ABSTRACT
Introduction: Multidrug resistant (MDR) Acinetobacter baumanii (A. baumannii) strains have emerged as novel nosocomial pathogens threatening patients’ lives, especially in intensive-care units (ICUs). This study aims to determine the prevalence of carbapenemase genes and CTX-M-15 and the resistance pattern of carbapenemase producing isolates.Methods: A total of 530 clinical specimens were collected from patients suffering from different infections, antibiotic susceptibility test was performed using kirby-bauer disk diffusion method. ESβL production was detected phenotypically by double-disc synergy test (DDST). Carbapenemase production was tested by Modified Hodge Test (MHT). Then, these isolates were tested for MBL detection by disc potentiation test. Carbapenemase encoding genes (VIM, IMP, GIM and SPM, OXA-51, OXA-23 and OXA-143) and CTX-M-15 were tested by polymerase chain reaction (PCR).Results: Out of 530 samples, 20 bacterial isolates were identified as A. baumannii from different infectious cases, 35% of isolates were ESBL-producers. Eleven isolates were resistant to imipenem (4 isolates) and meropenem (7 isolates). All carbapenem resistant isolates were MHT positive. Nine (45%) isolates were confirmed as A. baumannii by OXA-51 (all were carbapenem resistant). Distribution of IMP, VIM, GIM and SPM, OXA-23, OXA-143 and CTX-M-15 by PCR were 55, 50, 50, 25, 35, 45 and 33% respectively.Conclusion: The high prevalence of resistance genes and the resistance pattern of the isolates indicate that the detection of ESBLs and MBLs phenotypically and genotypically with the study of the resistance pattern of the isolates is critically important for the surveillance of drug resistance in the hospital environment.
1. Introduction
Acinetobacter baumannii (A. baumannii) is an aerobic Gram-negative bacilli and saccharide non-fermenter [Citation1]. It is widely spread in nature, hospital environments, skin surface of humans and organs such as intestinal, respiratory and urinary tract [Citation2]. It is considered the main pathogen that causes nosocomial infections as ventilator-associated pneumonia (VAP), endocarditis, bacteremia, wound infections, meningitis and urinary tract infections in immunocompromised patients and in patients underlying disease with prolonged hospitalization [Citation3] with arising prevalence in ICUs [Citation4]. The multidrug-resistant A. baumannii (MDRAB) which is resistant to at least three different classes of antimicrobial agents mainly beta lactams, aminoglycosides, fluoroquinolones, and carbapenems causes serious problems in various clinical settings worldwide [Citation5]. This organism has become increasingly resistant to broad-spectrum cephalosporin due to extended-spectrum β- lactamase produced by it.
Extended spectrum beta lactamases (ESBLs) are a class of group A beta lactamases which result in hydrolysis of first, second, and third-generation cephalosporins but are inhibited by beta-lactamase inhibitors like clavulanic acid [Citation6]. In Acinetobacter spp, ESBL genes identified in those species are mostly of VEB or PER types but TEM, SHV, GES, and CTX-M derivatives have also been reported [Citation7,Citation8].
The carbapenem antibiotic has been used in the management of hospital-acquired Gram-negative infections, because of their broad spectrum of activity and stability to hydrolysis by most of β lactamases, including ESBLs. Carbapenem resistance in A. baumannii is mainly due to the production of carbapenemases, especially OXA type carbapenem-hydrolyzing (class D) β-lactamases, which are either chromosomally located, like as blaOXA-51, which become expressed only when the insertion sequence ISAba1 element is inserted upstream of the gene, or acquired, mostly blaOXA-23-like, blaOXA-24-like and blaOXA-58-like subfamilies, and metallo-β-lactamases (class B; bla-IMP, bla-VIM, bla-NDM) [Citation5].
This study was carried out to determine the prevalence of ESBL, carbapenemase and MBL production in clinical isolates of A. baumannii from Minia hospitals phenotypically and genotypically.
1. Subjects and methods
1.1. Samples collection
Totally 530 clinical samples were obtained from patients with wound infection, ear infection, chest infection, burn infection, urinary tract infection, gastrointestinal infection and from patients resident in the intensive-care unit at Minia hospital, Minia, Egypt, throughout the period from August 2016 to January 2017.
1.2. Bacterial growth and identification
All samples were cultured on Herellea agar (Himedia, India) in aerobic conditions at 37°C for 24 h and then identified by conventional biochemical tests as citrate test, Catalase test, oxidase, Indole, oxidative fermentation test, fermentation of galactose, glucose, lactate, malonate, maltose, mannose, rhamnose, and xylose, arginine dihydrolase, histidine, leucine, malate, phenylalanine deaminase, and tyrosine hydrolysis tests. Also, A. baumannii was identified by their growth on Herellea agar (Himedia, India) showing pale lavender colonies [Citation9], followed by PCR amplification of blaOXA-51-like gene
1.3. Antimicrobial susceptibility testing
Antimicrobial susceptibility patterns were determined by disk diffusion method on Mueller–Hinton agar (MHA) (Biolab, Hungary) according to CLSI guidelines [Citation10]. The following antimicrobial disks (Mast Diagnostic, UK) were used Azlocillin (75 µg), ciprofloxacin (5 µg), ampicillin/sulbactam (20 µg), levofloxacin (5 µg) cefepime (30 µg), meropenem (10 µg), aztreonam (30 µg), imipenem (10 µg), polymyxin B (300 µg), colistin sulfate (10 µg), tigecycline (15 µg), tobramycin (10 µg), ceftazidime (30 µg), amoxicillin/clavulanic (20/10 µg), carbenicillin (100 µg), amikacin (30 µg), gentamicin (10 µg), piperacillin (100 µg), cefoperazone (75 µg).
1.4. Phenotypic detection of extended-spectrum beta-lactamases (ESBL) production by double-disc synergy test (DDST)
Detection of the ESBL production by A. baumannii strains was performed by placing discs of ceftazidime, cefotaxime, aztreonam, and cefepime (30 µg each) at a distance of 30 or 20 mm (center to center) from a disk containing AMC (amoxicillin 20 µg and clavulanic acid 10 µg). ESBL production was inferred when the cephalosporin zone was expanded by the clavulanate. Enhancement of zone of inhibition is indicative of the presence of an ESBL [Citation11].
1.5. Phenotypic detection of carbapenemases by
1.5.1. Modified hodge test (MHT)
The presence of carbapenemases in A. baumannii isolates was primarily detected using Modified Hodge test. The diluted culture of Escherichia coli ATCC 25,922 (0.5 McFarland standard) was swabbed on the surface of Mueller-Hinton agar plates in three different directions. Meropenem disc (10 μg) (Oxoid, Basingstoke, UK) was placed at the center of each plate. The tested isolates were streaked as a thin line from the edge of the meropenem disk to the edge of the plate. Bacterial growth was allowed for 18 h at 37°C. Indentation in the inhibition zone of E. coli or clove growth of E. coli around the meropenem disk revealed a positive MHT which indicates that this isolate is producing a carbapenemase [Citation12].
1.5.2. Disc potentiation test
Imipenem-resistant isolates were screened for the production of MBL. The double disk method was used to detect this enzyme. Colonies from overnight cultures on blood agar plates were suspended in Mueller-Hinton broth and the turbidity standardized to equal that of a bacterial concentration of 1:100 suspensions of the 0.5 McFarland standards. Then, the suspension was streaked onto Mueller-Hinton agar plates (Hi Media, Mumbai, India). A disc of Imipenem alone (10 μg) and Imipenem (10 μg) in combination with EDTA (750 μg/disc) was placed at the distance of 20 mm (Center to Center). After overnight incubation at 35°C, a ≥ 7 mm increase in the inhibition zone of diameter around Imipenem-EDTA discs, as compared to imipenem discs alone, interpreted as indicative of MBL production [Citation13].
1.6. Molecular detection of blaCTXM-15 like gene
The DNA template was prepared by boiling of suspension of bacterial pellet for 10 min and directly used in the polymerase chain reaction (PCR) assay [Citation14]. CTXM (15) gene was amplified using PCR as follows: DNA sample (1 µl) was added to PCR mix (12.5 µl) including 1 µl each of forward and reverse primers (CTX- F5ʹ-CGTCACGCTGTTGTTAGGAA-’3, CTX R5ʹ-ACGGCTTTCTGCCTTAGGTT-3ʹ). Total volume of the mixture was 25 µl for each sample. PCR conditions were set to 95°C denaturation for 5 min, 58°C annealing for 45 s, 72°C extension for 1 min and final extension step at 72°C for 1 min. A total of 35 cycles were run followed by separation of amplified product on 1.5% agarose gel stained with 10μg/ml ethidium bromide. DNA bands were visualized using UV transilluminator. 100-bp DNA ladder was used to assess the expected amplicon size (780 bp) [Citation15].
1.7. Amplification of carbapenemases encoding genes
The primers used to amplify the carbapenemases encoding genes (VIM, IMP, GIM, and SPM) are shown in and their sequence is previously published [Citation16]. DNA sample (1 µl) was added to PCR mix (12.5 µl) including 1 µl each of forward and reverse primers for each gene. Total volume of the mixture was 25 µl for each sample. PCR conditions were set to 95°C denaturation for 5 min, 56°C annealing for45 s, and 72°C extension for 1 min and final extension step was done at 72°C. A total of 35 cycles were run followed by running of amplified product on 1.5% agarose gel stained with 10 μg/ml of ethidium bromide. DNA bands were visualized as mentioned before.
Table 1. List of primers used in this study
1.8. Amplification of blaOxa-51, blaOxa-23 and blaOxa-143 like genes
Oxa-51, OXA-23 and OXA-143 primers () were used to partially amplify the gene encoding the intrinsic OXA-51-like enzymes and the acquired resistant genes of OXA-23 and OXA-143 gene, respectively [Citation17,Citation18]. The amplification conditions were, initial denaturation at 94°C for 5 min 30 cycles of 94°C for 25 s, 52°C for 40 s and 72°C for 50 s, and a final extension step at 72°C for 6 min.
2. Results
From totally 530 Clinical samples, just 20 isolates were identified to be A. baumannii (3.8%) according to cultural characteristics (pale lavender colonies on Herellea agar) and biochemical reaction results. The highest distribution of A. baumannii was presented in patients admitted to Intensive-care unit (ICU) (7.8%), followed by patients with wound infections (4.8%), and patients with burns (3.8%) ().
Table 2. Prevalence of A. baumannii isolated from different patients in relation to the type of infections
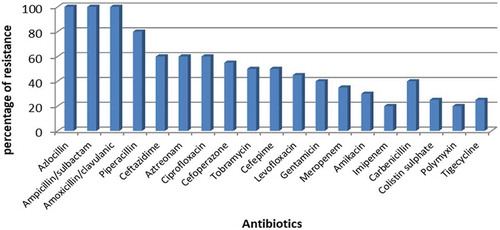
Among the 19 used antibiotics, A. baumannii was completely resistant to Ampicillin/sulbactam, amoxicillin/clavulanic and azlocillin, highly resistant to piperacillin, Aztreonam, ciprofloxacin (80%, 60%, and 60%, respectively). The lowest rate of resistance was shown against imipenem and polymyxin B (20% each) ().
All β-lactams resistant A. baumannii were tested for ESBLs production. It was found that 7 (35%) of A. baumannii isolates were positive for ESBLs production. These strains were collected from patients with wound infections (5 strains) and patients resident in intensive-care unit (two strains) ().
Table 3. Distribution of ESBLs and MBL production among A. baumannii strain
All imipenem (four isolates) and meropenem (seven isolates) resistant A. baumannii strains were positive for MHT in which carbapenemase production was detected by the appearance of the enhanced Escherichia coli ATCC 25,922 growth along with the tested organism that revealed a clover-leaf-like indentation. On the other hand, 10 isolates were positive for carbapenemase by disc potentiation test (four imipenem resistant and six meropenem resistant strains). Co-production of both ESBL and MBL was shown in 3/20 (15%) of isolates ().
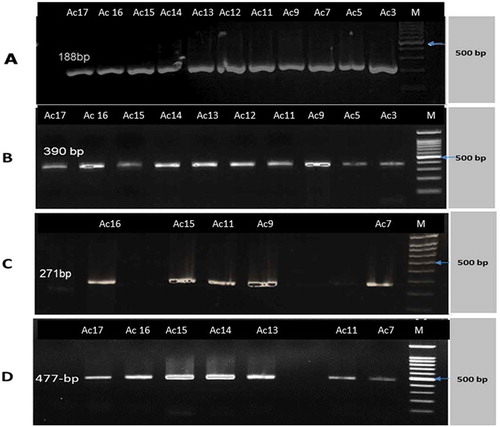
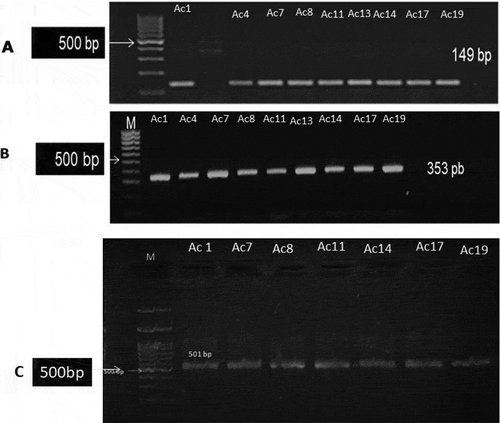
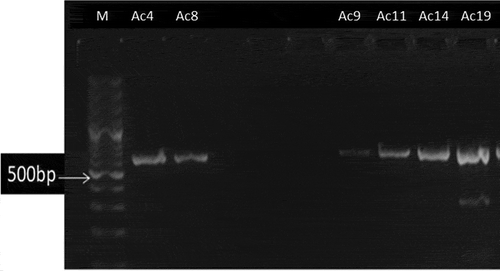
Out of 20 A. baumannii strains, only 6 (30%) of them harboring CTX-M-15 gene as shown in and . As shown in table (4), out of 20 isolates (showing pale lavender colonies on Herellea agar), eleven isolates (55%) of A. baumannii were positive for bla-IMP while bla-VIM and bla-GIM were found in 10 isolates. In addition, bla-SPM was found in five isolates (25%). Moreover, all bla−IMP positive isolates were positive for MHT. (a–d) show gel electrophoresis for the PCR amplicon of bla-IMP, bla-VIM,bla- SPM and bla-GIM respectively. Additionally, shows that out of 20 isolates showing colonies characteristics of A. baumannii on Herellea agar, 9 isolates harbored OXA-51 which confirming A. baumannii species. All OXA-51 positive strains were found to be positive for OXA-143 like gene while 7 isolates were positive for OXA-23. (a–c) showed gel electrophoresis for the PCR amplicon of OXA 51, OXA-23 and OXA-143. Results represented in showed that some isolates were resistant to all or most of the tested genes such as Strain no. Ac11 which was positive for all tested genes (CTX-M-15, Class B, and class D carbapenemases) showing resistance to 17 of the tested antibiotics but sensitive for tigecycline and colistin sulfate . Another strain, Strain no. Ac14 was positive for all the tested genes except bla-SPM showing resistance to 16 antibiotics but susceptible for imipenem, tigecycline, and polymxyin B.
Table 4. Distribution of the tested genes of ESBLs (CTX-M-15), Class B carbapenemases, Class D carbapenemases among the isolated A. baumannii, their resistance pattern and source of samples
Figure 2. PCR amplicons of A. baumannii strains CTX-M-15 gene. Lanes order is as follows: marker (100 bp); isolates number
3. Discussion
Acinetobacter baumannii is a common opportunistic pathogen present in health care setting worldwide. A. baumannii plays an important role in nosocomial infections due to its resistance against multiple classes of antibiotics [Citation19].
Analysis of our results indicated the occurrence of 3.8% A. baumannii infections in all Minia hospitals from clinical patient specimens. Our results closely nearer to that obtained by Safwat, Abdelwahab [Citation20] who reported that A. baumannii does not represent a major health hazard at MUH (Minia university hospitals), where it represented 5.4% and 5.8% of the clinical and environmental samples examined. On the other hand, Sadeghi, Khosravi [Citation21] showed higher percentage than our results (25.4%) while lower percentage (0.95%) was obtained by Kateete, Nakanjako [Citation22].
The majority of A. baumannii were isolated from patients resident in Intensive-care unit (7.8%) that agreed with results obtained by Petrova, Stanimirova [Citation23] who reported that the greatest number of isolates was obtained from the Intensive-Care Unit (ICU) and discussed that Acinetobacter baumannii was one of the main opportunistic pathogens of ICU infections. As the majority of patients in ICU are in poor health owing to various diseases including cardiovascular disease and diabetes. Also, they are required to remain in the ICU for a long period of time administering antibiotics with strong antimicrobial activity and wide antibacterial spectrum. Additionally, medical histories of patients including being on a ventilator machine and undergoing tracheostomy making them are at risk of obtaining opportunistic infections [Citation24].
In the present study, A. baumannii were completely resistant to azlocillin, Ampicillin/sulbactam and amoxicillin/clavulanic while 20% of isolates were resistant to imipenem and polymyxin B. During the last decade, the emergence of multi-drug resistant A. baumanii was reported due to the extensive use of antimicrobial agents worldwide in intensive-care units (ICUs). Carbapenems were considered the last resort for treating infections associated with multi-drug resistant strains due to its ability to be stable against ESBL and Ampc β-lactamases [Citation8]. Few years ago, carbapenem-resistant strains emerged due to the extensive use of these antibiotics. Furthermore, the inter-hospital dissemination of resistant strains in the absence of strict infection controls measures. Carbapenem resistance was found to be associated with high mortality rate and resistance to other classes of antibiotics such as aminoglycosides and quinolones [Citation25]. Dias, Resende [Citation26] reported that absolute resistance was shown by A.baumannii against imipenem, meropenem, ceftazidime, cefepime, and ciprofloxacin but in our study, low resistance rate for meropenem and imipenem, high resistance rate for ceftazidime and cefepime was observed. Furthermore, they showed that Tigecycline was the most effective antibiotics against A. baumannii but we reported that imipenem and colistin were the most effective antibiotic [Citation27].
Carbapenemases represent three classes of β-lactamases. The three classes are Ambler class A and D carbapenemase (serine carbapenemases) and class B carbapenemases (zinc dependent) which are inhibited by metal chelators, such as EDTA and are called metallo- β-lactamases (MBLs). Metallo-β-lactamases (MBLs) enzymes are able to hydrolyze all β-lactam antibiotics with the exception of monobactams. Genes encoding theses enzymes may be plasmid mediated or chromosomally mediated. The most common MBLs enzymes are belonged to VIM, IMP, SPM, GIM, SIM and NDM families [Citation28,Citation29]. On the other hand, ESBLs production is considered as important mechanisms of resistance and their association with MBLs increases the level of resistance. Phenotypic methods for the detection of ESBL and MBL that were used in this study were double‑disc synergy test (DDST), modified Hodge test (MHT), and combined disc diffusion test using imipenem or meropenem and EDTA.
High incidence of ESBLs production by A. baumannii was reported by many studies [Citation30–Citation32] but lower rate was shown in our study (35%). Carbapenem-resistant isolates were tested for carbapenemase enzyme production by MHT and Disc potentiation test, it was found that all (11 isolates) carbapenem-resistant isolates were MHT positive that agreed with results obtained by Petrova, Stanimirova [Citation23] who reported that out of 43 A. baumannii, 42 (97.7%) isolates were positive for carbapenemase production using MHT while 10 isolates were found to be positive by disc potentiation test (10/11, 90.9%). Many studies agreed with our results [Citation31,Citation33,Citation34]. Differences in the prevalence of ESBL and MBL-producing A. baumanni strains seem to be the result of the variation among different patients studied and different rates of antibiotic uses in different hospitals.
shows the distribution of the tested genes encoding ESBLs (CTX-M-15), Class B Ccarbapenemase (IMP, VIM, GIM, SPM like genes) and class D Oxacillinases (OXA-51, OXA-143, OXA-23 like genes) and their resistance pattern. As there are some oxacillinase genes are expressed naturally in A. baumannii, bla-OXA-51 is used for confirming A. baumannii [Citation17]. Our results showed that nine isolates were confirmed to be A. baumannii (bla-OXA-51 positive). All confirmed isolates were positive for OXA-143 and seven isolates were positive for OXA-23. Higher incidence rate of bla OXA-51 (94%) positive A. baumanii was reported by Shahcheraghi, Abbasalipour [Citation35] and Woodford, Ellington [Citation36] (93%). On the other hand, Safwat, Abdelwahab [Citation20] reported that blaoxa-51-like gene was found in 4/48 pus samples (8.3%) that did not show pale lavender colonies on herellea agar and in 8 pus samples (16.7%) showing pale lavendar colonies on herellea agar, which is lower than our result. It was found that the presence of OXA-51 has weak carbapenemase activity and it causes increase in A. baumanii when it was overproduced. So, A. baumanii resistance to carbapenem required the presence of the other acquired oxacillinases as OXA-23, OXA-143, OXA-24. OXA-143 enzyme (Class D carbapenem-hydrolyzing enzyme) in Acinetobacter baumannii was firstly described by Higgins, Poirel [Citation37]. Furthermore, Neves, Clemente [Citation38] found that OXA-23 was the most common acquired cabapenemase (51.2%) followed by OXA-143 (18.6%) among carbapenem-resistant Acinetobacter baumanii which confirm their important role in carbapenem resistance. In this study, OXA-143 was the most common followed by OXA-23. High prevalence of OXA-143 and OXA-23 was also reported by Antonio, Neves [Citation39] (58.3%) and Mostachio, Levin [Citation40] (76%). CTX-M-15 was found in 4/9 isolates that play a role in the increase in the level of resistance in association with other OXA genes. Hakemi Vala, Hallajzadeh [Citation41] detected that out of 28 Acinetobacter baumannii isolates, 3 (10.7%) were positive for CTX-M-15.
Our study reported that bla-IMP was the most prominent carbapenemase gene (55%) followed by bla−VIM, bla−GIM (50% each) and bla−SPM (25%) but lower percentage of isolates harboring bla-IMP was shown by Kazi, Nikam [Citation42]. Amudhan, Sekar [Citation43] indicated that out of the 116 A. baumannii isolates, 54 (46.5%) were bla VIM/blaIMP producers that are close to our study.
Safari, Mozaffari Nejad [Citation44] reported that 30% of 100 A. baumannii isolates were confirmed to harbor the blaVIM-family genes but the other genes including bla-IMP Family, bla- SPM-1, bla- SIM-1 and bla- GIM-1 were not be detected. On the other hand, lower percents for both bla-IMP (3.48%) and bla-IMP, bla-VIM (17.44%) were reported by . Also, Erfani, Yaghuobi [Citation45] reported higher percents for bla-VIM (60.4%) but they did not found any isolate that was positive for bla-IMP. It seems that the pattern of resistance over different years and the results of geographically different countries are effective factors in these variations.
Furthermore, we observed that there were some strains (strain no. 11, 14 and 17) that were isolated from the same source (Minia University hospital, ICU and Surgery unit) carrying CTX-M-15 gene in association with class B and class D β-lactamases. In addition, these isolates showed resistance to most of the tested antibiotics which is an alarm for the possibility of the dessimination of resistance among different bacteria in the hospital environment by the horizontal gene transfer in the absence of strict infection control measures.
4. Conclusion
Our results revealed a high level of antimicrobial resistance among the studied clinical isolates of A. baumanii. The prevalence of β-lactamase-producing isolates and their isolation from life-threatening infections is increasing at an alarming rate worldwide. Detection of ESBLs and MBLs phenotypically and genotypically with the study of the resistance pattern of these isolates is critically important for the surveillance of drug resistance in the hospital environment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Notes on contributors
Rehab M. Abd El-Baky
Rehab M. Abd El-Baky is a Associate professor of Microbiology and Immunology, Faculty of Pharmacy, Minia University, Mnia, Egypt and the Head of Microbiology and Immunology Department, Faculty of Pharmacy, Deraya University, Minia, Egypt. Specialize in studying the Virulence factors of Bacteria and antimicrobial resistance mechanisms.
Sara M. Farhan
Sara M. Farhan is a Postgraduate students and a demonstrator in the Microbiology and Immunology Department, Faculty of Pharmacy, Deraya University, Minia, Egypt.
Reham A. Ibrahim
Reham A. Ibrahim is a Lecturer of Microbiology and Immunology, Faculty of Pharmacy, Miinia University, Minia, Egypt and specialize in studying the epidemiology of infectious diseases and the mechanisms of antimicrobial resistance.
Khaled M. Mahran
Khaled M. Mahran is a Professor of General Surgery and Laparoscopic surgery, Faculty of Medicine, Minia University, Minia 61519 Egypt
Helal F. Hetta
Helal F. Hetta is a Lecturer of Microbiology and Immunology, Faculty of Pharmacy, Assuit University, Assuit, Egypt and a postdoctoral fellow in the Department of Internal Medicine, University of Cincinnati College of Medicine, Cincinnati, OH, USA, specialize in Medical Virology and Immunology.
References
- Giamarellou H, Antoniadou A, Kanellakopoulou K. Acinetobacter baumannii: a universal threat to public health? Int J Antimicrob Agents. 2008 Aug;32(2):106–119.
- Mishra SK, Rijal BP, Pokhrel BM. Emerging threat of multidrug resistant bugs–acinetobacter calcoaceticus baumannii complex and methicillin resistant Staphylococcus aureus. BMC Res Notes. 2013 Mar;15(6):98.
- Mahzounieh M, Khoshnood S, Ebrahimi A, et al. Detection of antiseptic-resistance genes in pseudomonas and acinetobacter spp. isolated from burn patients. Jundishapur J Nat Pharm Prod. 2014 May;9(2):e15402.
- Obeidat N, Jawdat F, Al-Bakri AG, et al. Major biologic characteristics of Acinetobacter baumannii isolates from hospital environmental and patients’ respiratory tract sources. Am J Infect Control. 2014 Apr;42(4):401–404.
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008 Jul;21(3):538–582.
- Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951.
- Naas T, Namdari F, Reglier-Poupet H, et al. Panresistant extended-spectrum beta-lactamase SHV-5-producing Acinetobacter baumannii from New York City. J Antimicrob Chemother. 2007 Nov;60(5):1174–1176.
- Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011 Dec;63(12):1061–1067.
- Herellea EC. (Acinetobacter) and Pseudomonas ovalis (P. putida) from frozen foods. Appl Microbiol. 1969 Jan;17(1):26–30.
- Wayne P. CLSI. Performance Standards for Antimicrobial Susceptibility. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and laboratory Standards Institute; 2019.
- Nagano N, Nagano Y, Cordevant C, et al. Nosocomial transmission of CTX-M-2 beta-lactamase-producing Acinetobacter baumannii in a neurosurgery ward. J Clin Microbiol. 2004 Sep;42(9):3978–3984.
- Noyal MJ, Menezes GA, Harish BN, et al. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res. 2009 Jun;129(6):707–712.
- Yong D, Lee K, Yum JH, et al. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002 Oct;40(10):3798–3801.
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol. 1987;56(1):2.4.1–2.4.5
- Ullah W, Qasim M, Rahman H, et al. CTX-M-15 and OXA-10 beta lactamases in multi drug resistant Pseudomonas aeruginosa: first report from Pakistan. Microb Pathog. 2017;105:240–244.
- Ellington MJ, Kistler J, Livermore DM, et al. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007 Feb;59(2):321–322.
- Brown S, Young H, Amyes S. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect. 2005;11(1):15–23.
- Higgins PG, Lehmann M, Seifert H. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2010;35(3):305.
- Perez F, Hujer AM, Hujer KM, et al. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007 Oct;51(10):3471–3484.
- Safwat D, Abdelwahab SF, Hasanen AM, et al. Prevalence of acinetobacter baumannii at minia university hospital. Egypt J Med Microbiol. 2011;20(1):63–68.
- Sadeghi P, Khosravi AD, Shahraki AH, et al. Identification of clinical isolates of Acinetobacter baumannii from Iran and study of their heterogeneity. J Chin Med Assoc. 2016;79(7):382–386.
- Kateete DP, Nakanjako R, Okee M, et al. Genotypic diversity among multidrug resistant Pseudomonas aeruginosa and Acinetobacter species at Mulago Hospital in Kampala, Uganda. BMC Res Notes. 2017;10(1):284.
- Petrova AP, Stanimirova ID, Ivanov IN, et al. Carbapenemase production of clinical isolates acinetobacter baumannii and pseudomonas aeruginosa from a Bulgarian University Hospital. Folia Med (Plovdiv). 2017;59(4):413–422.
- Ren G, Zhou M, Ding N, et al. Analysis on distribution features and drug resistance of clinically isolated Acinetobacter baumannii. Exp Ther Med. 2016 Sep;12(3):1715–1718.
- Djahmi N, Dunyach-Remy C, Pantel A, et al. Epidemiology of carbapenemase-producing enterobacteriaceae and acinetobacter baumannii in Mediterranean Countries. Biomed Res Int. 2014;2014:11.
- Dias VC, Resende JA, Bastos AN, et al. Epidemiological, physiological, and molecular characteristics of a brazilian collection of carbapenem-resistant acinetobacter baumannii and pseudomonas aeruginosa. Microb Drug Resist. 2017;23(7):852–863.
- Rossello J, Olona M, Campins M, et al. Investigation of an outbreak of nosocomial infection due to a multiply drug-resistant strain of Pseudomonas aeruginosa. J Hosp Infect. 1992 Feb;20(2):87–96.
- Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012 Jul;35(3):317–325.
- Singh H, Thangaraj P, Chakrabarti A. Acinetobacter baumannii: a brief account of mechanisms of multidrug resistance and current and future therapeutic management. J Clin Diagn Res. 2013 11/10 04/26/received 07/28/rev-request 09/18/accepted;7(11): 2602–2605.
- Farajnia S, Azhari F, Alikhani MY, et al. Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant acinetobacter baumannii isolates in North-West of Iran. Iran J Basic Med Sci. 2013 Jun;16(6):751–755.
- Owlia P, Azimi L, Gholami A, et al. ESBL- and MBL-mediated resistance in Acinetobacter baumannii: a global threat to burn patients. Infez Med. 2012 Sep;20(3):182–187.
- Jones RN, Biedenbach DJ, Sader HS, et al. Emerging epidemic of metallo-beta-lactamase-mediated resistances. Diagn Microbiol Infect Dis. 2005 Feb;51(2):77–84.
- Anwar M, Ejaz H, Zafar A, et al. Phenotypic detection of metallo-beta-lactamases in carbapenem resistant acinetobacter baumannii isolated from pediatric patients in Pakistan. J Pathog. 2016;2016:8603964.
- Easwaran S, Ramasamy R. Prevalence of metallo β lactamases producing pseudomonas spp. and acinetobacter spp. in a tertiary care teaching hospital. J Drug Discovery Ther. 2017;5(7):35–39.
- Shahcheraghi F, Abbasalipour M, Feizabadi M, et al. Isolation and genetic characterization of metallo-beta-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011 Jun;3(2):68–74.
- Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006 Apr;27(4):351–353.
- Higgins PG, Poirel L, Lehmann M, et al. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009 Dec;53(12):5035–5038.
- Neves FC, Clemente WT, Lincopan N, et al. Clinical and microbiological characteristics of OXA-23- and OXA-143-producing Acinetobacter baumannii in ICU patients at a teaching hospital, Brazil. Braz J Infect Dis. 2016 Nov 01;20(6):556–563.
- Antonio CS, Neves PR, Medeiros M, et al. High prevalence of carbapenem-resistant Acinetobacter baumannii carrying the blaOXA-143 gene in Brazilian hospitals. Antimicrob Agents Chemother. 2011 Mar;55(3):1322–1323.
- Mostachio AK, Levin AS, Rizek C, et al. High prevalence of OXA-143 and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. isolates in Brazil. Int J Antimicrob Agents. 2012 May;39(5):396–401.
- Hakemi Vala M, Hallajzadeh M, Hashemi A, et al. Detection of Ambler class A, B and D ss-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters. 2014 Mar 31;27(1):8–13.
- Kazi M, Nikam C, Shetty A, et al. Dual-tubed multiplex-PCR for molecular characterization of carbapenemases isolated among Acinetobacter spp. and Pseudomonas spp. J Appl Microbiol. 2015 May;118(5):1096–1102.
- Amudhan MS, Sekar U, Kamalanathan A, et al. bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J Infect Dev Ctries. 2012 Nov 26;6(11):757–762.
- Safari M, Mozaffari Nejad AS, Bahador A, et al. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J Biol Sci. 2015 Jul;22(4):424–429.
- Erfani Y, Yaghuobi S, Fallah F, et al. Detection of bla NDM-, bla VIM, and bla IMP genes in multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa from clinical isolates in Tehran hospitals. Int J Adv Biotechnol Res. 2017;8(2):500–506.