Abstract
Taxon delimitation in Pleurotus eryngii species-complex was investigated using an independent loci-based approach. A collection of 47 Pleurotus strains was analyzed through nucleotide sequences of the internal transcribed spacer region (ITS1, 5.8S and ITS2) and M13 minisatellite fragments. A sub-sample made of 12 clones representative of the ITS phylogenetic clusters was sequenced to elucidate variations in the elongation factor 1 alpha gene (EF1-α). A unique clade made of three evolutionary units, i.e. Chinese nebrodensis, Sicilian nebrodensis and eryngii-ferulae from the Mediterranean region, was revealed. The evolutionary genetic divergences were highly correlated (r = 0.86) for both genes. The average number of base substitutions per site was higher for EF1-α gene (0.013) than for ITS (0.001). M13 minisatellite polymorphism marked a clear association between strains and geographic origin but its resolution in taxa delimitation was low. Data from ITS and EF1-α genes supported the hypothesis that the species-complex is a unique gene-pool. Within the species-complex, the nebrodensis type is characterized by a level of nucleotide substitutions lower than as expected for its ranking at a species level.
Introduction
Pleurotus eryngii species-complex includes widely distributed basidiomycetes occurring in Europe, the Mediterranean region and Asia, specific to natural and semi-natural grasslands (Festuco–Brometea and Thero–Brachypodietea associations), and also grown as edible mushrooms on artificial substrates mainly in Europe, Israel, China and Japan.
Systematics of P. eryngii (DC.:Fr.) Quèl based on morphological (Linnaeus Citation1758), biological (Mayr Citation1942), and geographical and ecological (van Valen Citation1976) data lead to different classifications of the P. eryngii populations. Moreover, within the species-complex, taxa were grouped in botanical varieties (Inzenga Citation1863; Quelét Citation1872; Lanzi Citation1894; Hilber Citation1982; Mou et al. Citation1987; Lewinsohn et al. Citation2002; Venturella et al. Citation2002), sub-species (Zervakis and Balis Citation1996) or species (Candusso and Basso Citation1995).
The P. eryngii species-complex classification has been driven by the “host specificity” approach considering potential host plants, all belonging to the Apiaceae and Asteraceae families, on which the fungus performs as a weak parasite on stems and roots (Zervakis et al. Citation2001a; Lewinsohn et al. Citation2002). Based on this approach, P. eryngii (DC: Fr.) Quél. var. eryngii has been associated to root residues of Eryngium campestre L., P. eryngii var. ferulae Lanzi to Ferula communis L. subsp. communis, P. eryngii var. tingitanus Lewinshon et al. to Ferula tingitana, P. nebrodensis (Inzenga) Sacc. to Cachrys ferulacea (L.) Calestani, P. eryngii var. elaeoselini Venturella et al. to Elaeoselinum asclepium (L.) Bertol. subsp. asclepium, P. eryngii var. thapsiae Venturella et al. to Thapsia garganica L. and P. eryngii var. touliensis Mou to Ferula sinkinagensis Shen (Lewinsohn et al. Citation2002; Venturella et al. Citation2000, Citation2002; Kawai et al. Citation2008).
In a greenhouse-replicated trial, De Gioia et al. (Citation2005) found lower morphological differences among the a priori classified vars. ferulae, eryngii, and major differences with respect to the nebrodensis type. Taking into account morphological, biological and molecular data, the nebrodensis type has also been classified as a distinct species (Zervakis et al. Citation2001b).
Incomplete reproductive barriers are still present between taxa belonging to the P. eryngii species-complex (Zervakis and Balis Citation1996; Taylor et al Citation2000; Kawai et al. Citation2008; Rodriguez-Estrada et al Citation2010). Mating experiments performed under in vitro conditions showed that host-specific varieties were not isolated reproductively from each other and it was argued that mating between different strains could contribute to the homogenisation of genetic diversity within the species-complex (Hilber Citation1982; Vilgalys et al. Citation1993; Zervakis and Balis Citation1996). In addition, no definitive data is yet available about the ecological performance of straw distribution and establishment in natural habitats, genetic isolation by distance, degree and type of natural gene flow. To track host-specific biotypes, genets or genetically related sub-populations, a priori knowledge of the precise link between the experimental specimen and its host plant is relevant. Under these circumstances, species delimitation within the P. eryngii gene-pool is complicated.
Markers-assisted genomic analysis significantly increased the resolution of genetic relatedness studies within and among broader groups of extended kin (Vilgalys and Sun Citation1994; Zervakis et al. Citation1994, Citation2001b; Reverberi et al. Citation2000; Zhang et al. Citation2000, Citation2006; Venturella Citation2002; Urbanelli et al. Citation2002, Citation2003, Citation2007; Della Rosa et al. Citation2004; De Gioia et al. Citation2005; Ro et al. Citation2007; Kawai et al. Citation2008), as well as phylogenetic classification at a greater evolutionary depth (Zervakis et al. Citation1994, Citation2004; Gonzales and Labarère Citation2000; Taylor et al. Citation2000; Albertó et al. Citation2002; Rodriguez-Estrada et al. Citation2010). The degree of genetic separation between the supposed host specific ferulae and eryngii, however, is still controversial, since different authors (Zervakis et al. Citation2001b; Urbanelli et al. Citation2002, Citation2007; De Gioia et al. Citation2005) reported phylogenetic relationships at the level of extended kinship within local groups or sub-populations, rather than phylogenetic structures at intermediate or great evolutionary depths.
Several phylogenetic analyses using sequence data from the SSU rRNA mitochondrial gene or nuclear ITS DNA have shed light on inter- and intra-specific classification in Pleurotus (Vilgalys and Sun Citation1994; Gonzales and Labarère Citation2000; Albertó et al. Citation2002). These investigations showed that P. eryngii is more related to P. ostreatus (Jacq.: Fr.) Kummer and P. populinus Hilber and Miller than to other Pleurotus species. Within the P. eryngii gene pool, the nebrodensis type is the most diverging population and the extent of genetic variation does not clearly indicate whether its differentiation is at a “species” or at a “variety” level.
Different nuclear ribosomal RNA genes were extensively exploited in molecular fungal systematics: the small subunit RNA (17-18S) (Bruns et al. Citation1992; Swann and Taylor Citation1995; Guarro et al. Citation1999; Gonzales and Labarère 2000); large subunit RNA (25–28S) (Hibbett Citation1992; Hopple and Vilgalys Citation1999; Moncalvo et al. Citation2000; Maruyama et al. Citation2005; Padamsee et al. Citation2008); internal transcribed spacer (ITS) (White et al. Citation1990; O'Donnell Citation1992; Gardes and Bruns Citation1993; Chen et al. Citation2001; Park et al. Citation2004; Tuchwell et al. Citation2005; Zhang et al. Citation2006; Rodriguez-Estrada et al. Citation2008) and the intergenic spacer (IGS), including 5S RNA (Hibbett Citation1992; Appel and Gordon Citation1996; James et al. Citation2001; Alejandro and Todd Citation2005; Zhang et al. Citation2006; Geiser et al. Citation2007). ITS sequence genetic information performed as the best to discriminate within and between species in several living organisms including fungi.
Pleurotus molecular systematics based on both ITS and IGS sequence variation revealed two groups (P. eryngii var. ferulae and nebrodensis collected on Ferula sinkiangensis) within the Chinese germplasm (Zhang et al. Citation2006). In addition, Kawai et al. (Citation2008) used ITS sequences to propose the taxonomic position of the Chinese Pleurotus eryngii “Bai-Ling Gu” at variety level (P. eryngii var. touliensis). Furthermore, phylogenetic relationships have been inferred among varieties of P. eryngii using both ITS region and β–tubulin gene. Either allelic polymorphisms within the β-tubulin gene or lack of variation in the ITS region has been detected between the varieties eryngii and ferulae (Rodriguez-Estrada Citation2008; Rodriguez-Estrada et al Citation2008). Thus, these genes proved to be unable to provide a proof for species delimitation within the P. eryngii species-complex. Nonetheless, different genes can provide specific genetic information to resolve different biological problems. Among these genes, the translation elongation factor (EF1-α), involved in ribosomal protein synthesis in eukaryotes, was previously used for phylogeny studies on several basidiomycetes (Matheny et al. Citation2007), ascomycetes, such as Fusarium species (Kristensen et al. Citation2005; Bentley et al. Citation2006; Maphosa et al. Citation2006; Steward et al. Citation2006; Dupont et al. Citation2007; Mansfield and Kuldau Citation2007), on arbuscular mycorrhizal fungi (Helgason et al. Citation2003), and Puccinia and Uromyces spp. (Van der Merwe et al. Citation2007). Even so, this gene is still less exploited for Pleurotus genus (Marongiu et al. Citation2005; Mang Citation2008; Mang and Figliuolo Citation2008; Rodriguez-Estrada et al. Citation2010).
According to Avise and Ball (Citation1990), the phylogenetic species concept can contribute to a significant advance in species recognition if the magnitudes of phylogenetic patterns and the historical and reproductive reasons for such patterns are integrated as interpretative tools. Within this context, the goal of this study was to provide additional evidence about taxon delimitation within the P. eryngii gene-pool by focusing on ITS and EF1-α gene sequence analysis along with the M13 minisatellite polymorphisms.
Materials and methods
Strains studied
The fungal strains used in this study were characterised by a brown or white pileus colour, and had different geographic origins – Apulia Region (South Italy), Sicily (Mediterranean Area), Sardinia (Mediterranean Area) and China, and their preliminary classification (vars. eryngii, ferulae and nebrodensis) was based on the host plant ().
Table 1. Pleurotus eryngii sample core (47 strains) characterized via molecular analyses. The a priori classification, based on overall morphology ideotypes, was: type nebrodensis, ferulae and eryngii. The phylogenetic sub-clusters based on ITS and EF-1α gene sequences were: A = P. nebrodensis (Sicily), B = P. nebrodensis (China) and C = P. eryngii (including the a priori type ferulae and eryngii from Apulia, Sardinia and Sicily). All sequences were generated in this study (for details see GenBank accession numbers)
All cultures were maintained on malt extract agar medium (5% malt extract, 1.5% agar), stored at 4°C in vials and the vouchers kept in the fungal culture collection (De Gioia et al. Citation2005). Samples were analyzed with both ITS gene and M13 marker. A group composed of 12 strains, each one nested within the ITS clusters, was used to clone the EF1-α gene. Furthermore, the selected set of clones was composed of at least one specimen from each eco-geographic origin and representative of the a priori defined taxon of the P. eryngii species-complex. The P. ostreatus nucleotide sequences of the ITS region and EF1-α gene (accession numbers EU520255 and AY883432, respectively) retrieved from the GenBank database (NCBI) were used as reference for a species within the genus for phylogenetic analyses.
DNA isolation
DNA was isolated from freshly harvested mycelium scraped off the surface of a growing colony as described in De Gioia et al. (Citation2005).
ITS amplification and sequencing
The entire region of nuclear rDNA composed of ITS1 and 2 spacers and including also the 5.8S RNA gene was amplified using the universal primers: ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), as reported in White et al. (Citation1990). The same oligos were used as sequencing primers. Each PCR reaction, containing in a 50-μl reaction volume 1X PCR buffer, 2 mM MgCl2, 0.25 mM dNTPs, 1 U Taq Phusion™ High-Fidelity DNA polymerase (Finnzymes, Finland), 0.2 μM of each primer and 20 ng of purified total DNA template, was carried out at least twice. Polymerase chain reactions were performed with I-Cycler thermalcycler (BioRad, Hercules, CA, USA) and consisted of the following steps: 30 s at 98°C, 10 s at 98°C for 30 cycles, 30 s at 50°C and 30 s at 72°C, 5 min at 72°C. Amplified DNA fragments were separated in a 1% agarose gel for further analyses. The ITS amplicons yield was sufficient to allow direct sequencing. PCR products were purified and concentrated using standard protocols (Sambrook et al. Citation2001). DNA was dried by lyophilization, resuspended in 5 μl of sterile water, and then used as template in a sequencing grade reaction performed with BigDye® Terminator v.1.1 Cycle Sequencing Kit (Figliuolo and Di Stefano Citation2007). Sequencing products were cleaned with Centri-Sep columns (Princeton Separations, Inc., Princeton, NY, USA), lyophilized with Hetosicc apparatus and diluted in formamide. Sequencing was achieved with AB3130 Genetic Analyzer (Applied Biosystems, Inc., 2005). Sequence alignments and SNPs discovery were performed with SeqScape v. 3.1. Program (Applied Biosystems, Inc., 2005). Sequences have been deposited in GenBank/NCBI (accession numbers FJ904724–FJ904770; ). Alignments were carried out using the CLUSTALW (progressive alignment methods) into the MEGA 4.0 (Tamura et al. Citation2007) Evolutionary Genetic Analysis software.
EF-1α gene amplification, cloning and clones analyses
The EF-1 α gene region was amplified using Pleurotus genomic DNA as a template and the overlapping degenerated EF1-α primer sequences (Rehner Citation2001). Primer combination 526F (forward: 5′-GTCGTYGTYATYGGHCAYGT-3′) × 1567R (reverse: 5′-ACHGTRCCRATACCACCRATCTT-3′) was used to amplify 1000 bp of the EF1-α gene. PCR reaction conditions were: 1 min at 98 °C; 10 s at 98 °C for 30 cycles, 30 s at 65 °C, 15 s at 72 °C, 5 min at 72 °C. The amplicons from single locus were directly cloned into PCR Zero Blunt Vector using the Zero Blunt PCR Cloning Kit (Invitrogen USA). Twelve positive colonies were cultured into Luria broth media plus antibiotic (kanamycin 50 μg/ml). Subsequently, plasmid DNA was extracted from each positive colony using a PureLink HQ Mini Plasmid Purification Kit (Invitrogen). The EF1-α gene sequencing was carried out using M13 sequencing primers. Sequencing grade reaction preparation, chromatogram analysis and SNPs discovery were performed as reported above for the ITS region. The EF1-α gene nucleotide sequences deposited in GenBank/NCBI have accession numbers FJ904771–FJ904782 ().
Phylogenetic analyses
Nucleotide sequences were analyzed with MEGA 4.0 software, treating gaps as missing data, excluding ambiguous positions (complete deletion option) and using the CLUSTAL alignment method. Following sequence alignment, the evolutionary distances interpreted as units of the number of base substitutions per site were computed using the Maximum Composite Likelihood method (Tamura et al. Citation2004). Codon positions included were 1st+2nd+3rd+Noncoding. ITS and EF1-α data were analyzed both separately and in combination. For each gene, the evolutionary history was inferred using either the Neighbor-Joining (NJ) method (Saitou and Nei Citation1987) or the Maximum Parsimony (MP) method (Eck and Dayhoff Citation1966). A bootstrap consensus un-rooted tree (cut-off value 70%) was inferred from 1000 replicates to represent the evolutionary history of the taxa (Felsenstein Citation1985). The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Since the clustering outputs from the two clustering methods were concordant, only the NJ trees have been presented in this paper. Before combining the ITS and EF1-α loci across 12 different specimens for phylogenetic analyses, the relationship strength between the evolutionary distances of the above genes was tested. Using regression analysis (Proc reg in SAS Software) between the two matrices of genetic distances, the regression coefficient between ITS and EF1-α evolutionary distances and the r 2 value were computed with SAS v. 9.1 2002-2003 (SAS Institute Inc., Cary, NC, USA).
Minisatellite M13 amplification and analysis
Minisatellite variation analysis was carried out using the universal primer M13 (GAGGGTGGCGGTTCT) and amplifying the repetitive DNA genomic sequence (Latouche et al. Citation1997). PCR reaction was performed as indicated by Williams et al. (Citation1990) with minor modifications (25 μl final volume of PCR reaction, containing 1× PCR buffer, 2 mM MgCl2, 0.25 mM dNTPs, 1 U Taq DNA polymerase (Invitrogen), 0.2 μM M13 universal primer and 20 ng of genomic DNA template). Amplifications were performed with I-Cycler thermalcycler (BioRad) and consisted of the following steps: 2 min at 95°C, 1 min at 94°C for 35 cycles, 1 min at 45°C, 1 min at 72°C, 5 min at 72°C. Finally, amplified DNA fragments were separated in a 2% agarose gel and visualized with ethidium bromide.
M13 fragment analysis
Each M13 marker band was visually detected and relative mobility was recorded using a binary code. To calculate genetic distance indices, each band, at a given level of molecular weight, was interpreted as a single allele and each genotype as a single haplotype. Faint bands were not scored. The Dice's genetic diversity index (GD), measuring the number of unshared bands from multiple loci profile for each genotypic pairwise comparison (Nei and Li Citation1979), cluster analysis (Proc cluster, Average method) and tree drawing (Proc tree in SAS Software), were computed (SAS v. 9.1, 2002–2003).
Results
ITS and EF-1α nucleotide sequence variation
The PCR reaction performed with ITS1/4 universal primers yielded a single amplicon of about 700 bp allowing direct sequencing of the ITS region. A set of 47 ITS region nucleotide sequences were obtained and analysed () along with the 762-bp ITS sequence of P. ostreatus (EU520255) retrieved from GenBank (NCBI). The average content in four bases of the ITS region for the 47 strains belonging to P. eryngii species-complex was: T = 29.7%, C = 22.4%, A = 26.1% and G = 21.8%, with a transitions/transversions ratio equal to 1.4. The final alignment contained 615 nucleotide sites, of which 598 sites were constant, 17 were variable. Of 17 variable sites, 15 were parsimony-informative and two singletons (parsimony-uninformative). Namely, zero variable sites were recorded in nebrodensis type from China (three strains), one site was recorded in nebrodensis from Sicily (six strains) and six sites were registered in eryngii-ferulae types from Apulia and Sardinia (38 strains). For the outgroup strain P. ostreatus (EU502055), 461 sites were constant and 154 were variable.
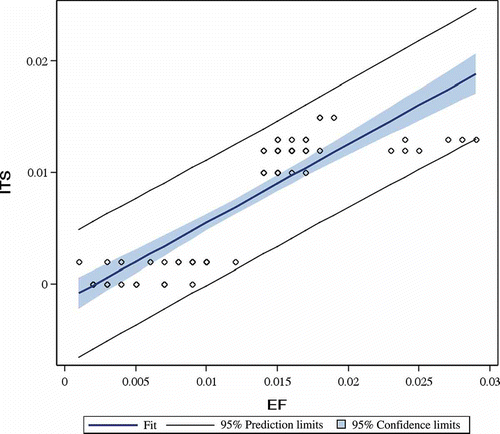
The 12 EF1-α gene sequences obtained in this study were of ∼1000 bp (). The clones average content in four bases was T = 23.9%, C = 27.5%, G = 23.6% and A = 25.0%, with a transitions/transversions ratio equal to 3.4. The final alignment contained over 973 nucleotide sites, of which 937 sites were constant, 35 were variable with 22 parsimony-informative sites. For the outgroup strain P. ostreatus (AY883432), the number of variable sites increased to 57. The distribution of the variable sites over the EF1-α clone set was partitioned between the gene-pools as follows: six sites in nebrodensis from Sicily and 15 sites were observed in eryngii-ferulae types. The average evolutionary divergence for P. eryngii species-complex, without considering P. ostreatus, was higher for EF-1α (d = 0.013) than for ITS (d = 0.001) (), as shown by the linear regression analysis of pairwise evolutionary genetic distances, computed using the common set of twelve strains for both genes (). Also, divergence within nebrodensis was higher (d = 0.002 for ITS and d = 0.009 for EF-1α) than within eryngii-ferulae types (d = 0.000 for ITS and d = 0.007 for EF-1α) (; and ). The divergence within the nebrodensis is consistent with the inter-continental geographic distance between Chinese and Sicilian strains. On the other hand, the eryngii-ferulae strains from the Mediterranean area showed a reduced within gene-pool divergence (; and ). The average evolutionary divergence between nebrodensis and eryngii-ferulae types was d = 0.002 for ITS and d = 0.017 for EF-1α (). Including P. ostreatus, the overall mean genetic distance for the ITS was 0.028 and for the EF-1α was 0.016 ().
Table 2. Average evolutionary divergence calculated via the maximum composite likelihood method for the ITS (615 bp) and EF1-α (973 bp) genes analyzed separately using as population P. eryngii species-complex without (top of the table) or with P. ostreatus
Figure 1. Evolutionary relationships among 47 strains of P. eryngii species-complex based on 584-bp ITS gene sequence. The evolutionary history was inferred using the Neighbor-Joining method and the analyses were conducted in MEGA4. The bootstrap consensus tree inferred from 1000 replicates and bootstrap values greater than 70% are shown. One ITS sequence (EU520255) belonging to P. ostreatus taken from the NCBI GenBank was included as species reference within the Pleurotus genus.

Figure 2. Evolutionary relationships among 13 taxa of P. eryngii species-complex based on EF1-α gene (∼600 bp) sequence variation. The evolutionary history was inferred using the Neighbor-Joining method and the analyses were conducted in MEGA4. The bootstrap consensus tree inferred from 1000 replicates and bootstrap values greater than 70% are shown. One EF1-α gene sequence (AY883432) belonging to P. ostreatus taken from the NCBI GenBank was included as species reference within the Pleurotus genus.
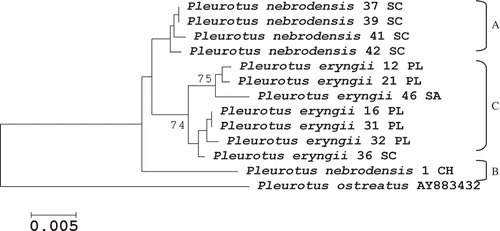
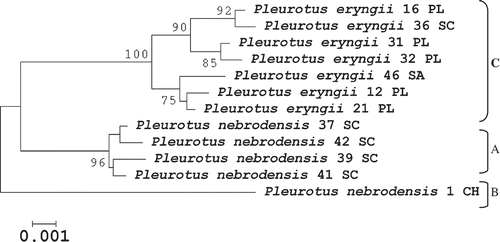
Figure 4. Evolutionary relationships among 12 Pleurotus taxa belonging to the P. eryngii species-complex based on both ITS and EF-1 (1574 bp) sequence variation. The evolutionary history was inferred using the Neighbor-Joining method and the analyses were conducted in MEGA4. The bootstrap consensus tree inferred from 1000 replicates and bootstrap values greater than 70% are shown.
The pairwise genetic distances at interspecific level (P. ostreatus vs. nebrodensis or P. ostreatus vs. eryngii-ferulae) for both ITS and EF-1α genes were ∼0.6 and ∼0.05, respectively. These values are larger than the distance between nebrodensis and eryngii-ferulae by a factor of ∼300 for EF-1α and ∼2.5 for ITS ().
Evolutionary trees based on ITS and EF1-α nucleotide variation
A unique clade of P. eryngii species-complex composed of three sub-groups consistently separated from P. ostreatus, which performed as a distinct species, was obtained using the evolutionary molecular ITS distances. Namely, the sub-clades were nebrodensis type from Sicily (A), highly supported with 82% of bootstrap values, nebrodensis type from China (B), also highly supported with the 97% of bootstrap values, and eryngii-ferulae group from Mediterranean region (group C), with a bootstrap value lower than 70% (, and ). The eryngii-ferulae sub-population from Apulia, Sardinia and Sicily showed two variants of the pileus colour (brown and white) () and genetic homogeneity for their ITS sequences, as shown in .
The NJ analysis of the EF1-α gene yielded a topology close to the ITS cluster, in which all taxa were grouped together into a unique clade, made of three sub-groups, the same as for ITS (A, B and C). Once more, the EF-1α sequence of P. ostreatus, based on its nucleotide sequence variation, was placed outside of the P. eryngii species-complex gene-pool as a different species within the genus (). The EF1-α gene, owing to its higher level of divergence, which was supported by high values of bootstrap (>70%), was better than ITS in separating the Mediterranean strains of the eryngii-ferulae gene-pool.
Given the congruent topology of the clusters separately obtained with ITS and EF1-α genes ( and ) and to increase our phylogenetic analyses resolution, both genes were analyzed over a common dataset of 12 strains. A significant linear correlation (slope = 0.86; p < 0.001) was found between the two types of evolutionary genetic distances (). The final alignment of the ITS and EF1-α genes consisted of 1588 sites, of which 1537 sites were constant, 50 were variable and 32 parsimony-informative. Nucleotide sequence variation of the ITS and EF1-α combined genes allowed a steady phylogeny reconstruction. Once more, three sub-groups (A, B and C) were found inside the gene-pool clade (). The highest significance level (100% bootstrap value) was observed for the eryngii-ferulae types sub-clade, which was composed of all strains from different Italian regions (Apulia, Sardinia and Sicily). The sub-clade made of nebrodensis type strains originated from Sicily was highly supported (96%). As expected, the most diverging genotype was the nebrodensis strain from China. Branching inside the sub-clades was also supported by high values of the bootstrap ().
Minisatellite M13 length polymorphism
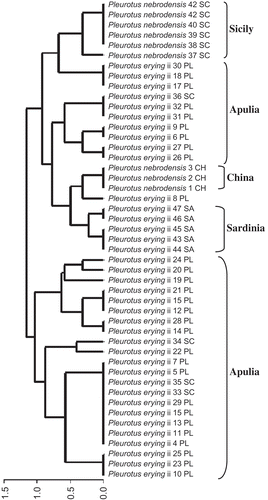
The pattern of M13 produced fragments of size ranging from 520 to 1300 bp. Minisatellite M13 clustering based on Dice's genetic distance showed that sub-groups of the P. eryngii species-complex were mainly structured according to the geographic origin of strains.
Nevertheless, the cluster included strains a priori classified as type nebrodensis from Sicily (which formed a sub-group) and strains classified as nebrodensis type from China. Strains from Sardinia (clustered into the same sub-group) and some strains from Apulia region formed the last sub-group of this cluster.
The second cluster was made of strains from Apulia region and contained three sub-groups showing various sub-clusters. The only exception was represented by a few Sicilian strains (33 SC, 34 SC and 36 SC) that were included in the same cluster as those from Apulia (P. eryngii and P. ferulae types) and did not belong to the nebrodensis type ().
Discussion
In the 19th Century, the P. eryngii species-complex was classified into botanical varieties (nebrodensis, eryngii and ferulae) (Inzenga Citation1863; Lanzi Citation1894 and Quelet 1872). Since then, many methods have been employed to understand taxonomy within this group, usually leading to different results. Most authors have reported the within gene-pool distinction at variety level (Hilber Citation1982; Mou et al. Citation1987; Venturella et al. Citation2000, Citation2002; Lewinsohn et al. Citation2002; De Gioia et al. Citation2005; Zhang et al. Citation2006; Ro et al. Citation2007; Kawai et al. Citation2008). In contrast, Urbanelli et al. (Citation2002, Citation2003) divided the vars. eryngii and ferulae into two distinct biological species. In addition, the white coloured nebrodensis type was considered a different species in many studies (Candusso and Basso Citation1995; Zhang et al. Citation2000, Citation2006; Zervakis et al. Citation2001b; Urbanelli et al. Citation2007). So far, molecular data seems to have supported the splitting of the gene-pool rather than its lumping. Nonetheless, our preliminary findings, based on intra-specific genetic distances within P. eryngii species-complex (De Gioia et al., Citation2005) and phylogeny (Mang and Figliuolo Citation2008; Mang Citation2008), support the presence of a single species not yet differentiated in sub-species. The same findings were recently presented by Rodriguez-Estrada et al. (Citation2010).
Our results clearly indicate that, at molecular and morphological levels, the P. eryngii species-complex can be divided in two main gene-pools: eryngii-ferulae type and nebrodensis type. The former group performs as a unique, homogenous gene-pool, and genetic differences between regional geographic origins can be dictated by recently evolved pedigree relationships, as demonstrated by the M13 marker. On the contrary, sequence variation displayed by the ITS and EF-1α genes, that predates the recent ancestry, operates as a taxonomic value marker. However, the higher rate of substitution observed within the species-complex for EF-1α was no longer noticed at species level when P. ostreatus was used as a reference.
Furthermore, phenotypes showed that some morphological markers, such as the absence or the presence of melanin, are not yet completely fixed within the gene-pool. In this study, strains labelled as P. eryngii 31_PL and P. eryngii 32_PL (eryngii-ferulae group), have white basidiomata colour. Our results agree with those of Zhang et al (Citation2006), which proved that the white basidiomata colour is not exclusively linked to var. nebrodensis but may also be found in some var. ferulae in China.
The nebrodensis type is the most differentiated sub-population at either molecular or morphological levels, but we consider that the number of fixed mutations is still too low to differentiate the lineage at species level within the genus. Thus, the phylogenetic species concept should integrate the magnitude of the threshold of nucleotide substitutions required for species differentiation. We also found that the nebrodensis group (either Sicilian or Chinese) had two fixed single nucleotide substitutions over the ITS region that could be used as a future molecular diagnostic tool. It is likely that the nebrodensis type from China is the most diverging gene-pool owing to the isolation by distance. This consideration is supported in this work by the average evolutionary divergence between the groups within the species-complex and the species within the genus. For instance, for ITS gene, the distance between nebrodensis and eryngii-ferulae is 0.002, while between P. ostreatus and nebrodensis it is 0.602, and a similar trend is obtained when comparing the P. ostreatus with the eryngii-ferulae types. However, a statistical molecular threshold for species recognition within the Pleurotus genus is not available. Vilgalys and Sun (Citation1994) showed that the levels of nucleotide substitution for the ITS dataset can reach 0.41 in the case of eight different species of the Pleurotus genus. Sreenivasaprasad et al. (Citation1992) considered that 6% variation in the ITS nucleotide sequence is acceptable as a threshold for species delineation of Colletotrichum, while Vinnere (Citation2004) illustrated that variation among isolates that are clustering in the Colletotrichum acutatum sensu lato can reach 9.6%, using the same genetic region. Zervakis and Balis (Citation1996) demonstrated that the percentage of compatible matings between ecotypes was variable (∼45% in pairing-analysis including nebrodensis and ∼60% between vars. eryngii and ferulae). It seems that a genetic differentiation is underway, probably promoted by ecological forces (climate and biological substrates) and geographic isolation. In addition, the in situ biological and genetic performance of the P. eryngii is still unclear given that it is not possible to exclude saprotrophism or growth preference of the mycelia straw also on different host residues of the poly-specific herbaceous coat composing the natural and semi-natural Festuco–Brometalia prairies. In several cases, the supposed host plants (e.g. Ferula communis and Eryngium campestris) are present in the same habitat (Forte et al. Citation2005).
De Gioia et al. (Citation2005) showed that variation for morphological traits and molecular markers within and among Mediterranean populations of P. eryngii was able to identify the population genetic structure of P. eryngii species-complex. This can be explained by the out-crossing performance associated with a high genetic compatibility within and between a priori defined eryngii and ferulae types. Besides, in different reports, the nebrodensis type was later classified as different vars. touliensis and elaeoselini (Inzenga Citation1863; Venturella et al. Citation2000; Kawai et al. Citation2008).
In conclusion, the results of this study, based on the multi-locus analysis, provided sufficient molecular evidence separating macroevolution (ITS and EF-1α genes) from regional patterns of recent variation (M13 marker) and supported a single evolutionary unit represented by P. eryngii. The overall molecular analyses clarified species delineation in the P. eryngii gene-pool and demonstrated that a separation of P. nebrodensis from P. eryngii at a species level would require a higher degree of evolutionary divergence. Also it has been confirmed that the eryngii and ferulae types are belonging to the same taxon (P. eryngii). Our conclusions support the hypothesis that, within the species P. eryngii, the nebrodensis type is better ranked at a variety level.
Acknowledgements
The authors thank Professor G.L. Rana (Università degli Studi della Basilicata, Potenza, Italy) for providing the Chinese Pleurotus strains.
References
- Albertó , EO , Petersen , HR , Hughes , KW and Lechner , B. 2002 . Miscellaneous notes on Pleurotus . Persoonia , 18 : 55 – 69 .
- Alejandro , R and Todd , W. 2005 . Evolution of a large ribosomal RNA multigene family in filamentous fungi: Birth and death of a concerted evolution paradigm . Proc Natl Acad Sci USA , 102 ( 14 ) : 5084 – 5089 .
- Appel , DJ and Gordon , TR. 1996 . Relationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA . Mol Plant–Microbe Interact. , 9 : 125 – 138 .
- Avise , JC and Ball , RM. 1990 . Principles of genealogical concordance in species concepts and biological taxonomy . Oxford Surv. Evol. Biol. , 7 : 45 – 67 .
- Bentley , AR , Cromey , MG , Farrokhi-Nejad , R , Leslie , JF , Summerell , BA and Burgess , LW. 2006 . Fusarium crown and root rot pathogens associated with wheat and grass stem bases on the South Island of New Zealand . Aust Plant Pathol. , 35 ( 5 ) : 495 – 502 .
- Bruns , TD , Vilgalys , R , Barns , SM , Gonzalez , D , Hibbett , DS , Lane , DJ , Simon , L , Stickel , S , Szaro , TM , Weisburg , WG and Sogin , ML. 1992 . Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences . Mol Phylogenet Evol. , 1 : 231 – 241 .
- Candusso , M and Basso , MT. 1995 . Comparative analysis of Pleurotus eryngii and P. nebrodensis . Documents Mycologique , 100 : 119 – 128 .
- Chen , YC , Eisner , JD , Kattar , MM , Rassoulian-Barrett , SL , Lafe , K , Bui , U , Limaye , AP and Cookson , BT. 2001 . Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts . J Clin Microbiol. , 39 : 4042 – 4051 .
- De Gioia , T , Sisto , D , Rana , GL and Figliuolo , G. 2005 . Genetic structure of the Pleurotus eryngii species-complex . Mycol Res. , 109 ( 1 ) : 71 – 80 .
- Della Rosa , V , Capuccio , I , Fanelli , C and Urbanelli , S. 2004 . Isolation and characterization of microsatellite markers in two basidiomycete species: Pleurotus eryngii and P. ferulae . Premier Note. Mol Ecol Notes , 4 : 271 – 273 .
- Dupont , J , Jacquet , C , Dennetière , B and Lacoste , S. 2007 . Invasion of the French Paleolithic painted cave of Lascaux by members of the Fusarium solani species complex . Mycologia , 99 ( 4 ) : 526 – 533 .
- Eck , RV and Dayhoff , MO. 1966 . Atlas of Protein Sequence and Structure , Silver Springs, MD : National Biomedical Research Foundation .
- Felsenstein , J. 1985 . Confidence limits on phylogenies: An approach using the bootstrap . Evolution , 39 : 783 – 791 .
- Figliuolo , G and Di Stefano , D. 2007 . Is single bulb producing garlic Allium sativum or Alllium ampeloprasum? . Sci Hortic. , 114 ( 4 ) : 243 – 249 .
- Forte , L , Perrino , EV and Terzi , M. 2005 . Le praterie a Stipa austroitalica Martinovsky’ ssp. austroitalica dell'Alta Murgia (Puglia) e della Murgia Materana (Basilicata) . [The prairies with Stipa austroitalica Martinovsky’ ssp. austroitalica from “Alta Murgia” (Apulia) and “Murgia Materana” (Basilicata)] Fitosociologia , 42 : 83 – 103 . in Italian
- Gardes , M and Bruns , TD. 1993 . ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts . Mol Ecol. , 2 : 113 – 118 .
- Geiser , DM , Gueidan , C , Miadlikowska , J , Lutzoni , F , Kauff , F , Hofstetter , V , Fraker , E , Schoch , CL , Tibell , L , Untereiner , WA and Aptroot , A. 2007 . Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae . Mycologia , 98 : 1054 – 1065 .
- Gonzalez , P and Labarère , J. 2000 . Phylogenetic relationships of Pleurotus species according to the sequence and secondary structure of the mithocondrial small-subunit rRNA, V4, V5, V6 and V9 domains . Microbiology , 146 : 209 – 221 .
- Guarro , J , Gene , J and Stchigel , A. 1999 . Developments in fungal taxonomy . Clin Microbiol Rev. (12) , 3 : 454 – 500 .
- Helgason , T , Watson , IJ and Young , PW. 2003 . Phylogeny of the Glomerales and Diversisporales (Fungi Glomeromycota) from actin and elongation factor 1 alpha sequences . FEMS Microbiol Lett. , 229 : 127 – 132 .
- Hibbett , DS. 1992 . Ribosomal DNA and Fungal Systematics . Trans Mycol Soc Jpn. , 33 : 533 – 566 .
- Hilber , O. 1982 . Die Gattung Plerurotus (Fr.) Kummer. Bibliotheca Mycologica 87 , 464 S Vaduz, , Germany : J. Kramer .
- Hopple , JS and Vilgalys , R. 1999 . Phylogenetic Relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly . Mol Phylogenet Evol. , 13 ( 1 ) : 1 – 19 .
- Inzenga , G. 1863 . Nuova specie di agarico del Prof . Giuseppe Inzenga. Giornale Reale Istituto Incoraggiamento Agricoltura Siciliana Palermo , 1 : 161 – 164 .
- James , TY , Moncalvo , JM , Li , S and Vilgalys , R. 2001 . Polymorphism at the ribosomal DNA spacers and its relation to breeding structure of the widespread mushroom Schizophyllum commune . Genetics , 157 ( 1 ) : 149 – 61 .
- Kawai , G , Babasaki , K and Neda , H. 2008 . Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu” it belongs to Pleurotus eryngii (DC.:Fr.) Quèl. and evolved independently in China . Mycoscience , 49 : 75 – 87 .
- Kristensen , R , Torp , M and Kosiak-Jensen , A. 2005 . Phylogeny and toxigenic potential is correlated with Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences . Mycol Res. , 109 ( 2 ) : 173 – 186 .
- Lanzi , M. 1894 . I funghi della provincia di Roma , Vol. IX , Roma : Memorie dell'accademia pontificia dei nuovi lincei . Pt. II
- Latouche , GN , Daniel , HM , Lee , OC , Mitchell , TG , Sorrel , TC and Meyer , W. 1997 . Comparison of use of phenotypic and genotypic characteristics for identification of species of the anamorph genus Candida and related teleomorph yeast species . J Clin Microbiol. , 35 ( 12 ) : 3171 – 3180 .
- Lewinsohn , D , Wasser , SP , Reshetnikov , SV , Hadar , Y and Nevo , E. 2002 . The Pleurotus eryngii species-complex in Israel: distribution and morphological description of a new taxon . Mycotaxon , 81 : 51 – 67 .
- Linnaeus , C. 1758 . Systema naturae per regna tria naturae: secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis(in Latin), Holmiae (Laurentii Salvii) , 10th Vol. 1-2 , 1384
- Mang , S. 2008 . Genome analysis of Pleurotus species-complex using gene sequences with taxonomic value [PhD dissertation] , 129 [Basilicata (IT)] : University of Basilicata . Available from: University of Basilicata Library
- Mang , S and Figliuolo , G. 2008 . “ Phylogenetic relationships among Pleurotus eryngii specimens from different eco-geographic sites of the Mediterranean Region ” . In Proceedings of the 6th International Conference on Mushroom Biology and Mushroom Products 29th September- 3rd October , Germany : Bonn .
- Mansfield , MA and Kuldau , GA. 2007 . Microbiological and molecular determination of mycobiota in fresh and ensiled maize silage . Mycologia , 99 ( 2 ) : 269 – 278 .
- Maphosa , L , Wingfield , BD , Coetzee , MPA , Mwenje , E and Wingfield , M. 2006 . Phylogenetic relationships among Armillaria species inferred from partial elongation factor 1-alpha DNA sequence data . Aust J Plant Pathol. , 35 : 513 – 520 .
- Marongiu , P , Maddau , L , Frisulo , S and Marras , F. 2005 . “ A multigene approach for the taxonomic determination of Pleurotus eryngii isolates ” . In Mushroom Biology and Mushroom Products , Edited by: Tan , Q , Zhang , J , Chen , M , Cao , H and Buswell , JA . 89 – 91 . Shanghai, , China : Shanghai Xinhua Printing Co .
- Maruyama , T , Kawahara , N , Yokoyama , K , Makino , Y , Fukiharu , T and Goda , Y. 2005 . Phylogenetic relationship of psychoactive fungi based on rRNA gene for a large subunit and their identification using the TaqMan assay (II) . Forens Sci Int. , 163 : 51 – 58 .
- Mayr , E. 1942 . Systematics and the Origin of the Species from the Viewpoint of a Zoologist , 334 New York : Columbia University Press .
- Matheny , PB , Wang , Z , Manfred , B , Curtis , JM , Lim , YW , Nilsson , RH , Hughes , KW , Hofstetter , V , Ammirati , JF , Schoch , CL , Langer , E , Langer , G , McLaughlin , DJ , Wilson , AW , Froslev , T , Ge , Z-W , Kerrigan , RW , Slot , JC , Yang , Z-L , Baroni , T , Fischer , M , Hosaka , K , Matsuura , K , Seidl , MT , Vauras , J and Hibbett , DS. 2007 . Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi) . Mol Phylogenet Evol. , 43 : 430 – 541 .
- Moncalvo , JM , Lutzoni , FM , Rehner , SA , Johnson , J and Vilgalys , R. 2000 . Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences . Syst Biol. , 49 : 278 – 305 .
- Mou , C , Cao , Y and Ma , J. 1987 . A new variety of Pleurotus eryngii and its cultural characters (in Chinese) Acta Mycol Sin , 6 : 153 – 156 .
- Nei , M and Li , WH. 1979 . Mathematical model for studying genetic variation in terms of restriction endonucleases . Proc Natl Acad Sci USA , 76 : 5269 – 5273 .
- Nei , M and Kumar , S. 2000 . Molecular Evolution and Phylogenetics , New York : Oxford University Press .
- O'Donnell , K. 1992 . Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambuccinum (Giberella palicuris) . Curr Genet. , 22 : 213 – 220 .
- Padamsee , M , Matheny , B , Dentinger , BTM and McLaughlin , DJ. 2008 . The mushroom family Psathyrellaceae: Evidence for large scale polyphyly of the genus Psathyrella . Mol Phylogenet Evol. , 46 : 415 – 429 .
- Park , HG , Ko , HG , Kim , SH and Park , WO. 2004 . Molecular identification of Asian isolates of medicinal mushroom Hericium erinaceum by phylogenetic analysis of nuclear ITS rDNA . J Microbiol Biotechnol. , 14 : 816 – 821 .
- Quélet , L. 1872 . Les Champignons du Jura e des Vosges Montbéliard (France): H . Barbier. , : 320
- Rehner S. 2001. Primers for Elongation Factor 1-α (EF1-α). Insect Biocontrol Laboratory: USDA, ARS, PSI. [email protected] (http://[email protected])
- Reverberi , M , Fanelli , C , Della Rosa , V , Marras , F , Maddau , L , Fabbri , AA and Urbanelli , S. 2000 . “ Molecular and physiological characterisation of the Pleurotus eryngii complex ” . In Proceedings of the 5th Congress of the European Foundation for Plant Pathology 37 – 40 .
- Ro , H-S , Kim , SS , Ryu , JS , Jeon , C-O , Lee , TS and Lee , H-S. 2007 . Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics . Mycol Res , 3 : 710 – 715 .
- Rodrigues-Estrada AE, 2008. Molecular phylogeny and increases of yield and the antioxidants selenium and ergothioneine in basidiomata of Pleurotus eryngii. [Ph.D. dissertation] .The Pennsylvania State University. http://etda.libraries.edu./theses/approved/WorldWideFiles/ETD-2822/RodriquezEstradaDissertation2008.pdf (http://etda.libraries.edu./theses/approved/WorldWideFiles/ETD-2822/RodriquezEstradaDissertation2008.pdf)
- Rodriguez Estrada , AE , Royse , DJ and Jimenez-Gasco , MM . 2008 . Nucleotide sequence polymorphisms of the partial β-tubulin gene in two varieties of Pleurotus eryngii . Mushroom Sci. , 17 : 83 – 96 .
- Rodriguez Estrada , AE , Jimenez Gasco , MM and Royse , DJ. 2010 . Pleurotus eryngii species complex: Sequence analysis and phylogeny based on partial EF1α and RPB2 genes . Fung Biol. , 114 : 421 – 428 .
- Saitou , N and Nei , M. 1987 . The neighbor-joining method: A new method for reconstructing phylogenetic trees . Mol Biol Evol. , 4 : 406 – 425 .
- Sambrook , J and Russell , DW. 2001 . Molecular Cloning: A Laboratory Manual , 3rd , Vol. 2 , Cold Spring Harbor, NY : Cold Spring Harbor Laboratory Press .
- Sreenivasaprasad , S , Brown , AE and Mills , PR. 1992 . DNA sequence variation and interrelationships among Colletotrichum isolates causing strawberry anthracnose . Physiol Mol Plant Pathol. , 41 : 265 – 281 .
- Steward , JE , Kim , MS , James , RL , Dumroese , RK and Klopfenstein , NB. 2006 . Molecular characterization of Fusarium oxysporum and Fusarium commune isolates from a conifer nursery . Phytopathology , 96 : 1124 – 1133 .
- Swann , EC and Taylor , JW. 1995 . Phylogenetic perspectives on basidiomycete systematics: evidence from the 18S rRNA gene . Can J Bot. , 73 : S862 – S868 .
- Tamura , K and Nei , M. 1983 . Estimation of the number of nucleotide substitutions in the control region of mithocondrial DNA in humans and chimpanzees . Mol Biol Evol. , 10 : 512 – 526 .
- Tamura , K , Nei , M and Kumar , S. 2004 . Prospects for inferring very large phylogenies by using the neighbor-joining method . Proc Natl Acad Sci USA , 101 : 11030 – 11035 .
- Tamura , K , Dudley , J , Nei , M and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Mol Biol Evol. , 24 : 1596 – 1599 .
- Taylor , JW , Jacobson , DJ , Kroken , S , Kasuga , T , Geiser , DM , Hibbett , DS and Fisher , MC. 2000 . Phylogenetic species recognition and species concepts in fungi . Fung Genet Biol. , 31 : 21 – 32 .
- Tuchwell , DS , Nicholson , MJ , McSweeney , CS , Theodorou , MK and Brookman , JL. 2005 . The rapid assignment of ruminal fungi to presumptive genera using ITS1 and ITS2 RNA secondary structures to produce group-specific fingerprints . Microbiology , 151 : 1557 – 1567 .
- Urbanelli , S , Fanelli , C , Fabbri , AA , Della Rosa , V , Maddau , L , Marras , F and Reverberi , M. 2002 . Molecular genetic analysis of two taxa of the Pleurotus eryngii complex: P. eryngii (DC. Fr.) Quèl. var. eryngii and P. eryngii (DC. Fr.) Quèl. var. ferulae . Biol J Linn Soc. , 75 : 125 – 136 .
- Urbanelli , S , Della Rosa , V , Fanelli , C , Fabbri , AA and Reverberi , M. 2003 . Genetic diversity and population structure of the Italian fungi belonging to the taxa Pleurotus eryngii (DC.:Fr.) Quèl. and P. ferulae (DC.: Fr.) Quèl . Heredity , 90 : 253 – 259 .
- Urbanelli , S , Della Rosa , V , Punelli , F , Porretta , D , Reverberi , M , Fabbri , AA and Fanelli , C. 2007 . DNA-fingerprinting (AFLP and RFLP) for genotypic identification in species of the Pleurotus eryngii complex . Appl Microbiol Biotechnol. , 74 : 592 – 600 .
- Van Valen , L. 1976 . Ecological species, multispecies, and oaks . Taxon , 25 : 233 – 239 .
- van der Merwe , M , Ericson , L , Walker , J , Thrall , PH and Burdon , JJ. 2007 . Evolutionary relationships among species of Puccinia and Uromyces (Pucciniaceae, Uredinales) inferred from partial protein coding gene phylogenies . Mycol Res. , 111 : 163 – 175 .
- Venturella , G. 2000 . Typication of Plerurotus nebrodensis . Mycotaxon , 75 : 229 – 231 .
- Venturella , G , Zervakis , G and La Rocca , S . 2000 . Pleurotus eryngii var. elaeoselini var. nov. from Sicily . Mycotaxon , 76 : 419 – 427 .
- Venturella , G , Zervakis , G and Saitta , A. 2002 . Pleurotus eryngii var. thapsiae var. nov. from Sicily . Mycotaxon , 81 : 69 – 74 .
- Vilgalys , R and Sun , BL. 1994 . Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences . Proc Natl Acad Sci USA , 91 : 4599 – 4603 .
- Vilgalys , R , Smith , A , Sun , B and Miller , OK. 1993 . Intersterility groups in the Pleurotus ostreatus complex from the continental United States adjacent to Canada . Can J Bot. , 71 : 113 – 128 .
- Vinnere O. 2004. Approaches to Species Delineation in Anamorphic (mitosporic) Fungi: A Study on Two Extreme Cases. [PhD dissertation]. Acta Universitatis Upsaliensis, [Uppsala University]. 72 p. ISSN 1104-232X ISBN 91-554-5832-7 http://www.uu.se/, [email protected] (http://www.uu.se/, [email protected])
- White , TJ , Bruns , T , Lee , S and Taylor , J. 1990 . “ Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics ” . In PCR protocols. A Guide to Methods and Applications , Edited by: Innis , MA , Gelfand , DH , Sninsky , JJ and White , TJ . 315 – 322 . New York : Academic Press .
- Williams , JGK , Kubelik , AR , Livak , KJ , Rafalski , JA and Tingey , SV. 1990 . DNA polymorphisms amplified by arbitrary primers are useful as genetic markers . Nucl Acid Res , 18 : 6531 – 6535 .
- Zervakis , G and Balis , C. 1996 . A pluralistic approach in the study of Pleurotus species with emphasis on compatibility and physiology of the European morphotaxa . Mycol. Res. , 100 ( 6 ) : 717 – 731 .
- Zervakis , G , Sourdis , J and Balis , C. 1994 . Genetic variability and systematics of eleven Pleurotus species based on isozyme analysis . Mycol Res. , 98 ( 3 ) : 329 – 341 .
- Zervakis , G , Katsaris , P , Ioannidou , E , Lahouvaris , E and Philippoussis . 2001a . Facultative biotrophic basidiomycetes produce high quality edible mushrooms . Phytopathologia Mediterranea , 40 : 190
- Zervakis , G , Venturella , G and Papadopoulou , K. 2001b . Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species-complex as determined by RAPD analysis, isozyme profiles and ecomorphological characters . Microbiology , 147 : 3183 – 3194 .
- Zervakis , G , Moncalvo , JM and Vilgalys , R. 2004 . Molecular phylogeny, biogeography and speciation of the mushroom species Pleurotus cystidiosus and allied taxa . Microbiology , 150 : 715 – 726 .
- Zhang , J , Huang , C and Li , C. 2000 . The cultivation of Pleurotus nebrodensis in China . Proceedings of the 5th International conference on Mushroom Biology and Mushroom Products , 12 ( Suppl. ) : 350 – 353 .
- Zhang , J , Huang , C , Ng , TB and Wang , HX. 2006 . Genetic polymorphism of ferula mushroom growing on Ferula sinkiangensis . Appl Microbiol Biotechnol , 71 : 304 – 309 .