ABSTRACT
Malaria remains a significant public health challenge and is of global importance. Imported malaria is a growing problem in non-endemic areas throughout the world and also in Qatar due to a massive influx of migrants from endemic countries. Antimalarial drug resistance is an important deterrent in our fight against malaria today. Molecular markers mirror intrinsic antimalarial drug resistance and their changes precede clinical resistance. Thus, in the present study, molecular markers of sulphadoxine-pyrimethamine (Pfdhfr and Pfdhps) and artemisinin (PfATPase6 and Pfk13) were sequenced to determine the drug resistance genotypes among 118 imported P. falciparum isolates in Qatar, between 2013 and 2016. All the isolates had mutant Pfdhfr alleles, with either double mutant (51I/108N) (59.3%) or triple mutant (51I, 59R and 108N) (30.6%) genotypes. I164L substitution was not found in this study. In case of Pfdhps, majority of the samples were carriers of either single (S436A/ A437G/ K540E) mutant (47.2%) or double (S436A/K540E, A437G/K540E, K540E/A581G) mutant (39.8%). A single novel point mutation (431V) was observed in the samples originated from Nigeria and Ghana. Polymorphisms in PfATPase6 were absent and only one non-synonymous mutation in Pfk13 was found at codon G453A from a sample of Kenyan origin. High levels of sulphadoxine-pyrimethamine resistance in the present study provide potential information about the spread of antimalarial drug resistance and will be beneficial for the treatment of imported malaria cases in Qatar.
Introduction
More than a century ago after its discovery, and more than six decades of efforts to control it, malaria remains a serious health problem in most tropical and subtropical countries. The World Health Organization (WHO) estimates that 3.3 billion people are at risk of malaria infection, 219 million clinical cases and 435,000 deaths globally in 2017 [Citation1]. Qatar has been free from local malaria transmission since 1970 [Citation2], however, an influx of migrant workers from malaria-endemic countries, representing a major threat for re-introduction of local drug resistance malaria transmission [Citation3–Citation6]. In addition to the flow of imported cases, the receptivity and risk of malaria reintroduction are evident by the presence of the mosquito vectors, Anopheles stephensi and An. multicolor [Citation4,Citation7,Citation8]. This situation is similar to that in other countries of the Arabic Peninsula [Citation9]. Currently, according to the national guidelines, artemisinin-based combination therapies (ACT) and artesunate/doxycycline or quinine/clindamycin is recommended for uncomplicated and complicated/severe P. falciparum malaria, respectively. However, chloroquine (CQ) followed by primaquine is the first-line regimen for the treatment of P. vivax malaria in Qatar. Moreover, mixed infections are usually treated as P. falciparum malaria.
Resistance to the antifolate sulphadoxine-pyrimethamine (SP) has been found to be associated with key point mutations in the dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) genes. The level of resistance to SP has been shown to increase with the accumulation of mutations at codons 16, 51, 59, 108 and 164 as the Pfdhfr inhibit its activity, and the parasite becomes resistant to pyrimethamine. While mutations at codons 436, 437, 540, 580 and 613 of Pfdhps reduce the substrate binding capacity and confer resistance to sulphadoxine [Citation10,Citation11].
The exact mechanism of action of ACTs is still a matter of debate but, recent emergence of P. falciparum artemisinin (ART) resistance in southeast Asia threatens the global efforts to control malaria. Hitherto, several studies provide compelling evidence that ACTs act by selectively inhibiting sarcoplasmic/endoplasmic reticulum Ca++-ATPase ortholog in P. falciparum (PfATPase6) protein, believed to be the primary target (at codon S769N) for ART, but is not yet fully confirmed [Citation12–Citation15]. In recent years, mutations in the propeller region of a kelch protein (K13) were associated with ART-resistant isolates [Citation16,Citation17], which was further confirmed by in vitro and ex vivo studies [Citation18].
Monitoring drug resistance and pattern of mutations is essential for early detection and subsequent preventions of the spread of drug resistance. Hence, there is a need for monitoring antimalarial drug resistance in Qatar for prompt management of imported P. falciparum malaria, as majority of immigrant workers come from malaria-endemic countries, namely India, Pakistan and Sudan [Citation3,Citation4,Citation6]. Therefore, the present study was conducted to determine the prevalence of point mutations involved in antimalarial resistance genes Pfdhfr, Pfdhps (SP) and PfATPase6, K13 (ART) among imported P. falciparum isolates in Qatar. The study’s findings will generate a new understanding of the disease as well as baseline information on antimalarial drug resistance among imported P. falciparum in Qatar.
Materials and methods
Study area, sample collection and ethical approval
The present study was carried out in Doha, Qatar, during January 2013–September 2016. Details of the study’s population characteristics have previously been described [Citation19]. The venous blood samples from 583 symptomatic patients attending the Hamad General Hospital (HMC), Doha, Qatar were collected in sterile vacutainer and screened for malaria parasites by microscopy, and species identification was confirmed by species-specific nested PCR [Citation20]. Socio-demographics information including age, gender, nationality, treatment and clinica was collated using a structured questionnaire and from patient’s medical records. The ethical clearance for patient’s participants and sample collections was obtained from Hamad Medical Corporation and Weill Cornell Medicine-Qatar Institutional Review Board (Protocol no. – 14–00097).
DNA extraction, nested PCR and sanger sequencing
The genomic DNA of P. falciparum was extracted using QIAamp DNA blood mini kit as per the manufacturer’s instructions (Qiagen, CA, USA).
Alleles of the Pfdhfr, Pfdhps genes (for SP resistance) and K-13 propeller, PfATPase6 (for ART resistance) were determined using nested PCR followed by sanger sequencing, as previously described protocols with some minor modifications [Citation21]. Primers for primary and nested PCR, and PCR cycling conditions are listed in the supplementary material [Citation21]. The spanning codons 51–164 of Pfdhfr gene, codons 425–640 of Pfdhps gene codons 653–797 of PfATPase6 and codon 427–702 of K-13 propeller were amplified by PCR. In brief, primary PCR was performed with 2 μL extracted genomic DNA (5ng/μL), 12.5 μl of 2X Master mix (Applied Biosystems, CA, USA), 1μM each primer and the mixture was made up to 25 μL volume with nuclease-free water (Ambion, CA, USA). The PCR products were purified using MinElute PCR Purification Kit according to the manufacturer’s instructions (Qiagen, CA, USA). Purified products were sequenced using the Sanger method (Genewiz INC, NJ, USA) with respected-nested primers. Obtained DNA sequences were edited manually in BioEdit9 and aligned using ClustalW10. Multiple Sequence Alignment (MSA) was performed to see intraspecific variation if any, among the sequences. The presence or absence of mutations was assessed by comparing each sequence to the P. falciparum 3D7 reference strain for dhps (PF3D7_0810800), dhfr (PF3D7_0417200), ATPase6 (PF3D7_0106300) andk13 (PF3D7_1343700). In order to determine the association between SNPs (present in Pfdhfr and Pfdhps) in the population, both inter and intragenic linkage disequilibrium (LD) tests were performed using the Haploview Software [Citation22].
Statistical analysis
Statistical analyses were performed using IBM SPSS version 22.0 (Armonk, NY: IBM Corp). Frequencies of alleles and genotypes among parasite groups were compared using the X [Citation2] test. The P values <0.05 were considered statistically significant. Unadjusted odds ratios (ORs) of mutant alleles among different parasite origins along with their 95% confidence intervals (CI) were calculated using Univariate logistic regression model.
Results
A total of 583 patients were examined by microscopy followed by PCR for malaria between January 2013 and October 2016 in Hamad General Hospital and Al-Khor Hospital, 448 (76.8%) were found to be positive for malaria parasites. The majority were infected with P. vivax (n = 318 [71%]), followed by P. falciparum (n = 118 [26.3%]) and mixed P. falciparum and P. vivax (n = 12 [2.7%]) were less frequent.
The majority of malaria patients were male, with a mean age of 33 years (95% CI 30.0–35.0). The vast majority of P. falciparum cases originated from East Africa (60%, n = 71) followed by the Indian subcontinent (20.3%, n = 24), West and Central Africa (17.8%, n = 21) and less frequently from Oman or Yemen and Saudi Arabia (1.7%, n = 2) (). The origin of the majority of patients of P. vivax was from the Indian-subcontinent (81.8%) followed by East Africa (10.7%) and different parts in Africa (6%).
Prevalence of pfdhfr alleles and genotypes
Out of 118 P. falciparum isolates, 108 (91.5%) were successfully amplified, sequenced and analyzed for Pfdhfr gene. All the isolates were mutant either single, double, triple and/or quadruple. Most of the sequenced isolates showed a mutation at codons N51I (89.8%), C59R (36.1%) and S108N (94.4%). However, no mutation was seen at codon I164L (). In addition, four novel mutations were detected at codons 185, 178, 183 and 181.
Table 1. Frequency (%) and number (n) of the Pfdhfr and Pfdhps mutations.
Out of 108 samples genotyped, a large proportion (59.3%) of the isolates carried the double mutants genotype, while the triple mutant genotypes were detected in 30.6% isolates. However, the prevalence of quadruple mutant genotypes was only 1.9% (). The frequency of triple mutation was high in West and Central Africa, and quadruple mutation was found only in the East Africa isolates ()
Figure 2. Mutation rates in P. falciparum genes (Pfdhfr,Pfdhps) that confer SP resistance from the isolates collected from East Africa, West and Central Africa and Indian subcontinent.
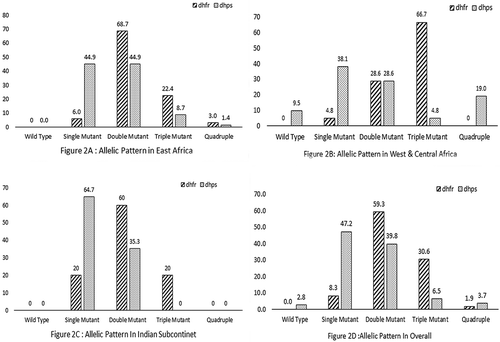
Based on the geographical location, eight genotypes were detected (ACICSILYT, ACNCNILYT, ACICNILYT, ACNRNILYT, ACIRNILYT, ACICNISYT, ACIRNILYA and ACIRNILHT) in 67 East Africa isolates. A majority of the samples (65.7%) harbored the double mutants ACICNILYT followed by triple mutants ACIRNILYT (20.9%) and single mutant ACICSILYT (4.5%) genotype. While among the isolates from the Indian subcontinents, seven different genotypes (ACNCSILYT, ACICSILYT, ACNCNILYT, ACNRNILYT, ACICNILYT, ACNCNILHT and ACIRNILYT) were found. Of these, 30%, 25% and 5% harbored double mutant genotypes (ACNRNILYT, ACICNILYT and ACNCNILHT), respectively. The triple mutant genotype ACIRNILYT were detected in 20%. The single mutant genotypes (ACNCNILYT, ACICSILYT and ACNCSILYT) were carried by 10%, 5% and 5% isolates, respectively. However, in West and Central Africa, we observed only four genotypes (ACICSILYT, ACICNILYT, ACIRNILYT and ACICNILHT) and most of the isolates (62%) carried triple mutant (ACIRNILYT) followed by double mutant (ACICNILYT) (28.6%) genotypes (). Further analysis revealed that triple mutants were significantly higher in West and Central Africa as compared to East Africa and Indian subcontinents (p=0.001).
Table 2. The frequency distribution of genotypes for Pfdhfr and Pfdhps haplotypes among different nationalities.
Prevalence of pfdhps alleles and genotypes
Sequencing of the Pfdhps gene was successful for 108 (91.5%) isolates and overall 2.8% harbored wild type alleles only, while the single mutant (S436A/ A437G/ K540E) allele was observed in 47.2%, double mutant (S436A/K540E, A437G/K540E, K540E/A581G) alleles in 39.8%, triple mutant (A437G/K540E/A581G) allele in 6.5%, quadruple in 2.8% and 0.9% had quintuple (431V/S436A/A437G/A581G/613S) mutant alleles (). No triple or quadruple mutation was observed in the Indian subcontinent. A majority of quadruple mutant genotypes was observed from West and Central Africa origin. Furthermore, mutations at 540E, 437G and 581G were frequent, occurring at a prevalence of 65.7%, 52.8% and 26.9%, respectively. Moreover, we found a single novel point mutation (431V) in six (5.6%) isolates, of these five were from Nigeria and one from Ghana (). The mutation frequencies of the double mutant alleles (540E/581G) were detected in 14.8% isolates (437G/540E) in 19.4%, 1.8% for both (436A/437G) and (431V/436A) and only 0.9% for both (436A/540E) and (436A/581G). The triple mutant alleles (437G, 540E, 581G), quadruple mutant alleles (431V, 436A, 437G, 581G) and quintuple mutant alleles (431V, 436A, 437G, 581G, 613S) were detected in 6.5%, 2.8% and 0.9% of isolates, respectively. Interestingly, significant quadruple mutants were observed in West and Central Africa as compared to East Africa and Indian subcontinents (p=0.001).
Furthermore, the sulphadoxine (wild type) Pfdhps genotype is SAKAA positioning 436, 437, 540, 581 and 613. In comparison to the wild type genotype (SAKAA), seven genotypes (AAKAA, SGKAA, SAEAA, AAEAA, SGEAA, SGEGA and SAEGA) were detected in 69 successfully sequenced samples from East Africa. The prevalence of SAEAA genotype was higher (34.8%) followed by SGEAA (26.1%), SAEGA (17.4%), SGKAA and SGEGA (8.7% of each) genotype. In the Indian subcontinents, of the 17 successfully sequenced isolates, five genotypes (SGKAA, SAEAA, SAKGA, SGEAA and SAEGA) were detected. Higher prevalence of single mutant SGKAA (47.1%) was noted followed by SAEGA and SGEAA (17.6% of each) and 11.8% SAEAA genotype. In the case of West and Central Africa, eight genotypes (SGKAA, VAGKGA, VAGKGS, VAAKAA, AGKAA, AAKGA, SGEGA and SAEGA) were detected in 21 successfully sequenced isolates. The single mutant genotype (SGKAA) was more prevalent and observed in 38.1% of the isolates,14.3% had VAGKGA, 9.5% of each had AGKAA and VAAKAA. Additionally, only single mutant genotype (SGKAA) was observed in GCC isolate ().
Polymorphisms of pfatpase6 and pfk13 genes
A total of 113 and 110 P. falciparum samples were successfully genotyped for PfATPase6 and propeller region of Pfk13 genes, respectively. All isolates carried the wild type PfATPase6 gene. Furthermore, in Pfk13, the frequency of mutations associated with ART resistance reported in Southeast Asia was not observed. However, only one non-synonymous mutation at codon G453A was identified in isolate imported from a sample of Kenyan origin.
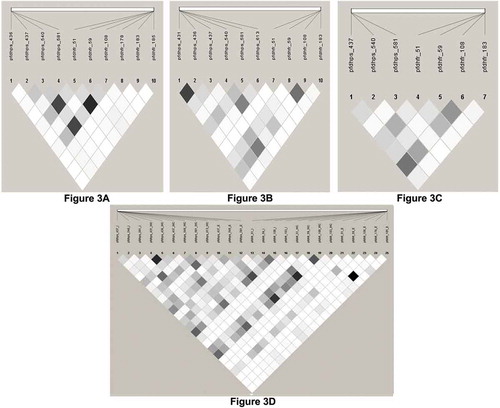
The Pair-wise LD estimation to assess the selection of P. falciparum resistant markers revealed a total of 37 possible pairs of SNPs with fine statistically significance intragenic and intragenic associations were found between different geographic regions from a possible 154 points ().
Figure 3. Linkage disequilibrium (LD) between pairs of SNPs located in Pfdhfr, Pfdhps genes implicated in SP drug resistance in imported P. falciparum cases in Qatar. The intra and inter-genic association between genes for the isolates collected from East Africa, West and Central Africa and Indian subcontinent were determined using the LD plot. The strength of LDbetween the SNPs was determined from the association of statistical significance by calculating the r2 values and represented by the darkness of the boxes.
Discussion
Globally, emergence and spread of antimalarial drug resistance of P. falciparum malaria is an important deterrent in our fight against malaria. Recent reports of tolerance/resistance to artemisinin derivatives and spread of drug-resistant strains is a major concern, in particular, the long-term effectiveness of artemisinin-combination therapy (ACT) in Southeast Asia, Indian subcontinents and Africa [Citation23]. Antimalarial drug resistance data is not available in Qatar and it has been well established that human migration can interact with natural features to establish drug resistance in new areas. Thus, there is a need to monitor antimalarial drug resistance in the migrant population and also in native Qatari visiting endemic regions to stall the potential spread of this deadly infection in Qatar. Recently, we determined the current extent of chloroquine resistance among imported P. falciparum malaria and found a high prevalence of CQ resistance patterns [Citation19]. In the present study, the genetic polymorphism in the antimalarial drugs resistance genes (Pfdhfr, Pfdhps, PfATPase6 and Pfk13) was investigated among imported P. falciparum isolates in Qatar.
Resistance to SP was first reported from the Thai-Cambodian border in the 1960s [Citation24] and has spread to several parts of Southeast Asia, South America and Africa [Citation25–Citation27]. Resistance is likely to progress geographically and in intensity at a rising rate, becoming fixed in the population if proper and timely measures are not taken to interrupt it.
In this study, all the isolates were found mutated and had at least one of 51I, 59R and 108N of Pfdhfr alleles, while no mutation was seen at codon 164L. Overall, the double mutant genotype (51I/108N) and triple mutant genotype (51I, 59R, 108N) were found to be more prevalent among the imported cases, which are in agreement with well-established SP resistance found in Africa and Southeast Asia [Citation28,Citation29]. Moreover, the double mutant genotype (51I/108N) was predominant from East African isolates than other areas because the majority of the isolates were from Sudan origin. The double mutation was prevalent in Sudan in 2007 [Citation30] and still maintains the double mutant genotype with an emerging triple mutation [Citation31]. Triple mutants (51I, 59R, 108N), which associated with high-level pyrimethamine resistance [Citation10,Citation11], were almost fixed in the African population [Citation32,Citation33]. High level of triple mutant genotypes was also found in the present study among isolates originates from Nigeria and Cameroon, which is in agreement with other studies [Citation32,Citation33]. Among the Indian sub-continent isolates, the double (59R/108N and 51I/108N) mutants genotypes were the most frequent genotypes followed by triple (51I/59R/108N) mutant genotypes, which are in agreement with previous reports from India [Citation21,Citation27,Citation34–Citation36].
Sulphadoxine resistance is mediated by a single point mutation at codon 437G of the Pfdhps gene. Higher levels of sulphadoxine resistance are associated with double mutation at codons 437G/581G and triple mutations at codons 436A/437G/613S [Citation37]. In the present study, 52.8% isolates had the key mutation at codon 437G, whereas none of the isolates has key mutations at both double (437G/581G) and triple mutations [436A/437G/613S]. Approximately, 90% isolates from East African origin had double mutation whereas quadruple mutation arose in 19% isolates (Nigerian origin) where triple and quadruple mutations are commonly prevalent [Citation33]. Single point mutation at codon 437G was higher in Indian subcontinents isolates followed by West & Central Africa and East Africa. These findings corroborate with earlier reports from India [Citation21,Citation34,Citation35]. Additionally, we found a novel mutation at codon 431V in five isolates imported from Nigeria and one isolate imported from Ghana, which corroborates with previous findings of imported malaria infections in the UK, which also originated from Nigeria [Citation38] and also pregnant women from Cameroon [Citation39], suggesting that this mutation is emerging especially in West Africa. Recently, mutation at 431V in combination with 436A, 437G, 581G and 613S was found to be the most common in a Nigerian cohort [Citation40].
The presence of key mutations in Pfdhfr and Pfdhps genes along with the emergence of new mutations and quadruple mutations at dhfr locus and quadruple/quintuple mutations in dhps locus are important findings which might be the cause for multidrug resistance of antifolate drugs. The emergence of these novel mutations might have enhanced the level of antifolate resistance to P. falciparum hence it should be taken into consideration when using the combination of antifolate drugs with artemisinin derivatives nationwide and also where the double mutant genotype of Pfdhfr (ACICNILYT) is common [Citation21,Citation41,Citation42].
The artemisinin-combination therapy (ACT) is recommended by the WHO as the first-line treatment for P. falciparum infections in all endemic regions [Citation1] and also in non-endemic countries like Qatar [Citation4]. In spite of these efforts, there have been sporadic reports of an in-vivo and an in-vitro artemisinin resistance during the last few years [Citation43–Citation45]. However, the mechanism of artemisinin resistance is still under investigation. Subsequently, ACT resistance is considered a major risk to public health in Southeast Asia and Sub-Saharan Africa, where the burden is serious. The mutations of Pfk13-propeller are well established in Southeast Asia, whereas only limited polymorphisms of Pfk13-propeller are detected in Africa [Citation16,Citation46]. Recently, in China, polymorphism in Pfk13 was reported in returned migrant workers from Africa [Citation47]. Thus, there is now a clear need to acquire a greater understanding of Plasmodium parasites’ genotypic resistance surveillance with k13-propeller polymorphism. In the present study, k13 propeller sequence analysis of P. falciparum parasites from different malaria-endemic areas in East Africa, Indian sub-continent and West & Central Africa did not detect the predicted artemisinin-resistant genotypes, but we observed only one non-synonymous mutation at codon G453A in a Kenyan isolate. Although antimalarial drug resistance originates from the greater Mekong sub-region and subsequently spread worldwide, still we do not have any functional mutation from sub-Saharan Africa [Citation48–Citation50]. No mutations in PfATPase6 were found in the current study, which is in agreement with previous reports from India and Africa, where PfATPase6 polymorphisms rarely reported [Citation51–Citation55]. Another marker Pfmdr1 is also considered as quinine, mefloquine, lumefantrine and ACT resistant markers [Citation56] and recently we have reported only 28% mutant or mixed genotype at N86 codon using the same samples [Citation19].
Although the molecular findings were not further matched with treatment outcome and in-vitro drug sensitivity of the falciparum isolates, which does not allow us to make a correct evaluation about the level of sensitivity of P. falciparum isolates, including the possible role of the novel Pfdhps, Pfdhfr genotypes because of the limitations of the present study. But at the same time, many studies have proved that the quadruple Pfdhfr/Pfdhps combined haplotype (59R + 108N/437G + 540E) are directly associated with either adequate clinical and parasitological response, early treatment failures or late parasitological failures to SP [Citation11,Citation35,Citation57]. Similarly, the reduced efficacy of artemisinin associated with the presence of the functional mutation in the K-13 gene [Citation44,Citation45].
In conclusion, the present study shows a high prevalence of resistance patterns of SP drugs to P. falciparum in imported cases of malaria, which might be helpful for the enrichment of molecular surveillance of antimalarial resistance. In addition, the emergence of new mutations in Pfdhfr and Pfdhps genes requires further study to monitor the changes in the frequency of mutation. In addition, the current findings also raise a concern about the potential effectiveness of the current first line of drug, i.e. ACT. Hence, the baseline data on antimalarial drug resistance will help policymakers in updating national antimalarial guidelines for the treatment of imported malaria cases in Qatar.
Availability of data and materials
Yes - all data are fully available without restriction and from the corresponding author on reasonable request.
Supplemental Material
Download MS Word (15.4 KB)Acknowledgments
This publication was made possible by a grant from the Qatar National Research Fund (NPRP-5-098-3-021). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the WCM-Q. The authors would like to thank Dr. Musaed Saad Al-Samawi, Dr. Peter Cameroon and Para-medical staff of Emergency Room, HMC, Doha, Qatar for helping in sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- WHO. Malaria world report, 2018. [cited 2019 Feb 2]. Available from: http://www.who.int/malaria/publications/world_malaria_report_2016/report/en/.
- A global brief on vector-borne diseases. Geneva: World Health Organization Headquarters; 2014. [cited 2019 Feb 04. Available from: https://apps.who.int/iris/bitstream/handle/10665/111008/WHO_DCO_WHD_2014.1_eng.pdf
- Al-Kuwari MG. Epidemiology of imported malaria in Qatar. J Travel Med. 2009;16:119–122.
- Khan FM, Lutof AK, Yassin MA, et al. Imported malaria in Qatar: A one-year hospital-based study in 2005. Travel Med Infect Dis. 2009;7:111–117.
- Bansal D, Acharya A, Bharti P, et al. Distribution of mutations associated with antifolate and chloroquine resistance among imported plasmodium vivax in the state of Qatar. Am J Trop Med Hyg. 2017;97:1797–1803.
- Farag E, Bansal D, Chehab MAH, et al. Epidemiology of malaria in the state of Qatar, 2008-2015. Mediterr J Hematol Infect Dis. 2018;10:e2018050–e.
- Kardousha MM First report of some adult mosquitoes captured by CDC gravid traps from north-eastern Qatar. Asian Pac J Trop Dis. 2016; 6:100–105.
- Snow RW, Amratia P, Zamani G, et al. The malaria transition on the Arabian Peninsula: progress toward a malaria-free region between 1960-2010. Adv Parasitol. 2013;82:205–251.
- Simon B, Sow F, Al Mukhaini SK, et al. An outbreak of locally acquired Plasmodium vivax malaria among migrant workers in Oman. Parasite. 2017;24:25.
- Cowman AF, Morry MJ, Biggs BA, et al. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113.
- Plowe CV, Kublin JG, Doumbo OKP. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist Updat. 1998;1:389–396.
- Cui L, Wang Z, Jiang H, et al. Lack of association of the S769N mutation in Plasmodium falciparum SERCA (PfATP6) with resistance to artemisinins. Antimicrob Agents Chemother. 2012;56:2546–2552.
- Afonso A, Hunt P, Cheesman S, et al. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob Agents Chemother. 2006;50:480–489.
- NN, C GP, Chakraborty C, et al. Mechanism of artemisinin resistance for malaria PfATP6 L263 mutations and discovering potential antimalarials: an integrated computational approach. Sci Rep. 2016;6:30106.
- Nguetse CN, Adegnika AA, Agbenyega T, et al. Molecular markers of anti-malarial drug resistance in Central, West and East African children with severe malaria. Malar J. 2017;16:217.
- Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55.
- Straimer J, Gnädig NF, Witkowski B, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431.
- Takala-Harrison S, Jacob CG, Arze C, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis. 2015;211:670–679.
- Acharya A, Bansal D, Bharti PK, et al. Molecular surveillance of chloroquine drug resistance markers (Pfcrt and Pfmdr1) among imported Plasmodium falciparum malaria in Qatar. Pathog Glob Health. 2018;112:57–62.
- Singh B, Bobogare A, Cox-Singh J, et al. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692.
- Patel P, Bharti PK, Bansal D, et al. Prevalence of mutations linked to antimalarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: A malaria elimination point of view. Sci Rep. 2017;7:16690.
- Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265.
- Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467.
- Björkman A, Phillips-Howard PA. The epidemiology of drug-resistant malaria. Trans R Soc Trop Med Hyg. 1990;84:177–180.
- WHO. Global report on antimalarial drug efficacy and drug resistance 2000–2010 [cited 2019 Feb 14] Available from: http://www.who.int/malaria/publications/atoz/9789241500479/en/index.html
- Shah NK, Dhillon GP, Dash AP, et al. Antimalarial drug resistance of Plasmodium falciparum in India: changes over time and space. Lancet Infect Dis. 2011;11:57–64.
- Sarmah NP, Sarma K, Bhattacharyya DR, et al. Antifolate drug resistance: novel mutations and haplotype distribution in dhps and dhfr from Northeast India. J Biosci. 2017;42:531–535.
- Sibley CH, Hyde JE, Sims PF, et al. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588.
- Nair S, Williams JT, Brockman A, et al. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003;20:1526–1536.
- Al-Saai S, Kheir A, Abdel-Muhsin AM, et al. Distinct haplotypes of dhfr and dhps among Plasmodium falciparum isolates in an area of high level of sulfadoxine-pyrimethamine (SP) resistance in eastern Sudan. Infect Genet Evol. 2009;9:778–783.
- Adeel AA, Elnour FA, Elmardi KA, et al. High efficacy of artemether-lumefantrine and declining efficacy of artesunate + sulfadoxine-pyrimethamine against Plasmodium falciparum in Sudan (2010–2015): evidence from in vivo and molecular marker studies. Malar J. 2016;15:285.
- Moyeh MN, Njimoh DL, Evehe MS, et al. Effects of drug policy changes on evolution of molecular markers of Plasmodium falciparum resistance to chloroquine, amodiaquine, and sulphadoxine-pyrimethamine in the south west region of Cameroon. Malar Res Treat. 2018;2018:7071383.
- MA O, US S, HA A, et al. Molecular epidemiology and evolution of drug-resistant genes in the malaria parasite Plasmodium falciparum in southwestern Nigeria. Infect Genet Evol. 2018;66:222–228.
- Mohapatra PK, Sarma DK, Prakash A, et al. Molecular evidence of increased resistance to anti-folate drugs in Plasmodium falciparum in North-East India: a signal for potential failure of artemisinin plus sulphadoxine-pyrimethamine combination therapy. PLoS One. 2014;9:e105562.
- Mishra N, Srivastava B, Bharti RS, et al. Monitoring the efficacy of antimalarial medicines in India via sentinel sites: outcomes and risk factors for treatment failure. J Vector Borne Dis. 2016;53:168–178.
- Kumar A, Moirangthem R, Gahlawat SK, et al. Emergence of sulfadoxine-pyrimethamine resistance in Indian isolates of Plasmodium falciparum in the last two decades. Infect Genet Evol. 2015;36:190–198.
- Matondo SI, Temba GS, Kavishe AA, et al. High levels of sulphadoxine-pyrimethamine resistance Pfdhfr-Pfdhps quintuple mutations: a cross sectional survey of six regions in Tanzania. Malar J. 2014;13:152.
- Sutherland CJ, Fifer H, Pearce RJ, et al. Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the United Kingdom. Antimicrob Agents Chemother. 2009;53:3405–3410.
- Chauvin P, Menard S, Iriart X, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in Yaoundé, Cameroon: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2015;70:2566–2571.
- Oguike MC, Falade CO, Shu E, et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitol Drugs Drug Resist. 2016;6:220–229.
- NkoliMandoko P, Rouvier F, Matendo Kakina L, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in the democratic republic of the Congo: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2018;73:2704–2715.
- Lynch CA, Pearce R, Pota H, et al. Travel and the emergence of high-level drug resistance in Plasmodium falciparum in southwest Uganda: results from a population-based study. Malar J. 2017;16:150.
- Jambou R, Legrand E, Niang M, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963.
- Witkowski B, Khim N, Chim P, et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob Agents Chemother. 2013;57:914–923.
- Hott A, Tucker MS, Casandra D, et al. Fitness of artemisinin-resistant Plasmodium falciparum in vitro. J Antimicrob Chemother. 2015;70:2787–2796.
- Menard D, Khim N, Beghain J, et al. KARMA Consortium. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–2464.
- Yang C, Zhang H, Zhou R, et al. Polymorphisms of Plasmodium falciparum k13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis. 2017;17:560.
- Kamau E, ampino S, Amenga-Etego L, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–1355.
- Mita T, Culleton R, Takahashi N, et al. Little Polymorphism at the K13 propeller locus in worldwide plasmodium falciparum populations prior to the introduction of artemisinin combination therapies. Antimicrob Agents Chemother. 2016;60:3340–3347.
- Oboh MA, Ndiaye D, Antony HA, et al. Status of artemisinin resistance in malaria parasite plasmodium falciparum from molecular analyses of the Kelch13 gene in southwestern Nigeria. Biomed Res Int. 2018;2018:2305062.
- Mugittu K, Genton B, Mshinda H, et al. Molecular monitoring of Plasmodium falciparum resistance to artemisinin in Tanzania. Malar J. 2006;5:126.
- Menegon M, Talha AA, Severini C, et al. Frequency distribution of antimalarial drug resistance alleles among Plasmodium falciparum isolates from Gezira State, central Sudan, and Gedarif State, eastern Sudan. Am J Trop Med Hyg. 2010;83:250–257.
- Li J, Chen J, Xie D, et al. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int J Parasitol Drugs Drug Resist. 2016;6:54–59.
- Saha P, Guha SK, Das S, et al. Comparative efficacies of artemisinin combination therapies in Plasmodium falciparum malaria and polymorphism of pfATPase6, pfcrt, pfdhfr, and pfdhps genes in tea gardens of Jalpaiguri District, India. Antimicrob Agents Chemother. 2012;56:2511–2517.
- Sharma J, Dutta P, Khan SA, et al. Detection of point mutation in Plasmodium falciparum ATPase6 gene associated with artemisinin resistance from Assam and Arunachal Pradesh. J Vector Borne Dis. 2014;51:282–285.
- Kavishe RA, Paulo P, Kaaya RD, et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J. 2014; 13: 264.
- Nagesha HS, GJ C, AI S, et al. Mutations in the pfmdr1, dhfr and dhps genes of Plasmodium falciparum are associated with in-vivo drug resistance in West Papua, Indonesia. Trans R Soc Trop Med Hyg. 2001;95:43–49.