ABSTRACT
Global health security is constantly under threat from infectious diseases. Despite advances in biotechnology that have improved diagnosis and treatment of such diseases, delays in detecting outbreaks and the lack of countermeasures for some biological agents continue to pose severe challenges to global health security. In this review, we describe some of the challenges facing global health security and how genome editing technologies can help overcome them. We provide specific examples of how the genome-editing tool CRISPR is being used to develop new tools to characterize pathogenic agents, diagnose infectious disease, and develop vaccines and therapeutics to mitigate the effects of an outbreak. The article also discusses some of the challenges associated with genome-editing technologies and the efforts that scientists are undertaking to mitigate them. Overall, CRISPR and genome-editing technologies are poised to have a significant positive influence on global health security over the years to come.
Introduction
The global community suffers from a range of known health burdens as well as unanticipated outbreaks that place an additional stress on health systems. With the high degree of connectivity across political, economic, and geographical boundaries, these outbreaks are able to grow rapidly from local to global concerns.
Diseases such as malaria, TB, and HIV are endemic across large regions of the world and are responsible for a disproportionate degree of the global health disease burden[Citation1]. Additionally, influenza poses a perennial challenge that can be exacerbated by the emergence of a novel strain for which people do not have any preexisting immunity. Outbreaks of diseases that emerge suddenly and unexpectedly, such as Ebola or H1N1 influenza, have proven difficult to contain. The emergence of SARS-CoV-2 in China in 2019, and its rapid spread around the world, is a stark reminder of the risks posed by zoonotic diseases that are highly transmissible once they jump into human populations. Overall, the global health community lacks a comprehensive kit of diagnostic, preventative, and therapeutic tools for mitigating the effects of these diseases.
The rapid development of genome-editing tools, including TALENs, ZFNs, and CRISPR-Cas, that are able to programmatically target highly specific sequences of DNA or RNA provide a powerful new method of addressing global health challenges [Citation2]. CRISPR-Cas in particular has become widely used for specific targeting and cleavage of DNA or RNA, with many potential applications in biomedicine including studying the host–pathogen relationship, editing the host genome for pathogen resistance, detecting pathogens, and directly targeting pathogens for therapeutic purposes [Citation3].
In this article, we describe threats to global health security, analyze the challenges to identifying, diagnosing, and treating disease outbreaks, explore the opportunities that CRISPR-based tools are likely to provide for addressing those challenges, and discuss obstacles to using CRISPR to strengthen global health security.
Threats to global health security
Global health security is vulnerable to three types of biological threats in addition to the endemic diseases and global health burdens described above:
Outbreak of a well-known agent with a high capacity for dispersion and/or harm
The World Health Organization (WHO) maintains a list of priority diseases that pose special risks to global health security. In 2018, that list included Crimean-Congo hemorrhagic fever (CCHF), Ebola and Marburg viruses, and Lassa fever [Citation4]. These high priority diseases are determined based on a number of factors, but the most important are their infectivity, severity, transmissibility, and lack of available diagnostics and/or medical countermeasures. For example, CCHF exhibits a high mortality rate, is transmissible by both tick bite and contaminated bodily fluids, and there are no effective treatments or vaccines available [Citation5].
A large outbreak with one of these agents would most likely occur through spillover from an animal reservoir into the human population. For example, the 2014 outbreak of Ebola in West Africa is believed to have originated from contact between an infected bat and a young boy, who spread the disease into the human population [Citation6].
Outbreak of a known agent with new properties
Another type of biological threat is a new variant of an existing agent that has not previously been observed by the global health community. These threats can be generated either by natural evolution or intentional manipulation. Considering first natural evolution, these variants can appear as a response to environmental pressure or from the natural propensity of certain pathogens to mutate quickly. The influenza virus exemplifies the risks posed by rapid, naturally occurring mutations and this virus has caused multiple global outbreaks in humans and birds including the 2009 H1N1 pandemic [Citation7,Citation8] and 2004 outbreak of highly pathogenic H5N1 avian influenza [Citation9]. The 2015 Zika outbreak in the Americas is another illustrative example, where an infection that was generally considered mild obtained mutations that allowed it to spread rapidly and lead to cases of Guillain–Barré syndrome and microcephaly [Citation10]. Another looming problem for the global health community is the rise of antimicrobial resistance as bacteria evolve to become resistant to antibiotics [Citation11].
Separate from natural evolution, the intentional manipulation of a pathogen’s genetic material could also pose a potential source of a biological outbreak. Given the risk of natural evolution generating viruses with increased virulence or transmissibility, ‘gain of function’ research purposefully generates such strains for the research and risk assessment purposes to contribute to global health preparedness. For example, two controversial studies in 2012 demonstrated that avian influenza could become transmissible between ferrets – the standard model for human infection and transmission in influenza research [Citation12,Citation13]. A pathogen could also be engineered with enhanced virulence, transmissibility, or resistance to medical countermeasures for use as a weapon.
Outbreak of an unknown agent
A final challenge to global health security is the emergence of a completely novel agent. This novel infectious disease could be the result of either a completely undiscovered pathogen, or an organism misclassified as nonpathogenic. In the 2018 WHO list of priority diseases, this type of threat was simply titled ‘Disease X’[Citation4], as a broadly encompassing term for any disease that could pose an epidemic threat but is currently unknown to science. Recent examples of the appearance of novel threats include, among others, the appearance of Legionnaires’ disease in 1976 [Citation14], the SARS outbreak in China in 2003 [Citation15], the identification of Middle East Respiratory Syndrome coronavirus (MERS-CoV) in 2012 [Citation16], and the emergence of SARS-CoV-2 in China in 2019 [Citation17].
Challenges in identifying, diagnosing, and treating outbreaks
With the variety of agents that pose potential global health risks, mitigating these risks is a major challenge. While the logistical challenges associated with implementing new technological solutions across various geographic and political borders are considerable, the development of new technologies is integral to efforts to strengthen global health security. The ability to identify, diagnose, and treat infections is critical to minimizing the harm done by known global health burdens and unexpected outbreaks. For example, leading up to the WHO declaration of the global Ebola epidemic in 2014, a lack of comprehensive testing led to the misdiagnosis of many early cases and hid how widespread the epidemic had become before the official declaration [Citation18]. This section discusses some of the major challenges in developing technologies to identify, diagnose, and treat emerging and pandemic diseases.
Identifying and diagnosing new threats
Each of the three potential sources of biological outbreaks described above presents a different level of difficulty in identifying the threat present. The spread of a well-known agent is easier to identify since a wealth of information is likely available about the agent’s composition, its mechanisms of replication, and the symptoms of infection. Developing tools to identify new threats from mutated or modified agents faces increased challenges relative to well-known or wild-type agents, given that mutations could interfere with the ability to detect the pathogen if they occur in regions critical for developed assays. Thus, development of robust assays that can account for genetic insertions and deletions, genetic drift, and rearrangements are essential for early detection of a potential outbreak.
The greatest challenge in identifying the causative agent of an outbreak occurs when the agent is not already considered a biothreat. If the agent is a completely unknown species or strain, a wide panel of assays are required to narrow down the type of threat and additional sequencing, biochemistry, and microbiology may be necessary to fully evaluate the agent. The challenge is somewhat different if the threat is from a manipulated or mutated non-pathogen, as the symptoms of infection may be generally mild (as was the case with Zika [Citation10]) or resemble those of another known agent, especially if an unknown agent acquired pathogenic DNA or RNA from a known agent. Further, potential contamination by co-occurring organisms could lead to incorrect conclusions about the threat. A classic example is the misidentification of Haemophilus influenzae as the causative agent of ‘the flu’ before the discovery of the influenza virus [Citation19]. Technologies that can rapidly identify many potential threats are critical for a timely response to a potential outbreak.
Developing new vaccines and therapeutics
Generally, there are two major routes to treating infections. First, antibiotics and antivirals are small molecules that bind to a specific site in a biological molecule to disrupt the pathogen’s life cycle. Alternatively, vaccines or monoclonal antibodies are used to treat prophylactically (although some diseases have a period when vaccines are still effective after infection) by priming the human immune system to recognize a specific agent. Ideally, either course of treatment is designed to cover as many strains of an agent as possible, given that one of the biggest challenges in developing effective treatments is the tendency for pathogens to mutate and recombine.
Influenza is a good example of a pathogen that is difficult to treat with these approaches (). It is an RNA virus made up of eight unique genomic segments that can reshuffle during co-infection to drive diversity on top of the high mutation rate afforded by its RNA-dependent RNA polymerase [Citation20]. Over the course of roughly 80 years of research into influenza, two drugs were found to be most effective: amantadine and oseltamivir/zanamivir (better known as Tamiflu and Relenza, respectively) [Citation21]. However, overuse of amantadine has now led to the near-complete resistance of influenza to this drug, leaving only one class of drugs available to treat infections [Citation22]. Vaccines for influenza have been available since shortly after the virus was discovered, with improved strain monitoring and technology improving the effectiveness of the vaccine in more recent years. However, the vaccine’s effectiveness is still fairly low due to the mutation rate of the virus, either from wild strains developing escape mutants, or mutations occurring during the growth of the vaccine strain in eggs [Citation23]. Despite large worldwide investments into influenza research and the tools available to modern biology, mutation and recombination events present a major challenge in developing new vaccines and therapeutics for influenza.
Figure 1. The influenza viral replication pathway involves binding to the cell surface, encapsulation, release of the ribonucleic protein (RNP) complexes, transport of the RNPs to the nucleus, generation of new RNPs, packaging, and release. Currently, two drugs have been developed to inhibit influenza replication: amantadine (purple) that blocks RNP release and oseltamivir (blue) that blocks budding of new viruses. Cas13 has the potential to directly target viral RNAs to inhibit replication, although packaging in RNPs is likely to block access to the RNA for CRISPR targeting. Cas9 could potentially be used to edit the genome to the make the cell resistant to infection. Finally, CRISPR can also be used in a diagnostic setting to detect viral RNAs using techniques like SHERLOCK[Citation42] and DETECTR[Citation37]
![Figure 1. The influenza viral replication pathway involves binding to the cell surface, encapsulation, release of the ribonucleic protein (RNP) complexes, transport of the RNPs to the nucleus, generation of new RNPs, packaging, and release. Currently, two drugs have been developed to inhibit influenza replication: amantadine (purple) that blocks RNP release and oseltamivir (blue) that blocks budding of new viruses. Cas13 has the potential to directly target viral RNAs to inhibit replication, although packaging in RNPs is likely to block access to the RNA for CRISPR targeting. Cas9 could potentially be used to edit the genome to the make the cell resistant to infection. Finally, CRISPR can also be used in a diagnostic setting to detect viral RNAs using techniques like SHERLOCK[Citation42] and DETECTR[Citation37]](/cms/asset/36d7f9a0-39c8-42f4-8664-0ea848c635ce/ypgh_a_1880202_f0001_oc.jpg)
Introduction to genome-editing technologies
The recent development of genome-editing technologies, including TALENs [Citation24], ZFNs [Citation25], and CRISPR-Cas [Citation26], has fundamentally changed the direction of biomedical research by providing new tools that can accurately edit an organism’s genome, which may belong to a human, pathogen, or animal model. CRISPR-Cas9 in particular has been a major focus in biotechnology due to its potency, precision, and ease of use. The impact of CRISPR-Cas9 has been so pervasive that Emmanuelle Charpentier and Jennifer Doudna were awarded the 2020 Nobel Prize in Chemistry for its discovery only eight years prior [Citation27].
As a testament to the global reach of CRISPR, Addgene reports distributing over 180,000 CRISPR reagents to over 4,000 institutions across 87 different countries as of June 2020 [Citation28]. As of the end of 2017, nearly 1,500 patents originating in 28 countries had already been filed in relation to CRISPR [Citation29]. The number of filings has displayed exponential growth, with thousands of additional patents likely having been filed since then.
Another sign of increasing investment in CRISPR technologies is the increasing number of patents filed on CRISPR technologies by industry [Citation30]. Between 2013 and 2015 alone, over USD 600 million had been raised via venture capital or public markets for applications of Cas9 [Citation31]. The majority of companies working with CRISPR at this time were focused on either human therapeutics or biotechnology research, with some other applications in industrial biotech or agriculture [Citation30]. Since 2015, additional companies with interests in applying CRISPR to global health challenges have emerged [Citation32].
While this discussion focuses on Class 2 CRISPR systems (those only containing one editing enzyme such as Cas9) due to their widespread adoption and ease of use, many of the applications described below could also be theoretically enabled by TALENs, ZFNs, and Class 1CRISPR systems (whose editors are multiprotein complexes) as well, albeit with increased difficulty.
Overview of Class 2 CRISPR-Cas editors
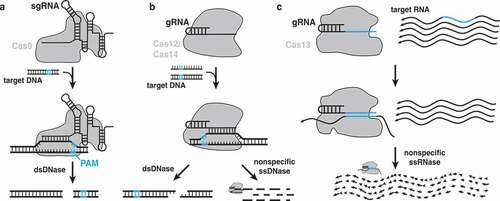
In recent years, CRISPR effectors including Cas9, Cas12, Cas13, and Cas14 have been used to develop a plethora of tools useful for basic science as well as biomedical research. Each of these four Cas proteins is somewhat different, although each is a single large protein that uses a guide RNA to direct it to a specific nucleic acid target sequence, unlike Class 1systems that use a multiprotein complex. Cas9 cleaves double-stranded DNA to leave blunt ends [Citation33–35] ()). Cas12 recognizes single or double-stranded DNA to activate a nuclease that can leave a staggered cut (with double-stranded DNA) or nonspecifically degrade single-stranded DNA [Citation36–39] ()). Cas13 is a nonspecific RNase that is activated when it matches its guide RNA to a target [Citation40–44] ()). Cas14, also known as Cas12f, functions the same way as other Cas12 enzymes [Citation45,Citation46] ()). Cas9 in particular has seen major use in the rapid development of new animal disease models, drastically cutting the time and cost of development. Between Cas9, Cas12, and Cas13, many engineered forms exist that may contain one or more of the following modifications: complete or partial abrogation of nuclease activity (dCas) [Citation47], fluorescent tagging [Citation48], ability to be activated by chemical or light stimuli [Citation49,Citation50], and the ability to recruit chromatin remodeling factors [Citation51]. These modifications have formed the basis of a number of new tools whose applications are discussed further below.
Figure 2. Differences between Cas9, Cas12/Cas14, and Cas13. a) The Cas9 protein binds to two RNAs that can be fused into a single guide RNA (sgRNA). The Cas9 RNP complex recognizes a DNA sequence matching the spacer sequence next to a matching protospacer adjacent motif (PAM) sequence, unwinds the DNA, and performs a blunt cut near the PAM sequence. b) Cas12 binds a small guide RNA (gRNA) that recognizes a double-stranded DNA target, unwinds the DNA, and cuts either both strands with a staggered cut distal from the PAM sequence. Binding to a DNA target, either single- or double-stranded, can also activate nonspecific DNAse activity that cleaves ssDNA nonspecifically. Cas14 functions similarly. c) Cas13 forms a complex with its gRNA to find and bind a matching RNA sequence, which activates a nonspecific nuclease that nonspecifically cleaves all surrounding single-stranded RNA
Technologies for editing genomic DNA
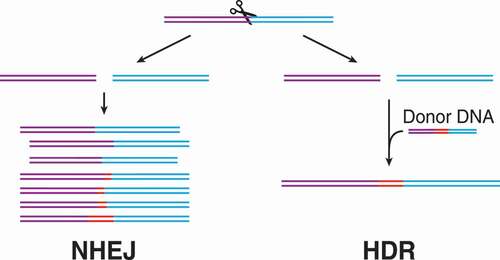
Much of the excitement surrounding CRISPR technologies comes from its use as a genome-editing tool and its potential for developing new treatments for genetic disease. In DNA editing applications, base changes are created by the way that eukaryotic cells repair their DNA after a cleavage event. Two possible modes of DNA repair occur: 1) non-homologous end joining (NHEJ) in which the cell simply tries to ‘paste’ the broken DNA strands back together, typically resulting in insertions or deletions, and 2) homology directed repair (HDR) in which the cell uses a homologous donor sequence to determine what bases are missing/damaged in the break [Citation52] (). The two repair mechanisms result in fundamentally different outcomes. NHEJ frequently results in nonsense or missense mutations, while HDR can either fully repair a double-strand break or introduce specific insertions or deletions. HDR requires the use of another, homologous DNA sequence to serve as a blueprint for repair. Thus, only supplying a Cas protein and guide RNA will usually result in NHEJ repair and lead to the silencing of a gene. To create intentional insertions, mutations, or deletions a DNA sequence must be supplied as well to serve as a donor to use the HDR pathway and avoid the random result of NHEJ repair, which competes with HDR. Both of these types of editing are employed to make genetic changes depending on the desired outcome [Citation52].
Figure 3. Differences between non-homologous end joining (NHEJ) and homology directed repair (HDR). After a double-stranded break, there are two major mechanisms of cell-mediated repair: NHEJ and HDR. If a donor DNA is present with homology to each side of the break, the HDR pathway will lead to templated repair. Thus, HDR can lead to highly specific editing for biotechnology applications. In competition with HDR is NHEJ, which sticks the broken DNA ends together, typically involving the generation of insertions or deletions (indels). These indels are usually disruptive to a gene if in a coding region
Alternatively, CRISPR effectors have been coupled with deaminases to allow individual bases to be edited without the need for a double-strand break [Citation53,Citation54] ()). These base editors can introduce specific point mutations into a DNA sequence without the need to rely on NHEJ or supply an extraneous template for HDR. Currently, a number of base editors exist using Cas9 [Citation55,Citation56] or Cas12 [Citation57] that are able to convert AT base pairs to GC or vice versa. Base editing in RNA molecules has also been demonstrated with Cas13 [Citation58], although there is disagreement in the field as to its effectiveness [Citation59].
Figure 4. Genome-editing methods without double-strand breaks. a) In base editing, a catalytically dead Cas9 or Cas9 nickase is fused to a phage Gam protein (green), cytidine deaminase (blue), and uracil glycosylase inhibitor (UGI; purple) to create one version of base editor [Citation56]. After binding to a DNA target, the deaminase accesses the unwound, single-stranded DNA and converts C→T (for cytidine deaminase) or A→I (for adenosine deaminase). The edited base is then either corrected back by the repair machinery or the opposite nucleotide is changed to correct the base pairing. The Gam protein and UGI are included to help bias the repair toward the desired edited product and minimize off-target effects. b) In prime editing, a Cas9 nickase is fused to a reverse transcriptase and carries a prime editing guide RNA (pegRNA). The pegRNA contains a tail with a reverse complement to the nicked strand and an editing template sequence. The pegRNA anneals to the nicked ssDNA, which then serves a primer for reverse transcriptase to extend the genomic DNA into the designed edit in the pegRNA tail. After Cas9 release, the excess DNA creates two possible flap overhangs, which can either be resolved to return to original sequence or include the designed edit
![Figure 4. Genome-editing methods without double-strand breaks. a) In base editing, a catalytically dead Cas9 or Cas9 nickase is fused to a phage Gam protein (green), cytidine deaminase (blue), and uracil glycosylase inhibitor (UGI; purple) to create one version of base editor [Citation56]. After binding to a DNA target, the deaminase accesses the unwound, single-stranded DNA and converts C→T (for cytidine deaminase) or A→I (for adenosine deaminase). The edited base is then either corrected back by the repair machinery or the opposite nucleotide is changed to correct the base pairing. The Gam protein and UGI are included to help bias the repair toward the desired edited product and minimize off-target effects. b) In prime editing, a Cas9 nickase is fused to a reverse transcriptase and carries a prime editing guide RNA (pegRNA). The pegRNA contains a tail with a reverse complement to the nicked strand and an editing template sequence. The pegRNA anneals to the nicked ssDNA, which then serves a primer for reverse transcriptase to extend the genomic DNA into the designed edit in the pegRNA tail. After Cas9 release, the excess DNA creates two possible flap overhangs, which can either be resolved to return to original sequence or include the designed edit](/cms/asset/59f362d9-f5c6-448d-bebc-880ba45dae0f/ypgh_a_1880202_f0004_oc.jpg)
Another method to introduce genome edits without a double-stranded break called ‘prime editing’ uses a Cas9 nickase fused to a reverse transcriptase and a prime editing guide RNA (pegRNA) to template specific base changes [Citation60] ()). The pegRNA contains a long tail with the reverse complement of the target DNA sequence at its end that serves as a primer for the reverse transcriptase after Cas9 binds. Programmed edits are introduced through changes in the bases of the pegRNA tail that are ultimately converted to DNA by the reverse transcriptase in a new DNA strand. The eukaryotic DNA repair machinery then repairs the nick, using either the original sequence or the new DNA strand created by reverse transcription. The use of reverse transcriptase helps overcome the accuracy issues associated with deaminase tethering and allows for more flexible editing designs.
Technologies for controlling genes without editing
A major challenge in genome editing is the possibility of permanent unwanted edits. To avoid this problem, researchers have developed other CRISPR tools that do not edit the genome permanently, but instead recruit chromatin remodeling proteins [Citation61,Citation62] or methyltransferases [Citation51] to alter expression levels of the CRISPR targeted gene. Depending on the method used, the alterations to gene expression levels can be long-lasting, resulting in phenotypic changes without the need for genotypic changes.
Another strategy to silence genes without a genotypic alteration is to target the mRNA product instead of the gene itself. As described above, Cas13 can be used to target individual mRNA transcripts for cleavage [Citation44,Citation63,Citation64] or base editing [Citation58]. Some work with Cas9 has demonstrated that certain homologs can also target RNA [Citation65,Citation66], including the hepatitis C virus in eukaryotic cells [Citation67].
Advantages of CRISPR-based genome editing
One of the biggest changes in biomedical research brought about by genome editing, and especially CRISPR, is the ability to create new cell and animal models quickly and efficiently. Unlike ZFN and TALEN technologies, CRISPR does not require redesign of the effector nuclease, only the guide RNA, which is much simpler. Synthesizing guides is relatively inexpensive, and many can be tested in a short period of time. Further, the high activity of Cas nucleases results in a higher probability of making a desired mutation or change, which shortens screening times and becomes more significant as the maturation time of the model organism increases [Citation68,Citation69]. The faster turnaround time of new model organisms benefits biological research as a whole, allowing for more appropriate testing environments and less time spent building research materials and more time collecting new data. This was demonstrated during the COVID-19 pandemic with the creation of a mouse model expressing the human angiotensin-converting enzyme 2 (hACE2) using CRISPR-Cas9 in place of the mouse version of the enzyme [Citation70]. The development of monoclonal antibodies (mAbs) and vaccines for prophylactic treatments can also benefit from CRISPR/Cas9 through increased rates of cell line generation as well as the ability to perform genomic screens to identify what epitopes are targeted by mAbs [Citation71].
Future potential for genome-editing solutions in global health security
In the years since the publication describing the basic mechanisms of Cas9 DNA cleavage [Citation27], new techniques and tools using CRISPR have been developed across all biological fields highlighting how pervasive this technology has become [Citation72]. This section describes several ways in which CRISPR can strengthen global health security developing new means for identifying and characterizing pathogenic threats, detecting and diagnosing infectious agents, and developing new treatments for these diseases.
Identifying and characterizing new threats
CRISPR has been used to develop a suite of tools that scientists can use to better understand new and existing pathogens. One of the most beneficial uses of CRISPR for pathogen research has been the use of CRISPR interference to identify host factors critical to an agent’s replication cycle. With CRISPR interference, a catalytically dead Cas9 (dCas9) fused to a chromatin remodeling protein is guided to the beginning of a gene to silence expression [Citation51]. By using a library of gRNAs covering the host organism’s genome, individual genes or gene clusters can be knocked down in parallel to identify which genes’ silencing leads to a survival phenotype [Citation73]. Thus, in one straightforward experiment, a list of genes involved in pathogen biogenesis can be obtained. Similarly, CRISPR interference knockdowns can be used to work out the function of an agent’s genes without needing to make recombinant virus or create transgenic cell lines or animals, both of which can be time-consuming.
It is worth noting here that many of these knockdown assays were previously possible using RNA interference (RNAi) pathways [Citation74], possibly more effectively than CRISPR in certain cases. CRISPR, however, affords the additional flexibility of being able to make inheritable genetic changes by switching to the wild-type Cas9 protein for genome editing or with epigenetic markers [Citation51]. These genetic changes can be made with a high degree of precision allowing for targeted gene knockouts to study the effect of host gene functions in the pathogen life cycle, which were previously much more laborious to create [Citation75]. A recent application of CRISPR knockout libraries for combating global health threats was seen during the COVID-19 pandemic. Multiple groups identified host factors involved in the biogenesis of Coronaviridae family viruses, providing new lists of potentially druggable targets for inhibiting this family of viruses [Citation76–78].
Diagnosing new threats
A major factor in containing outbreaks is the speed at which a potential outbreak can be detected and the causative agent can be identified in the affected population. A particularly useful application of CRISPR technology is to diagnose the presence of a specific pathogen in an environmental or clinical sample. There are two similar strategies that are currently employed, SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) [Citation42], which uses Cas13, and DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter) [Citation45], which uses Cas12. With SHERLOCK input, RNA or DNA from a sample containing a potential agent is amplified and converted to RNA with recombinase polymerase amplification (RPA) [Citation79]. The amplified RNA is then exposed to Cas13 with a guide RNA to probe for the presence of a specific sequence in the agent. If that sequence is present, the nonspecific RNase activity of Cas13 is activated ()), resulting in cleavage of a reporter RNA that releases a fluorescent signal. The study’s authors additionally showed that SHERLOCK can be reconstituted on paper for cheaper deployment into the field to detect Zika and Dengue virus in human samples [Citation42]. Other work building on the SHERLOCK platform demonstrated that many different pathogens could be detected at once by multiplexing CRISPR detection in nanoliter droplets [Citation80].
The workflow for DETECTR is essentially the same as SHERLOCK, except that Cas12 is used instead of Cas13 to recognize and cleave DNA sequences instead of RNA sequences [Citation37]. Using DETECTR, the authors demonstrated the ability to distinguish between two closely related strains of human papillomavirus (HPV).
Recent work describing paper-based agent detection [Citation81] is particularly exciting, and the recent use of Cas12 and Cas13 to detect RNA or DNA from potential agents is poised to have a major impact on outbreak response and general agent monitoring in the field [Citation42,Citation82]. Future improvements to increase the detection limit [Citation83], increase signal-to-noise strength, and shorten assay run time may make these types of paper assays a standard field method for agent detection. In a clinical setting, the use of CRISPR diagnostics would greatly benefit from the development of standardized testing devices. Such devices could become commonplace in hospitals and diagnostic labs in the near future for fast and sensitive pathogen detection and genotyping.
CRISPR-based diagnostics are now receiving their first test during the COVID-19 pandemic. Both the DETECTR and SHERLOCK platforms were used to develop specific diagnostics for SARS-CoV-2 within weeks of the WHO’s declaration of a public health emergency of international concern [Citation84,Citation85]. In early May 2020, SHERLOCK became the first CRISPR-based diagnostic tool to be authorized for emergency use during the pandemic by the U.S. Food and Drug Administration (FDA), with authorization of DETECTR following soon after in late August 2020. Both methods have been shown to be rapid, with SHERLOCK producing results within 90 minutes [Citation84] and DETECTR able to report results in approximately 30 minutes [Citation86]. In another major step forward, DETECTR has been made highly portable, and is now capable of using a cell phone camera in a compact laser device for quantitative detection in the field, opening the potential for increased throughput and rapid response times to outbreaks without the need for traditional laboratory equipment [Citation86].
Developing new treatments
The potential for genome editing, especially CRISPR technologies, to impact the development and use of new treatments is tremendous. Genome-editing technologies have a major advantage over traditional drugs in that they can target the genetic basis of a disease, and do not require the development time and cost to understand and block interactions between biological molecules with complex three-dimensional topography. CRISPR screens to identify key genes associated with agent replication mechanisms are likely to be increasingly useful for quickly identifying which factors of infection are important to consider when developing treatments [Citation75].
The first human recipient of ZFNs to treat Hunter Syndrome began a new period of genome editing in humans, and was followed shortly by a wave of clinical trials using CRISPR-Cas genome editors [Citation87]. Additional work on curing a variety of genetic diseases with CRISPR, such as muscular dystrophy [Citation88], is underway [Citation89]. This progress on treating genetic diseases can feed back to impact global health security efforts. For example, retroviruses like HIV can be removed from the population by excising the viral DNA from the genome, which has already been tested in mice [Citation90]. There is more work to be done, however, as concerns have been raised about the possibility of escape mutations driving further evolution of the virus [Citation91]. Alternatively, viral infections could be combated by editing the human genome to resist viral infections altogether. For example, genome editing could be used to truncate the CCR5 receptor to mimic the CCR5-Δ32 mutation that provides innate immunity to the virus [Citation92].
Directly targeting viruses is also a potential route to treating disease outbreaks [Citation93,Citation94]. CRISPR-based antivirals are particularly exciting due to the ease of generating large numbers of guide RNAs relative to the difficulty of developing small molecule drugs that pathogens will likely eventually evolve resistance to. The adaptation of a prokaryotic antiviral system (i.e. CRISPR) for use in humans for antiviral purposes is still in its early stages, but future successes could provide a completely new method for rapidly combating emerging viral diseases. We are also likely to see a large expansion in the use of RNA targeting and editing. Concerns over accidental edits to patient genomes and the degree of permanency implied make direct RNA targeting or editing very attractive.
Many of the agents on the WHO’s list of priority diseases and the CDC’s list of potential biological weapons are RNA viruses that do not convert to DNA, requiring direct RNA targeting to be cleaved. Coronaviruses, enveloped positive-sense RNA viruses, fall into this category as well. Spurred by the SARS-Cov-2 outbreaks, researchers demonstrated the potential use of Cas13 as a treatment for targeting and clearing the SARS-Cov-2 virus from human cells [Citation95]. These proof-of-concept experiments are key to providing a path toward the use of CRISPR for directly targeting viral pathogens.
Similar to its potential use in targeting viral infections, CRISPR has the potential to serve as a tool for treating antibiotic-resistant bacterial pathogens [Citation96]. Multiple studies have shown that antibiotic-resistance genes within resistant bacteria can be specifically targeted with CRISPR to selectively kill bacterial pathogens from a community [Citation97–100]. Appropriate delivery remains an issue though. While plasmid or phage vectors are effective in introducing DNA to bacterial populations, they could easily escape into the environment, and lipid-based protein delivery approaches are not yet effective enough to produce a strong effect [Citation98]. However, these early efforts into CRISPR-based ‘antibiotics’ provide us a starting point to look beyond small molecules for treating antibiotic-resistant bacterial infections.
Other uses for genome editing pertaining to global health security
Genome-editing technologies also have a number of relevant applications to global health security outside of the human body. While not covered in this review, gene drives have the potential to control vectors and minimize the possibility of certain outbreaks, either by eliminating the vector entirely or by editing the vector’s genome to remove its ability to carry a particular agent [Citation101].
Environmental surveillance systems could also be developed to monitor the presence of agents. Biosensing circuitry has been a major focus of the synthetic biology community [Citation102]. The incorporation of such biosensing circuitry into crop plants using CRISPR technologies could provide a real-time detection system for regional agents of concern.
Challenges and risk assessment for genome-editing applications
As with many major scientific advances, there are a number of challenges and risks associated with developing and using CRISPR gene-editing technologies, discussed below.
Scientific challenges
Progress in genome-editing research faces a number of the standard challenges expected in science, but three, in particular, are the most notable: measuring the level of unwanted editing, making editors resistant to naturally occurring mutations, and delivering genome editors.
First, measuring the degree of off-targeting (the level of unwanted editing) that occurs is fairly challenging. A number of techniques have been developed so far, such as TTISS [Citation103], GUIDE-seq [Citation104], CIRCLE-seq [Citation105], VIVO [Citation106], and Digenome-seq [Citation107], but the need to separate naturally occurring insertions, deletions, and mutations from those caused by gene-editing off-targets remains a major challenge of the field. While measuring off-target edits is challenging, there is simultaneously a large body of work aimed at improving editors to minimize off-target effects. Efforts to improve the accuracy of S. pyogenes Cas9 have been fruitful, resulting in a hyper-accurate Cas9 variant [Citation108], but took years of development that included other variants that had lower on-target activity [Citation109]. Both of these efforts are critical for reducing human health risks, as unwanted genome edits could potentially lead to a lethal or cancerous outcome. One solution to avoid accidental permanent DNA editing is to instead target RNA (as described above), which is temporary by its very nature.
Second, another challenge with genome-editing technology for treatments of biothreat agents is the high propensity of those agents to mutate. Indeed, directly targeting a pathogen creates the possibility of stimulating its intrinsic mutation rate [Citation110]. As previously stated, many of the agents identified by the CDC as biosecurity threats are RNA viruses, which as a group tend to rapidly mutate, making vaccine and traditional drug development difficult. Targeting specific strains, or incomplete clearing of virus from the host, could result in an artificial selection for viruses with mutations or sequences that avoid CRISPR targeting. The same would be true for other pathogens as well. The mutation of bases that are critical for CRISPR targeting, such as those in the seed region of the target sequence, or bases in the protospacer adjacent motif [Citation111] could potentially lead to inactive treatments. Care must be taken to choose guide RNAs that target the most conserved regions of a gene to avoid mutation issues, while working within the restrictions placed by the PAM sequence required by each effector. Similarly, the appearance of single nucleotide polymorphisms within the human population presents a challenge when designing prophylactic genome-editing treatments, as certain guide sequences many only result in proper targeting in a subset of the global population.
Third, the problem of delivery is probably the biggest scientific hurdle that must be cleared to make CRISPR genome editing a commonplace treatment. Small molecule drugs, when properly designed, have the advantage of high stability and ease of transport into cells. Biologics like Cas:gRNA complexes are susceptible to proteases and nucleases in the body, which can quickly degrade foreign biological molecules. A number of solutions have been proposed [Citation91], but are further complicated by the large size of most Cas effectors relevant for gene-editing [Citation112], which are more difficult to package into traditional vectors, like the adeno-associated virus [Citation113]. The need to deliver the Cas protein and gRNA to a large number of cells, but only a specific subset, while avoiding triggering the immune system complicates the delivery problem. Studies have demonstrated that a large fraction of patients tested had antibodies for the Cas9 proteins from Streptococcus pyogenes and Staphylococcus aureus, two commonly used Cas9 editors, as well as antigen-specific T-cells against SauCas9114. The authors point out that delivery methods where Cas editors are not exposed in the serum (such as those packaged in a delivery vehicle) are safe from antibodies. However, the challenge of avoiding the antigen-specific T-cells, which can clear cells that contain Cas gene editors if the degraded editor is presented on the cell surface [Citation114], still seemingly remains. Other work has demonstrated that improper preparation of the gRNAs for Cas9 or Cas12 canresult in an immune response [Citation115]. The problem of CRISPR packaging and delivery is likely to dominate the scientific challenges facing CRISPR genome-editing tools, but the efforts toward treating infectious disease with CRISPR-Cas will likely benefit from the lessons learned during development of other biological therapeutics.
Risks to human health and biosecurity associated with genome-editing tools
While genome editing has enormous potential for improving human health, there are risks associated with the intentional, reckless, or accidental misuse of this technology. At the same time that genome editing enables new discoveries and the creation of new tools to more precisely engineer living organisms, these advances could be used to cause harm, whether deliberately through the creation of a biological weapon, accidentally in the event of a biosafety failure that results in the escape of an engineered organism into the environment, inadvertently through the discovery of new knowledge or vulnerabilities, or recklessly through inappropriate conduct that harms the health of humans or the ecosystem [Citation116]. For example, in 2018, a Chinese researcher announced that he had secretly used CRISPR to edit human embryos with the intention of making them resistant to HIV infection [Citation117]. This experimentation with human germline editing was quickly and widely condemned as unethical by the scientific community, as leading researchers in the field, although divided, had previously agreed no edited zygote should be taken to term [Citation118–120]. After the birth of the edited babies was revealed, many leaders of the CRISPR field called for a worldwide moratorium on human germline editing until the social, ethical, and technical dimensions of human germline editing could be more fully addressed [Citation121]. However, no such moratorium has since materialized.
Genome-editing tools could also pose a threat to global health security if they are directly used as a weapon [Citation122]. The ability of genome-editing tools to delete, suppress, or amplify the expression of specific genes could be used to disrupt the normal functioning of specific physiological systems, preferentially target specific target populations with rare genetic mutations, or hijack the human microbiome to produce harmful biochemicals or manipulate the production of natural compounds. There is also the risk that genome editing could be used to create gene drives that could spread deleterious genes through animal or plant populations. However, developing such CRISPR-based weapons would require significant effort and would face the same challenges in achieving widespread dissemination as traditional biological weapons in addition to the new challenges in delivering CRISPR-based therapies to the correct part of the human body.
Conclusion
As the COVID-19 pandemic has demonstrated, unexpected biological threats can emerge suddenly and spread rapidly. These features place a premium on investments in technologies that can accelerate the development and deployment of diagnostics, vaccines, and therapeutics. Genome-editing technologies such as CRISPR have the potential to develop a range of new tools to characterize, diagnose, and treat new and existing pathogens. Of particular promise is the development of highly sensitive CRISPR-based detection platforms that promise to bring rapid surveillance of pathogens into the field. Since many of the emerging biological threats originate in developing countries that may lack access to state-of-the-art laboratories, developing new diagnostic tools that are cheap, readily portable, and require minimal lab equipment are key to addressing new biothreats in a timely manner. CRISPR-based treatments that directly target a pathogen or host-cell receptors are also a promising alternative to small molecule drugs or vaccines. CRISPR is also making it possible for scientists to conduct larger, faster, and more comprehensive experiments designed to better understand the key features of pathogens. In addition, CRISPR enables a faster and simpler process for creating more accurate animal models for studying a new disease. Several groups and scholars have called for government and international organizations to more fully integrate advances in the life sciences and biotechnology, such as CRISPR, into national and international efforts to enhance pandemic preparedness and response [Citation123–125]. If the benefits of CRISPR can be adequately balanced with the risks involved with its use, the biotechnology, public health, and medical communities will make great strides in strengthening global health security.
Acknowledgments
We thank Jennifer Weisman for her thoughtful feedback and David Relman, Edward Perello, and Sarah Denton for their support of the larger “Editing Biosecurity” research project with which this work was affiliated.
Disclosure statement
KEW is currently employed by Arbor Biotechnologies; the statements and opinions expressed in this work are solely attributable to the author.
Data Availability Statement
No data were generated as part of this work.
Additional information
Funding
References
- Medicine, N. A. of S., Engineering, and, Division, H. and M., Health, B. on G. & States, C. on G. H. and the F. of the U. Global health and the future role of the United States. (2017) doi:https://doi.org/10.17226/24737.
- Carroll D. Genome editing: past, present, and future. Yale J Biol Med. 2017;90(4):653–659.
- Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869.
- Mehand MS, Al-Shorbaji F, Millett P, et al. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. 2018;159:63–67.
- Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64(3):145–160.
- WHO Ebola Response Team. After ebola in West Africa–unpredictable risks, preventable epidemics. N Engl J Med. 2016;375:587–596.
- Smith GJD, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125.
- SteelFisher GK, Blendon RJ, Bekheit MM, et al. The public’s response to the 2009 H1N1 influenza pandemic. N Engl J Med. 2010;362(22):e65.
- Chen H, Smith GJD, Zhang SY, et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436(7048):191–192.
- World Health Organization. Zika virus, microcephaly and Guillain-Barré syndrome situation report. (2016).
- Spížek J, Novotná J, Řezanka T, et al. Do we need new antibiotics? The search for new targets and new compounds. J Ind Microbiol Biotechnol. 2010;37(12):1241–1248.
- Herfst S, Schrauwen EJA, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541.
- Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428.
- Jr WCW. Legionnaires disease: historical perspective. Clin Microbiol Rev. 1988;1(1):60–81.
- Zhao Z, Zhang F, Xu M, et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52(8):715–720.
- Zaki AM, Boheemen S, Van, Bestebroer T, et al. A. D. M. E. & Fouchier, R. A. M. isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820.
- Burki T. Outbreak of coronavirus disease 2019. Lancet Infect Dis. 2020;20(3):292–293.
- Goba A, Khan SH, Fonnie M, et al. An outbreak of ebola virus disease in the lassa fever zone. J Infect Dis. 2016;214(suppl 3):S110–S121.
- Smith WAndrews CH, Laidlaw PP, et al. Obtained from influenza patients. Lancet. 1933;222(5732):66–68.
- Samji T. Influenza A: understanding the viral life cycle. Yale J Biol Med. 2009;82:153–159.
- Barik S. New treatments for influenza. BMC Med. 2012;10(1):104.
- Hussain M, Galvin HD, Haw TY, et al. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist. 2017;10:121–134.
- Zost SJ Parkhouse K, Gumina ME et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U.S.A. 114, 12578–12583 (2017).
- Sanjana NE, Cong L, Zhou Y, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7(1):171–192.
- Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646.
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355.
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821.
- Tsang J, LaManna CM. Open sharing during COVID-19: CRISPR-based detection tools. Crispr J. 2020;3(3):142–145.
- Martin-Laffon J, Kuntz M, Ricroch AE. Worldwide CRISPR patent landscape shows strong geographical biases. Nat Biotechnol. 2019;37(6):613–620.
- Brinegar K, Yetisen AK, Choi S, et al. The commercialization of genome-editing technologies. Crit Rev Biotechnol. 2017;37(7):1–12.
- Erp PB, Van, Bloomer G, Wilkinson R, et al. The history and market impact of CRISPR RNA-guided nucleases. Curr Opin Virol. 2015;12:85–90.
- Murray J. The class of 2019: five synthetic biology companies to watch this year. synbiobeta. https://synbiobeta.com/the-class-of-2019-five-synthetic-biology-companies-to-watch-this-year/ (2019). Web. 15 Dec. 2020.
- Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–86 (2012).
- Jiang F, Doudna JA. CRISPR-cas9 structures and mechanisms. Annu Rev Biophys. 2017;46(1):505–529.
- Jiang F, Zhou K, Ma L, et al. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348(6242):1477–1481.
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771.
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes single-stranded DNase activity. Science. 2017;360:1–29.
- Swarts DC, Oost J van der, Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell. 2017;66(2):221–233.e4.
- Yan WX, Hunnewell P, Alfonse LE, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2018;363:eaav7271–9.
- East-Seletsky A, O'Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273.
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573–aaf5573.
- Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442.
- Smargon AA, Cox DBT, Pyzocha NK, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided rnase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630.e7.
- Yan WX, Chong S, Zhang H, et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;327-339.e5.
- Harrington LB, Burstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842.
- Karvelis T, Bigelyte G, Young JK, et al. PAM recognition by miniature CRISPR–Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48(9):5016–5023.
- Larson MH, Gilbert LA, Wang X, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–2196.
- Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491.
- Oakes BL, Nadler DC, Flamholz A, et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nature. 2015;34:646–651.
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200.
- Amabile A, Migliaria A, Capasso P, et al. Inheritable silencing of endogenous genes by hit- and-run targeted epigenetic editing. Cell. 2016;167(1):219–224.e14.
- Zaboikin M, Zaboikina T, Freter C, et al. Non-homologous end joining and homology directed dna repair frequency of double-stranded breaks introduced by genome editing reagents. PLoS ONE. 2017;12(1):e0169931.
- Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424.
- Hess GT, Tycko J, Yao D, et al. Methods and Applications of CRISPR-mediated base editing in eukaryotic genomes. Mol Cell. 2017;68(1):26–43.
- Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017;3(8):eaao4774.
- Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471.
- Li X, Wang Y, Liu Y, et al. Base editing with a Cpf1–cytidine deaminase fusion. Nat Biotechnol. 2018;36(4):324–327.
- Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;550:eaaq0180–15.
- Vogel P, Moschref M, Li Q, et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat Methods. 2018;15(7):535–538.
- Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157.
- Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451.
- Mandegar MA, Huebsch N, Frolov EB, et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18(4):541–553.
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280–284.
- Konermann S, Lotfy P, Brideau NJ, et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e14.
- Strutt SC, Torrez RM, Kaya E, et al. RNA-dependent RNA targeting by CRISPR-Cas9. eLife. 2018;7:e32724.
- Dugar G, Leenay RT, Eisenbart SK, et al. CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the campylobacter jejuni Cas9. Mol Cell. 2018;69(5):893–905.e7.
- Price AA, Sampson TR, Ratner HK, et al. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 112, 6164–6169 (2015).
- Smalley E. CRISPR mouse model boom, rat model renaissance. Nat Biotechnol. 2016;34(9):893–894.
- Ma D, Liu F. Genome editing and its applications in model organisms. Genomics Proteomics Bioinformatics. 2015;13(6):336–344.
- Sun S-H, Chen Q, Gu HJ, et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124–133.e4.
- Zotova A, Zotov I, Filatov A, et al. Determining antigen specificity of a monoclonal antibody using genome-scale CRISPR-Cas9 knockout library. J Immunol Methods. 2016;439:8–14.
- Thompson NC, Zyonts S. Who tries (and who succeeds) in staying at the forefront of Science. Social Science Research Network. 2017:1-52.
- Parnas O, Jovanovic M, Eisenhaure TM, et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162(3):675–686.
- Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3(2):145–158.
- Fellmann C, Gowen BG, Lin P-C, et al. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat Rev Drug Discov. 2016;16(2):89–100.
- Wang R, Simoneau CR, Kulsuptrakul J, et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2020;184(1):106-119.e14.
- Schneider WM, Luna JM, Hoffmann HH, et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2020;184(1):120-132.e14.
- Wei J, Alfajaro MM, DeWeirdt PC, et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2020184(1):76-91.e13.
- Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. Trends in analytical chemistry: TRAC. 2018;98:19–35.
- Ackerman CM, Myhrvold C, Thakku SG et al. Massively multiplexed nucleic acid detection using Cas13. Nature. 2020;582(7811):277-282.
- Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–1266.
- Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448.
- Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444.
- Joung J, Ladha A, Saito M, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. Medrxiv Prepr Serv Heal Sci 2020.05.04.20091231 (2020) doi:https://doi.org/10.1101/2020.05.04.20091231.
- Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874.
- Fozouni P, Son S, de León Derby MD, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2020;184(2):323-333.e9.
- Li H, Yang Y, Hong W, et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1–23.
- Lee K, Conboy M, Park HM, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1(11):889–901.
- Yang W, Tu Z, Sun Q, et al. CRISPR/Cas9: implications for modeling and therapy of neurodegenerative diseases. Front Mol Neurosci. 2016;9:30.
- Yin C, Zhang T, Qu X, et al. In vivo excision of HIV-1 provirus by SaCas9 and multiplex single-guide RNAs in animal models. Mol Ther. 2017;25(5):1168–1186.
- The SM. The CRISPR/Cas9 system: their delivery,in vivo and ex vivo applications and clinical development by startups. Biotechnol Prog. 2017;33(4):1035–1045.
- Pancino G, Saez-Cirion A, Scott-Algara D, et al. Natural resistance to HIV infection: lessons learned from HIV‐exposed uninfected individuals. J Infect Dis. 2010;202:345–350.
- Soppe JA, Lebbink RJ. Antiviral goes viral: harnessing CRISPR/Cas9 to Combat Viruses in Humans. Trends Microbiol. 2017;25(10):833–850.
- White MK, Hu W, The KK. CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov Med. 2015;19(105):255–262.
- Abbott TR, Dhamdhere G, Liu Y, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and Influenza. Cell. 2020;181(4):865–876.e12.
- Gholizadeh P, Kose S, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111–1121.
- Bikard D, Euler CD, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150.
- Kang YK, Kwon K, Ryu JS, et al. Nonviral genome editing based on a polymer-derivatized crispr nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjugate Chem. 2017;28(4):957–967.
- Gomaa AA, Klumpe HE, Luo ML, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5(1):e00928–13.
- Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32(11):1141–1145.
- Hammond A, Galizi R, Kyrou K, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature. 2016;34:78–83.
- Slomovic S, Pardee K, Collins JJ Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. U.S.A. 112, 14429–14435 (2015).
- Schmid-Burgk JL, Gao L, Li D, et al. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020;78(4):794–800.e8.
- Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature. 2015;33:187–197.
- Tsai SQ, Nguyen NT, Malagon-Lopez J, et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–614.
- Akcakaya P, Bobbin ML, Guo JA, et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;2:914.
- Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nature. 2015;12(3):237–243
- Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550(7676):407–410.
- Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495.
- Wang Z, Pan Q, Gendron P, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15(3):481–489.
- Makarova KS, Zhang F, Koonin EV. SnapShot: class 2 CRISPR-Cas Systems. Cell. 2017;168(1–2):328.e1.
- Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nature Rev Microbiol. 2017;15(3):169–182.
- Bak RO, Porteus MH. CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep. 2017;20(3):750–756.
- Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–254.
- Kim S, Koo T, Jee H-G, et al. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018;28(3):367–373.
- Kirkpatrick J, Koblentz GD, Palmer MJ, etl. Editing biosecurity: needs and strategies for governing genome editing. (2018).
- Cyranoski D CRISPR-baby scientist fails to satisfy critics. 13–14 (2018).
- Wang H, Li J, Li W, et al. CRISPR twins: a condemnation from Chinese academic societies. Nature. 2018;564(7736):345.
- Normile D Organizers of gene-editing meeting blast Chinese study but call for ‘pathway’ to human trials. (2018).
- Law, C. on S., Technology, and, Affairs, P. and G., Medicine, N. A. of S., Engineering, and & Olson, S. International summit on human gene editing: a global discussion. (2016) doi:https://doi.org/10.17226/21913.
- Lander ES, Baylis F, Zhang F, et al. Adopt a moratorium on heritable genome editing. Nature. 2019;567(7747):165–168.
- Gronvall GK. The scientific response to COVID-19 and lessons for security. Survival. 2020;62(3):77–92.
- Burwell SM, Townsend FF, Bollyky TJ, et al. Improving pandemic preparedness: lessons from COVID-19. https://www.cfr.org/report/pandemic-preparedness-lessons-COVID-19/pdf/TFR_Pandemic_Preparedness.pdf (2020). Web. 15 Dec. 2020.
- Board GPM A world in disorder. https://apps.who.int/gpmb/assets/annual_report/GPMB_AR_2020_EN.pdf; (2020). Web. 15 Dec. 2020.
- Biodefense, B. C. on. Diagnostics for biodefense: flying blind with no plan to land. https://biodefensecommission.org/wp-content/uploads/2020/11/Diagnostics-Special-Focus_final_web2-1.pdf (2020). Web. 15 Dec. 2020.
