ABSTRACT
Introduction
Essential Hypertension has been a great burden on public health services for a long time, with many life-threatening complications. Therefore, we decided to study Atrial Natriuretic peptide (ANP) gene expression as one of the most important blood pressure controlling genes, in order to use ANP gene as a potential diagnostic or therapeutic marker in the near future.
Methods
One hundred essential hypertensive patients and 100 normotensive controls were included. Study Subjects were subjected to ANP gene expression analysis, together with blood pressure measurement, Lab investigations, and BMI analysis.
Results
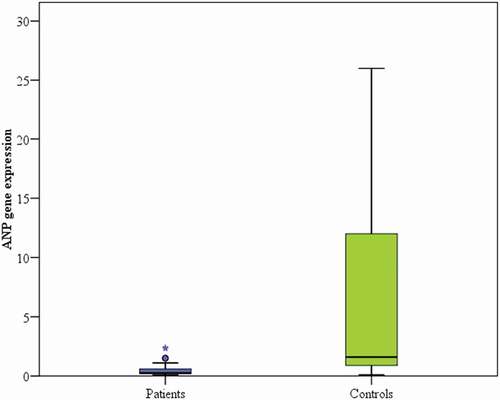
There was a statistical difference between ANP gene expression and blood pressure, with lower ANP gene expression level (median of 0.3) being present among hypertensive patients and higher ANP gene expression level (median of 1.6) among normotensive controls (p < 0.001).
Discussion
We proved that ANP gene expression to be low in essential hypertension patients compared with normotensive individuals.
KEYWORDS:
1. Introduction
Arterial Essential Hypertension (EH) is a big global medical problem of high impact on healthcare resources [Citation1,Citation2]. It was estimated in 2010 that around 1.13 billion people suffer from hypertension worldwide [Citation3]. EH represents 90% of the causes of increased blood pressure, with the remaining 10% due to secondary causes [Citation4,Citation5]. EH represents a major risk factor for serious cardiovascular and renal illnesses like heart failure, stroke, coronary heart disease, and atrial fibrillation [Citation2,Citation6]. Those suffering from EH have 2 to 3 folds higher risk for these cardiovascular illnesses compared to non-hypertensive subjects [Citation7]. Nowadays, despite the recent treatment modalities available for hypertension, it is estimated that less than one in five patients with hypertension is under control [Citation2].
EH has undetermined causes until now [Citation5]. It has been well known that EH is multifactorial in origin, with several genes interacting with various environmental and lifestyle factors that result in increased blood pressure in susceptible patients [Citation8,Citation9]. Several factors have been proved to be involved in the development of hypertension, such as increased cardiac output and plasma volume [Citation10], potentiated sympathetic activity [Citation11], hyperinsulinemia, and insulin resistance [Citation12], and nutritional factors such as high sodium consumption [Citation13].
As many causes are implicated in EH pathogenesis, it can be said that the renin-angiotensin-aldosterone system (RAAS) is the most important cause [Citation14,Citation15]. The RAAS certainly plays an essential role in water and electrolyte homeostasis in the body. RAAS is composed of renin, angiotensinogen, angiotensinogen converting enzyme, angiotensin II type 1 receptor, aldosterone synthetase, and atrial natriuretic peptide (ANP) [Citation16]. Renin cleaves angiotensinogen to angiotensin I (Ang I). Ang I is then converted to angiotensin II (Ang II) by angiotensin-converting enzyme (ACE) [Citation17]. Ang II then exerts its action via Ang II type 1 and type 2 receptors (AT1 and AT2, respectively). AT1 is the receptor involved in the classic pathway of RAAS and is, subsequently, responsible for the development of hypertension. Ang II actions include vascular constriction, salt and water retention, hypertrophy, aldosterone release and stimulation of Nicotinamide adenine dinucleotide phosphate (NADPH) oxidase leading to mitochondrial dysfunction and oxidative stress [Citation18,Citation19]. RAAS inhibitors, including ACE inhibitors, renin inhibitors, AngII type 1 receptor blockers (ARbs), have been introduced in hypertension treatment. But, although they are effective in controlling BP, their ability to prevent end-organ damage is limited and they have multiple side effects after prolonged use [Citation2,Citation20].
ANP is one of the essential components of the RAAS that controls blood pressure in EH [Citation21]. ANP is an endogenous cardiac hormone that regulates natriuresis, homeostasis, and vasodilatation that, in turn, lower blood pressure and blood volume [Citation22–25]. The ANP is initially secreted as an inactive pro-atrial natriuretic peptide (pro-ANP) that is converted into the active form of ANP by a transmembrane serine protease called Corin [Citation26,Citation27]. Activated ANP is a 28 amino-acid endogenous vasoactive peptide hormone, secreted predominantly by atrial cardiomyocytes [Citation23].
ANP acts through binding to guanylyl cyclase natriuretic peptide receptor-1 (NPR-1) [Citation28,Citation29]. NPR-1 stimulation leads to rapid conversion of intracellular GTP into cGMP increasing its intracellular levels leading to activation of cGMP-dependent phosphodiesterases, cGMP-dependent protein kinases (PKG-I and PKG-II), and cGMP-dependent ion channels promoting sodium excretion and inducing vasodilatation [Citation28–30].
ANP secretion is stimulated primarily by distension of atrial tissues in healthy persons [Citation31] and to a lesser extent in ventricular myocytes, if exposed to severe tension [Citation32]. The ventricular cells producing ANP rise significantly if the cardiac ventricles are pressure loaded and hypertrophied, as in the case of chronic hypertension. Therefore, ANP is considered a reliable marker of myocardial hypertrophy [Citation33].
The molecular gene encoding ANP is one of the most developmentally and transcriptionally regulated genes. On the molecular biology level of cardiac cells, ANP encoding gene is one of the essential cardiac genes that led to the development of recent and novel research on gene regulation in the cardiovascular system [Citation25]. In previous research, ANP gene polymorphisms were found to be affecting ANP gene expression levels and thus, linked to cardiovascular disturbances and illnesses like coronary heart disease and left ventricular failure [Citation34].
As the burden of hypertension and its complications are greatly increasing, as well as the multiple adverse effects of the available medications, despite their efficacy, we decided to study the role of ANP gene expression in patients with EH as a new promising diagnostic and therapeutic marker for EH in the future.
2. Patients and methods
Our case-control study was approved by the Medical Ethics Committee of Alexandria Faculty of Medicine. We recruited 100 patients newly diagnosed with essential hypertension from the Internal Medicine department at Alexandria University hospitals, Egypt. Exclusion criteria included secondary hypertension, previously diagnosed hypertensive patients on treatment, diabetes mellitus, and renal failure. One hundred normotensive subjects of matched age and sex to the patients’ group were recruited from the outpatients’ clinic as the control group. All study subjects signed an informed consent showing the nature and type of the study.
Full history taking and thorough physical examination were done on all study subjects. The blood pressure was measured 3 times in the resting state in the supine position, with the average of the last readings being recorded. Hypertension was defined as systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg [Citation35]. Routine laboratory tests were performed to all study subjects, as fasting plasma glucose (FPG), lipid profile, blood urea, serum creatinine, sodium, and potassium were assessed under overnight fasting conditions using Dimension RxL autoanalyzer. Height and weight were measured for all subjects to calculate body mass index (BMI). These tests and measurements were done because diabetes mellitus, hyperlipedemia, renal impairment, and obesity are well-known risk factors or associations of hypertension.
2.1. Genetic study
2.1.1. RNA extraction
RNA was isolated from the peripheral blood mononuclear cells according to the manufacturer’s protocol using QIAamp RNA Blood Mini Kits (Qiagen, USA). The quality and quantity of the extracted RNA were detected using Nanodrop 2000 spectrophotometer (Nanodrop Technologies, USA). The extracted RNA was stored at −80 C until further processing.
2.1.2. Reverse transcription
Complementary DNA (cDNA) was then synthesized from the previously extracted RNA using SensiFAST cDNA Synthesis Kit (Bioline, London, UK) following the manufacturer’s protocol for RNA reverse transcription. The PCR amplification was done on the SimpliAmp Thermal Cycler (Applied Biosystems, USA). The thermal profile was as follows: annealing at 25 C for 10 min, reverse transcription at 42 C for 15 min followed by inactivation at 85 C for 5 min.
2.1.3. ANP gene expression analysis
Quantitative detection of ANP gene expression level was carried out on Stratagene Mx3000P Real-time PCR system (Agilent, Germany) using presynthesized cDNA, Maxima SYBR Green PCR Master Mix (Thermo Scientific, USA) and sequence-specific primers. The reaction mix was prepared by adding 12.5 uL Maxima SYBR Green, 1 uL forward primer, 1 uL reverse primer, 5 uL template DNA and nuclease-free water of 5.5 uL to make a total volume of 25 uL. The forward primer for ANP gene was 5′-AGATAACAGCCAGGGAGGACAA-3′ and the reverse primer for ANP gene was 5′-AGGCGAGGAAGTCACCATCAA-3′. The thermal cycling conditions were as follows: initial denaturation step at 95°C for 10 min followed by 40 cycles that included 15 seconds at 95 C° for denaturation, 30 seconds at 60 C° for annealing and 30 seconds at 72 C° for an extension. 18S rRNA gene was used as the endogenous control with its expression being stable in all samples and independent of the analyzed variables. The forward primer for 18S rRNA gene was 5′-GTGGTGTTGAGGAAAGCAGACA-3′ and the reverse primer for 18S rRNA gene was 5′-TGATCACAGGTTCCACCTCATC-3′. Relative levels of ANP gene expression were calculated using the 2 −ΔΔCT method [Citation36].
2.1.4. Statistical analysis
IBM SPSS software version 20 was used for data analysis. Values of p˂0.05 were considered statistically significant. Quantitative data were expressed as mean ±standard deviation (SD). Student’s t-test was use for comparison between 2 groups of normally distributed data. Comparisons between groups for categorical variables were assessed using the chi-square test. Mann–Whitney test was used for comparing every two groups. Correlation between various parameters was calculated using Pearson’s test. ROC curve analysis was used to determine the sensitivity and specificity of ANP gene expression in differentiating hypertensives from normotensives.
3. Results
3.1. Demographic characteristics of hypertensive patients and normotensive controls
The baseline criteria of both groups of the study population can be checked in . The distribution of hypertensive patients and normotensive controls by age, gender, smoking status and BMI was similar and insignificant between the controls and the hypertensive patients. On the other hand, systolic and diastolic blood pressures were significantly increased among EH patients compared with controls (SBP: 161.7 ± 14.7 for patients vs 118.8 ± 5.6 for controls and DBP: 98 ± 6.5 for patients vs 79.5 ± 5.2 for controls) (p < 0.001). Regarding laboratory data, no statistical significance was detected between both study groups regarding blood glucose, blood urea, serum creatinine, total cholesterol, HDL-cholesterol, triglycerides, sodium, and potassium levels. Only LDL-cholesterol showed a significant difference between hypertensive patients and normotensive controls (99.4 ± 19 for patients vs 107.4 ± 17.4 for controls) (p < 0.001, ).
Table 1. Clinicopathological features of study subjects
3.2. ANP gene expression profile and its association with the clinical and laboratory findings
There was a statistical difference between ANP gene expression and blood pressure level, showing lower ANP gene expression level among hypertensive patients versus higher ANP gene expression level among normotensive controls (p < 0.001, , ).
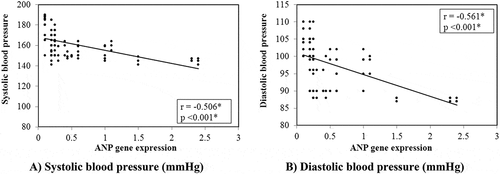
represents the scatter plot between SBP and ANP, and DBP and ANP gene expression levels among hypertensive patients. It showed that there is a negative correlation between ANP gene expression level and both SBP (r = −0.506, p < 0.001) and DBP (r = −0.561, p < 0.001).
Hypertensive patients were divided into two groups by the median value of the ANP gene expression into a high expression-ANP group and low expression-ANP group (). No statistical significance was observed between the different expression levels of ANP gene and other clinical variables, including age, gender, smoking status and BMI ().
Table 2. Relationship between ANP gene expression levels and different parameters in EH patients
There was an insignificant relationship between laboratory findings including blood glucose, blood urea, serum creatinine, LDL-cholesterol, HDL-cholesterol, triglycerides, sodium, potassium on one side and the different ANP gene expression levels on the other side, in both groups: patients and controls.
However, there was a significant correlation between SBP and DBP with the different expression levels of ANP gene expression (p < 0.001 and p < 0.001 respectively) ().
3.3. Analytical performance of ANP gene expression level
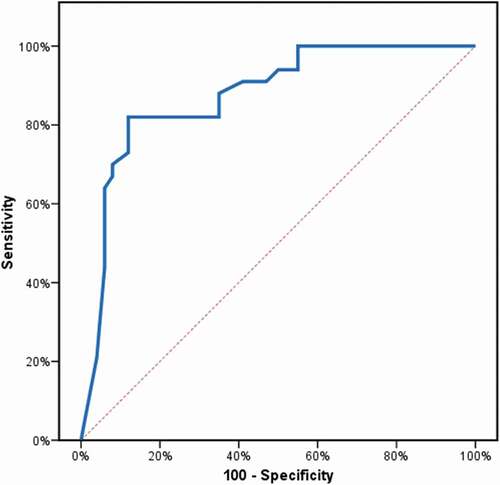
ROC curve analysis was used to determine the diagnostic performance of ANP gene expression in differentiating patients with hypertension from non-hypertensive controls. ANP expression at a cutoff value of ≤0.6 had 82% sensitivity and 88% specificity to detect increased blood pressure (, ).
Table 3. Agreement (sensitivity, specificity) for ANP gene expression to detect high A) SBP and B) DBP patients (vs control)
4. Discussion
Arterial Essential Hypertension (EH) represents a challenging public health issue worldwide [Citation1,Citation2]. It predisposes to major public health problems like cardiovascular illnesses, neurological sequelae, and renal impairment [Citation2,Citation6]. Therefore, we decided to carry out this study in order to find out whether gene expressions of related proteins like ANP would be of value in diagnosing and managing hypertension in an earlier stage before complications occur, or if it could value in choosing treatment modalities.
In this study, hypertensive patients showed lower expression of ANP gene than normotensive controls. As we know, The RAAS system helps regulate blood pressure and homeostasis within the human body through various mechanisms involving hormones and enzymes [Citation21]. RAAS is certainly involved in essential hypertension development [Citation21]. ANP, an essential component of the RAAS, has been shown to be involved in smooth muscle contraction through cGMP kinase, diuresis, and inhibition of RAAS, affecting salt and water retention and blood pressure [Citation37,Citation38]. ANP gene was previously proved to encode high blood pressure levels in animal models [Citation39].
In our study, we detected down-regulation of ANP gene expression in EH patients as compared to normotensive controls. We also noted a negative correlation between ANP gene expression and both SBP and DBP in EH patients. Therefore, we assumed that decreased expression level of ANP gene might be responsible for hypertension development among individuals. In concordance with our results, an earlier study on obese hypertensive patients revealed lower ANP gene expression levels compared to normal individuals [Citation40]. Similar results were also obtained from previous studies carried out on animals showing decreased ANP mRNA and protein levels in hypertension [Citation41]. A previous study showed similar results of low ANP gene expression of the mRNA in EH with the possibility of being a key factor in the disease progression [Citation21].
Wang et al. demonstrated that obese individuals had low ANP serum levels and obesity is a well-known risk factor for hypertension [Citation42]. A previous study on rats showed that gene expression of cardiac natriuretic peptide receptor-A was reduced on infusion of angiotensin II in hearts [Citation43].
The RAAS inhibitors as ACE inhibitors and Ang II type 1 receptor blockers (ARBs), despite being effective in reducing blood pressure, they do not prevent end-organ damage. This led to the exploration of alternative treatments for hypertension. Ghatage et al. discussed recent developments in RAAS therapy that might treat hypertension as well as improve end-organ damage. They shed lights on several peptides and non-peptides with better effects in hypertension treatment that are still under research, for instance, angiotensinogen siRNA and AT2 receptor stimulators [Citation2,Citation44,Citation45].
Another similar study conducted by Ghodsian and his colleagues studied ANP genetic analysis in EH. They screened genetic polymorphisms of ANP among the Malaysian population. They proved that ANP gene polymorphism was a risk factor for hypertension [Citation46]. Another study showed that ACE genetic polymorphisms were independent risk factors for hypertension [Citation47].
A recent study demonstrated that Ang II inhibited natriuretic peptide receptor 1 (Npr1) transcription by phosphorylating cAMP response element-binding protein (CREB) protein. This Npr1 gene transcription inhibition critically induced hypertension and cardiovascular dysfunction They recommended cotreatment with class I histone deacetylase (HDAC) inhibitors to reverse Ang II inhibitory effect on Npr1 expression [Citation48].
Some limitations of our study should be mentioned. These included small sample size and follow-up of the patients to demonstrate the effect of ANP gene expression on hypertension over time and with treatment.
In conclusion, our study showed that ANP gene expression was low in essential hypertension patients compared with normotensive individuals and that new antihypertensive medications are required in the future in order to treat hypertension as well as its complications especially on the cardiovascular system.
Data availability
The datasets used during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We sincerely thank all patients and healthy volunteers for their participation in the current study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Canale MP, Noce A, Di Lauro M, et al. Gut dysbiosis and western diet in the pathogenesis of essential arterial hypertension: a narrative review. Nutrients. 2021;13(4). DOI:https://doi.org/10.3390/nu13041162.
- Ghatage T, Goyal SG, Dhar A, et al. Novel therapeutics for the treatment of hypertension and its associated complications: peptide- and nonpeptide-based strategies. Hypertens Res. 2021;44(7):740–755.
- Mills KT, Stefanescu A, He J. The global epidemiology of hypertension. Nat Rev Nephrol. 2020;16(4):223–237.
- Bolívar JJ. Essential hypertension: an approach to its etiology and neurogenic pathophysiology. Int J Hypertens. 2013;2013:547809.
- Charles L, Triscott J, Dobbs B. Secondary hypertension: discovering the underlying cause. Am Fam Physician. 2017;96(7):453–461.
- Fuchs FD, Whelton PK. High blood pressure and cardiovascular disease. Hypertension. 2020;75(2):285–292.
- Lewington S, Clarke R, Qizilbash N, et al. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360(9349):1903–1913.
- Williams RR, Hunt SC, Hasstedt SJ, et al. Current knowledge regarding the genetics of human hypertension. J Hypertens Suppl. 1989;7(6):S8–13.
- Oparil S, Acelajado MC, Bakris GL, et al. Hypertension. Nat Rev Dis Primers. 2018;4:18014.
- Reisin E, Messerli FH. Obesity-related hypertension: mechanisms, cardiovascular risks, and heredity. Curr Opin Nephrol Hypertens. 1995;4(1):67–71.
- Rocchini AP, Moorehead CP, DeRemer S, et al. Pathogenesis of weight-related changes in blood pressure in dogs. Hypertension. 1989;13(6 Pt 2):922–928.
- Rocchini AP. Insulin resistance and blood pressure regulation in obese and nonobese subjects. Special lecture. Hypertension. 1991;17(6 Pt 2):837–842.
- Rocchini AP, Key J, Bondie D, et al. The effect of weight loss on the sensitivity of blood pressure to sodium in obese adolescents. N Engl J Med. 1989;321(9):580–585.
- Hall JE, Do Carmo JM, da Silva AA, et al. Obesity, kidney dysfunction and hypertension: mechanistic links. Nat Rev Nephrol. 2019;15(6):367–385.
- Paz Ocaranza M, Riquelme JA, García L, et al. Counter-regulatory renin-angiotensin system in cardiovascular disease. Nat Rev Cardiol. 2020;17(2):116–129.
- Poch E, González D, Giner V, et al. Molecular basis of salt sensitivity in human hypertension. Evaluation of renin-angiotensin-aldosterone system gene polymorphisms. Hypertension. 2001;38(5):1204–1209.
- Oparil S, Schmieder RE. New approaches in the treatment of hypertension. Circ Res. 2015;116(6):1074–1095.
- Mehta PK, Griendling KK. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am J Physiol Cell Physiol. 2007;292(1):C82–97.
- Kumar U, Wettersten N, Garimella PS. Cardiorenal syndrome: pathophysiology. Cardiol Clin. 2019;37(3):251–265.
- Nguyen Q, Dominguez J, Nguyen L, et al. Hypertension management: an update. Am Health Drug Benefits. 2010;3(1):47–56.
- Chandra S, Saluja D, Narang R, et al. Atrial natriuretic peptide and aldosterone synthase gene in essential hypertension: a case-control study. Gene. 2015;567(1):92–97.
- Pandey KN. Molecular and genetic aspects of guanylyl cyclase natriuretic peptide receptor-A in regulation of blood pressure and renal function. Physiol Genomics. 2018;50(11):913–928.
- Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N Engl J Med. 1998;339(5):321–328.
- Dzau VJ, Baxter JD, Cantin M, et al. Nomenclature for atrial peptides. N Engl J Med. 1987;316(20):1278–1279.
- Goetze JP, Bruneau BG, Ramos HR, et al. Cardiac natriuretic peptides. Nat Rev Cardiol. 2020;17(11):698–717.
- Ichiki T, Huntley BK, Heublein DM, et al. Corin is present in the normal human heart, kidney, and blood, with Pro–B-Type natriuretic peptide processing in the circulation. Clin Chem. 2011;57(1):40–47.
- Yan W, Wu F, Morser J, et al. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc Nat Acad Sci. 2000;97(15):8525–8529.
- Misono KS, Philo JS, Arakawa T, et al. Structure, signaling mechanism and regulation of the natriuretic peptide receptor guanylate cyclase. FEBS J. 2011;278(11):1818–1829.
- Pandey KN. Guanylyl cyclase/natriuretic peptide receptor-A signaling antagonizes phosphoinositide hydrolysis, Ca2+ release, and activation of protein kinase C. Front Mol Neurosci. 2014;7:75.
- Potter LR, Yoder AR, and Flora DR, et al. Natriuretic peptides: their structures, receptors, physiologic functions and therapeutic applications. Handb Exp Pharmacol. 2009;(191):341–366 doi: https://doi.org/10.1007/978-3-540-68964-5_15. PMID: 19089336; PMCID: PMC4855512. doi: https://doi.org/10.1007/978-3-540-68964-5_15. PMID: 19089336; PMCID: PMC4855512.
- Nose H, Takamata A, Mack GW, et al. Right atrial pressure and ANP release during prolonged exercise in a hot environment. J Appl Physiol (1985). 1994;76(5):1882–1887.
- Yan W, Sheng N, Seto M, et al. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274(21):14926–14935.
- Kessler-Icekson G, Barhum Y, Schaper J, et al. ANP expression in the hypertensive heart. Exp Clin Cardiol. 2002;7(2–3):80–84.
- Rubattu S, Bigatti G, Evangelista A, et al. Association of atrial natriuretic peptide and type A natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006;48(3):499–505.
- Carretero OA, Oparil S. Essential hypertension. Circulation. 2000;101(3):329–335.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Kiemer AK, Fürst R, Vollmar AM. Vasoprotective actions of the atrial natriuretic peptide. Curr Med Chem Cardiovasc Hematol Agents. 2005;3(1):11–21.
- Chen H, Levine YC, Golan DE, et al. Atrial natriuretic peptide-initiated cGMP pathways regulate vasodilator-stimulated phosphoprotein phosphorylation and angiogenesis in vascular endothelium. J Biol Chem. 2008;283(7):4439–4447.
- John SW, Krege JH, Oliver PM, et al. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science. 1995;267(5198):679–681.
- Dessì-Fulgheri P, Sarzani R, Tamburrini P, et al. Plasma atrial natriuretic peptide and natriuretic peptide receptor gene expression in adipose tissue of normotensive and hypertensive obese patients. J Hypertens. 1997;15(12 Pt 2):1695–1699.
- Kaganovsky E, Belkin V, Barhum Y, et al. Occurrence and distribution of atrial natriuretic peptide-containing cells in the left ventricle of hypertensive rats. Effect of antihypertensive treatment. Cell Tissue Res. 2001;303(1):57–67.
- Wang TJ, Larson MG, Levy D, et al. Impact of obesity on plasma natriuretic peptide levels. Circulation. 2004;109(5):594–600.
- Gopi V, Parthasarathy A, Umadevi S, et al. Angiotensin-II down-regulates cardiac natriuretic peptide receptor-A mediated anti-hypertrophic signaling in experimental rat hearts. Indian J Exp Biol. 2013;51(1):48–55.
- Arendse LB, Danser AHJ, Poglitsch M, et al. Novel therapeutic approaches targeting the renin-angiotensin system and associated peptides in hypertension and heart failure. Pharmacol Rev. 2019;71(4):539–570.
- Uijl E, Mirabito Colafella KM, Sun Y, et al. Strong and sustained antihypertensive effect of small interfering RNA targeting liver angiotensinogen. Hypertension. 2019;73(6):1249–1257.
- Ghodsian N, Ismail P, Ahmadloo S, et al. Genetic analysis of the atrial natriuretic peptide gene polymorphisms among essential hypertensive patients in Malaysia. Biomed Res Int. 2016;2016:6712529.
- Pinheiro DS, Santos RS, Jardim P, et al. The combination of ACE I/D and ACE2 G8790A polymorphisms revels susceptibility to hypertension: a genetic association study in Brazilian patients. PLoS One. 2019;14(8):e0221248.
- Arise KK, Kumar P, Garg R, et al. Angiotensin II represses Npr1 expression and receptor function by recruitment of transcription factors CREB and HSF-4a and activation of HDACs. Sci Rep. 2020;10(1):4337.