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530.
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
- Carithers LJ, Moore HM. The genotype-tissue expression (GTEx) project. Biopreserv Biobank. 2015;13(5):307–308.
- Blanc M, David F, Abrami L, et al. SwissPalm: protein palmitoylation database. F1000Res. 2015;4:261.
- Ciardiello C, Leone A, Lanuti P, et al. Large oncosomes overexpressing integrin alpha-V promote prostate cancer adhesion and invasion via AKT activation. J Exp Clin Cancer Res. 2019;38(1):317. .
- Conley A, Minciacchi VR, Lee DH, et al. High-throughput sequencing of two populations of extracellular vesicles provides an MRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2017;14(3):305–316.
- Freeman MR, Yang W, Di Vizio D. Caveolin-1 and prostate cancer progression. Edited by Jean-François Jasmin, Philippe G. Frank and Michael P. Lisanti. Landes Bioscience and Springer Science + Business Media. Advances in experimental medicine and biology. Vol. 729. 2012; Chapter 7. p. 95–110.
- Varkaris A, Katsiampoura AD, Araujo JC, et al. Src signaling pathways in prostate cancer. Cancer Metastasis Rev. 2014;33(2–3):595–606.
- Di Vizio D, Adam RM, Kim J, et al. Caveolin-1 interacts with a lipid raft-associated population of fatty acid synthase. Cell Cycle. 2008;7(14):2257–2267. .
- Haraszti RA, Didiot MC, Sapp E, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5(1):32570. .
- Zaballa M-E-E, van der Goot FG. the molecular era of protein s-acylation: spotlight on structure, mechanisms, and dynamics. Crit Rev Biochem Mol Biol. 2018;53(4):420–451.
- Zhou M, Wiener H, Su W, et al. VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J Cell Biol. 2016;214(4):445–458.
- Yam AY, Xia Y, Lin HTJ, et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15(12):1255–1262.
- Seo S, Baye LM, Schulz NP, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome Assembly. Proc Natl Acad Sci. 2010;107(4):1488–1493.
- Kim P, Park A, Han G, et al. TissGDB: tissue-Specific Gene Database in Cancer. Nucleic Acids Res. 2018;46(D1):D1031–D1038.
- Mikic I, Planey S, Zhang J, et al. A live cell, image-based approach to understanding the enzymology and pharmacology of 2-bromopalmitate and palmitoylation. Methods Enzymol. 2006;414:150–187.
- Dietzen DJ, Hastings WR, Lublin DM. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J Biol Chem. 1995;270(12):6838–6842.
- Liang X, Nazarian A, Erdjument-Bromage H, et al. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276(33):30987–30994.
- Romancino DP, Buffa V, Caruso S, et al. palmitoylation is a post-translational modification of alix regulating the membrane organization of exosome-like small extracellular vesicles. Biochim Biophys Acta - Gen Subj. 2018;1862(12):2879–2887.
- Korkmaz KS, Elbi C, Korkmaz CG, et al. Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J Biol Chem. 2002;277(39):36689–36696.
- Gomes IM, Maia CJ, Santos CR. STEAP proteins: from structure to applications in cancer therapy. Mol Cancer Res. 2012;10(5):573–587.
- Shibata T, Hadano J, Kawasaki D, et al. Drosophila TG-A transglutaminase is secreted via an unconventional golgi-independent mechanism involving exosomes and two types of fatty acylations. J Biol Chem. 2017;292(25):10723–10734.
- Levental I, Lingwood D, Grzybek M, et al. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A. 2010;107(51):22050–22054.
- Del Conde I, Shrimpton CN, Thiagarajan P, et al. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106(5):1604–1611.
- Wei H, Malcor JDM, Harper MT. Lipid rafts are essential for release of phosphatidylserine-exposing extracellular vesicles from platelets. Sci Rep. 2018;8(9987).
- De Gassart A, Géminard C, Février B, et al. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344.
- Valapala M, Vishwanatha JK. Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J Biol Chem. 2011;286(35):30911–30925.
- Yu C, Alterman M, Dobrowsky RT. Ceramide displaces cholesterol from lipid rafts and decreases the association of the cholesterol binding protein caveolin-1. J Lipid Res. 2005;46(8):1678–1691.
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta - Biomembr. 2011;1808(12):2981–2994.
