Abstract
Fungal genome annotation is the starting point for analysis of genome content. This generally involves the application of diverse methods to identify features on a genome assembly such as protein-coding and non-coding genes, repeats and transposable elements, and pseudogenes. Here, we describe tools and methods leveraged for eukaryotic genome annotation with a focus on the annotation of fungal nuclear and mitochondrial genomes. We highlight the application of the latest technologies and tools to improve the quality of predicted gene sets. The Broad Institute eukaryotic genome annotation pipeline is described as one example of how such methods and tools are integrated into a sequencing center's production genome annotation environment.
1. Introduction
Fifteen years ago heralded the first genome sequence of a free-living eukaryote, that of the fungal species Schizosaccharomyces cerevisiae (Goffeau et al. Citation1996). Since that time, 315 eukaryotic genomes have been sequenced and assembled, 108 of which are fungal (http://www.ncbi.nlm.nih.gov/genomes/static/gpstat.html). Much has been learned as a result of genome sequencing, especially in the area of fungal genomics, from mechanisms of fungal genome evolution, fungi-specific gene family innovations, and the genomic potential for sexual cycles (reviewed in Cuomo and Birren Citation2010). Another 47 fungal genome sequences are currently in progress and, with the recent revolution in genome sequencing technologies and marked decreases in sequencing costs, hundreds to thousands of fungal genome projects are currently in the planning stages.
Genome sequencing and assembly yields an enormous string of characters using only a four-letter DNA alphabet (G, A, T, C), which, by itself, is of very limited utility. These sequences may differ substantially, due to inherent properties such as GC content, repeat content and, if diploid, the rate of polymorphism. The goal of genome annotation is to decipher the four-letter code to identify the features of greatest biological importance, most notably the genes. Although the genome sequence substrates for annotation may vary, the overall strategies taken to decipher each code are quite similar. This review describes many of the bioinformatics methods and tools that can model fungal (and more generally, eukaryotic) genes and predict their functions. We end the review with a summary of how these methods are glued together into a more comprehensive annotation pipeline, as currently deployed at the Broad Institute.
2. Discovery of protein-coding genes
In addition to the advances in sequencing technology, gene-finding efforts have made significant leaps in accuracy and general utility. The importance of gene finding is reflected by the continual innovation of bioinformatics methods. Gene-finding software tools generally fall into one of two categories: sequence homology detection or ab initio gene prediction. Each strategy has advantages and disadvantages, as elaborated below, but when used together, they provide a powerful, robust method to identify the components of genes.
2.1. Sequence homology-based gene structure annotation methods
Homology-based gene finding methods are considered strong evidence to precisely localize and model gene structures where experimentally verified data is available or when conservation patterns can be inferred from alignments of genomes from related species. Specifics vary, but most tools align sequences from databases of protein or transcripts to the genome so that gaps are allowed at introns, and the consensus dinculeotide GT (or, to a lesser extent, GC) donor and AG acceptor splice sites of introns are preferred at internal alignment segment boundaries. These spliced alignments provide strong evidence for components of gene structures, in many cases fully resolving complete exons and introns, and at the very least, highlighting a candidate gene location.
2.1.1. Gene structure annotation using transcriptome sequences
Transcript sequences, when derived from the same organism whose genome is being sequenced, provide the most accurate form of evidence for resolving gene structures, as they are highly identical to the genome and precisely delineate intron–exon boundaries. These transcript sequences can include (1) expressed sequence tags (ESTs), sequences derived from single- pass Sanger sequencing reads of the 5′ or 3′ termini of cDNA clones, (2) full-length cDNAs (FL-cDNAs), and most recently, (3) cDNA sequences derived from next-generation short read transcriptome sequencing (termed RNA-Seq). FL-cDNA sequences provide the most useful substrate for gene structure annotation, as they ideally encode the transcriptional start site, all exons of a fully spliced transcript, and specify the polyadenylation cleavage site at the 3′ end. With accurate spliced alignments of a FL-cDNAs to the genome, all components of the gene structure are often revealed, including the open reading frame (ORF) and terminal untranslated regions (UTRs) of exons (Haas et al. Citation2002). Gene structures supported by FL-cDNAs are generally accepted as the gold standard for gene structure annotation; although there are some exceptions, this general rule holds true for the vast majority of cases. However, FL-cDNAs as currently generated by paired-end Sanger sequencing are too expensive to routinely obtain as a resource for genome annotation. Historically, EST sequences have served as a more cost-effective and, therefore, more routinely generated resource to support genome annotation. As with FL-cDNAs, alignments of ESTs to genome sequences can identify expressed genes and components of gene structures, such as intron/exon boundaries, but, due to the limited length of a single Sanger sequencing read, ESTs rarely resolve a gene structure in its entirety. Further improvements to the scale and cost of next generation sequencing methods may produce longer reads at reasonable cost in the near future. To take advantage of conventional technologies, several approaches have been developed, including PASA (Haas et al. Citation2003), ESTGenes (Eyras et al. Citation2004) and CallReferenceGenes (McGuire et al. Citation2008), to assemble multiple overlapping cDNA alignments into more full-length gene structures. These tools are able to generate multiple transcript models per gene when there differences in overlapping alignments that result from alternative splicing.
For cDNA alignments to be useful for direct gene structure annotation, they must be nearly identical in sequence to the target genome, which generally requires that the cDNAs derive from the same species. We have found cases where alignments with as little as 70–80% nucleotide identity have proved useful in cross-species alignments, but to a much lesser extent than less-divergent sequences. The Analysis and Annotation Tool (AAT) (Huang et al. Citation1997) is particularly adept at cross-species spliced transcript alignments, especially with highly divergent species. Many more software tools exist that are more specialized towards generating spliced alignments of transcript sequences, including but not limited to EST_GENOME (Mott Citation1997), sim4 (Florea et al. Citation1998), Spidey (Wheelan et al. Citation2001), BLAT (Kent Citation2002) and GMAP (Wu and Watanabe Citation2005). Due to the algorithmic differences underlying these tools, different spliced aligners generate slightly different but complementary results. Therefore, it is sometimes beneficial to apply more than one alignment tool (e.g. BLAT and GMAP) to the same data set and use metrics, such as percent identity and length of the transcript aligned coupled with splice dinucleotide consensus agreement, to choose the best quality alignment (Haas 2003–2007).
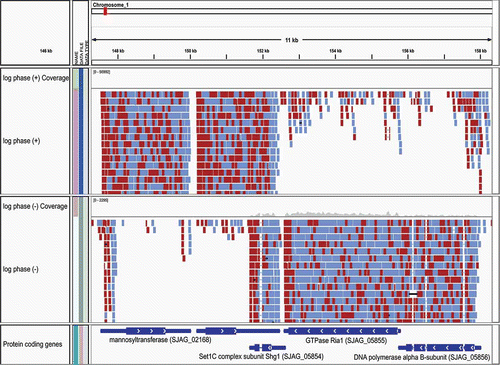
Next-generation sequencing of transcriptomes, termed RNA-Seq, has recently become a powerful tool for studying gene expression and annotating gene structure. This technology has rapidly advanced, such that strand-specific sequencing of tens to hundreds of millions of paired >100 base sequencing reads is now a routine and cost-effective operation. To be best leveraged for genome annotation, these short RNA-Seq reads (which are even shorter than ESTs) must first be assembled into more complete transcript structures. Two general approaches have been pursued for reconstructing transcripts from RNA-Seq data: (1) a ‘mapping first’ strategy in which the short reads are first aligned to the genome followed by local assembly of the alignments into more complete transcript structures; or (2) ‘assembly first’ de novo assembly of short reads to reconstruct transcript sequences, which are then aligned to the genome to resolve gene structures (reviewed in Haas and Zody (Citation2010)). The challenge of mapping millions of short RNA-Seq reads to a genome while accounting for reads that cross intron boundaries has led to the development of new specialized spliced alignment tools, including TopHat (Trapnell et al. Citation2009), GSNAP (Wu and Nacu Citation2010) and MapSplice (Wang et al. Citation2010), among others. With longer RNA-Seq reads (≥70 base Illumina reads), we find that the earlier-mentioned BLAT software also performs well for generating spliced alignments of RNA-Seq reads to fungal genomes. An example showing strand-specific Illumina RNA-Seq reads aligned to the Schizosaccharoymyces japonicus genome is shown in .
Figure 1. Strand-specific RNA-Seq reads aligned to the Schizosaccharomyces japonicus genome as viewed in the Broad's Integrative Genomics Viewer. Strand-specific RNA-Seq reads are shown aligning to the top strand (top) and bottom strand (center) separately. The left and right RNA-Seq paired fragment reads are colored red and light blue, respectively. The reference gene structure annotations for this 11-kb region of the genome is shown at bottom colored dark blue.
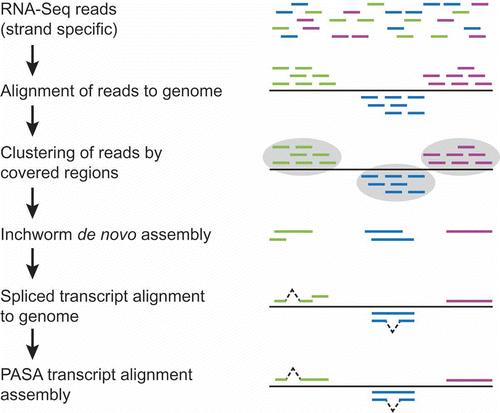
At the Broad Institute, we leverage a hybrid strategy that involves first mapping reads to the genome, followed by partitioning the reads and genomic regions into disjoint sets of sequence coverage, preferably in a strand-specific manner (complete pipeline illustrated as ). The reads within each coverage group are next de novo assembled into more complete transcript sequences (often full-length). These reconstructed transcripts, along with expression values inferred from the reads incorporated into each transcript, are then input into a PASA pipeline using its RNA-Seq mode (as more fully described in Rhind et al. (Citation2011)). PASA aligns these newly assembled transcripts to the genome using GMAP, filters invalid alignments and those transcripts more likely resulting as artifacts of the RNA-Seq assembly process, and reconstructs more complete transcripts using its alignment assembly algorithm. These reconstructed transcripts derived from the hybrid de novo and alignment assembly method provide a substrate for genome annotation that rivals the utility of full-length cDNAs but at much lower cost and with minimal sequencing effort required.
Figure 2. Hybrid approach to RNA-Seq-based transcript reconstruction leveraging genome alignment and de novo assembly. RNA-Seq reads are first aligned to the genome, then partitioned into disjoint regions of alignment coverage. Inchworm is leveraged to de novo assemble the read sequences into transcripts. The resulting transcripts are aligned to the genome using a conventional cDNA alignment tool, and PASA is leveraged to further assemble overlapping alignments and extract gene structure annotations.
2.1.2. Annotation of alternatively spliced transcripts
Alternative splicing, which allows multiple proteins to be expressed from a single gene, plays a pivotal role in numerous biological processes (see reviews in Keren et al. (Citation2010) and Stamm et al. (Citation2005)). One of the most remarkable cases of alternative splicing is found in the Dscam gene of Drosophila. This gene encodes several exons for which there are several mutually exclusive choices, yielding combinatorial complexity with the capacity to yield over 38,000 alternatively spliced transcripts and a corresponding variety of protein products (Schmucker et al. Citation2000). In addition to generating protein products with altered enzymatic functions, stability, or subcellular localizations, alternative splicing can post-transcriptionally regulate gene expression, targeting unproductively spliced transcripts to the nonsense-medicated decay (NMD) pathway (reviewed in Nicholson and Muhlemann (Citation2010) and Stalder and Muhlemann (Citation2008)).
Alternative splicing is found among all eukaryotic genomes where introns are prevalent; as more transcripts are sequenced, especially with the large amount of RNA-Seq data generated with the next generation of sequencing technologies, more evidence becomes available extending the repertoire of genes known to be alternatively spliced. The importance of alternative splicing is underscored by the finding that over 90% of human genes exhibit evidence of transcript diversity (Wang et al. Citation2008). Studies in plants indicate that around 20% of the expressed genes are alternatively spliced (Wang and Brendel Citation2006; Campbell et al. Citation2006). Fungi exhibit some alternatively spliced transcripts and, similarly to plants, retained introns dominate over other types of alternative splicing events such as cassette exons (McGuire et al., Citation2008). Since genome annotation provides the first insights into each organism's collection of genes, properly annotating these transcript variants and their encoded proteins is essential to producing a complete catalog of predicted transcripts.
Transcript isoforms derived from alternative splicing can be effectively modeled by several automated annotation systems (Haas et al. Citation2003; Eyras et al. Citation2004; Florea et al. Citation2005). PASA (Haas et al. Citation2003), in particular, was originally designed to automate the incorporation of expressed transcript alignments into existing eukaryotic gene structure annotations. In addition to adding UTR annotations to existing gene structure predictions that are otherwise consistent with the spliced alignments, PASA updates internally inconsistent regions of gene structures, and adds gene models for alternatively spliced genes as supported by the transcript data. PASA's underlying algorithm for assembling spliced transcript alignments is particularly well suited to the problem of alternative splicing (Campbell et al. Citation2006). Transcript alignments that overlap and have inconsistent intron positions are assembled into separate maximal alignment assemblies, and each is used independently to automatically model a distinct isoform for the corresponding gene.
2.1.3. Gene structure annotation based on protein sequence homology
Leveraging evidence of homology to sequences derived from divergent species is best achieved using protein conservation. Non-redundant comprehensive protein databases, provided by GenBank (Benson et al. Citation2005) or UniProt (Wu et al. Citation2006), yield some of the best annotation resources, readily applicable to any previously uncharacterized genome. Protein sequence alignments to genomes, when found above the ‘twilight zone’ of percent identity (30%), can yield convincing support for partial gene structures. Often, these alignments will not extend to start or stop codons, and so the evidence for gene structures is more centrally located.
A widely used protein homology-based gene-modeling tool is GeneWise (Birney et al. Citation2004a), which combines protein alignment and gene prediction into a single statistical model via a paired hidden Markov model (HMM). A typical spliced protein alignment program will report only an alignment, without regard for an intact open reading frame. GeneWise provides a gene prediction based on protein homology, which can, in some cases, serve as a standalone structural gene annotation. Because, as stated earlier, protein alignments do not often model the termini of coding regions, GeneWise predictions often lack start or stop codons, instead providing partial gene structures that correspond to the internal regions of the protein sequence. Because GeneWise requires known protein sequences as input, its utility is restricted to finding genes with previously described homologs; it is unable to predict novel genes. GeneWise plays an essential role in the protein-centric automated gene annotations provided by Ensembl (Birney et al. Citation2004b). The Broad automated gene annotation pipeline uses TBLASTN to find top protein hits first, then uses GeneWise to generate spliced gene models from these hits. Instead of using the GeneWise predictions as final genome annotations, at the Broad Institute, GeneWise results are used as evidence to be combined with other evidence sources to generate consensus gene predictions, as described further below.
In addition to GeneWise, the annotation pipeline used at the Broad Institute also incorporates the AAT package (Huang et al. Citation1997) to generate sensitive spliced protein alignments to eukaryotic genomes. Although its rigorous dynamic programming alignment algorithm is relatively slow in comparison to GeneWise, we often find results to be especially useful where sequence similarity is low. The multiple alignment utility included in AAT shows all protein and transcript spliced genome alignments together, highlighting valuable evidence for exon boundaries and chosen splice sites (). Other tools that generate spliced alignments of both proteins and transcripts to the genome are exonerate (Slater and Birney Citation2005) and GeneSeqer (Usuka et al. Citation2000).
Figure 3. Spliced nucleotide and protein alignments infer intron structures. A section of AAT Alignments of homologous protein and EST sequences to the Neurospora crassa gene (shown as query) for alkaline phosphatase (NCU01376). This region of the alignment unambiguously identifies an intron within the gene structure; consensus splice sites are shown in bold.

Short peptides identified by mass spectroscopy are an additional valuable source of evidence that enable and augment gene finding efforts, e.g. Schizosaccharomyces pombe (Bitton et al. Citation2011). While these peptide data provide strong support for the presence of a gene and the reading frame of the gene at that location, the short peptides derived from mass spectroscopy experiments generally do not provide sufficient data to resolve complete gene structures. However, confirmation that specific transcripts are translated, particularly very small transcripts encoding short ORFs, is invaluable. The use of proteomics data for genome annotation is reviewed in (Ansong et al. Citation2008).
2.2. Ab intio gene prediction
Ab initio gene prediction programs, which solely rely on the genome sequence under study, are an essential part of the genome annotation process (reviewed in Brent and Guigo (Citation2004), Do and Choi (Citation2006) and Zhang (Citation2002)). At the heart of ab initio gene predictors are statistical models, often hidden Markov models (HMM), which are trained to find features of genes, such as exons, splice sites, start and stop codons, introns, and the noncoding DNA found between genes. A generalized hidden Markov model (GHMM) is a more complex type of HMM that can model gene structures with intron and exon lengths tuned to known feature length distributions. The input to such software is simply the string of letters that defines a genome sequence, and the output is the coordinates of gene structures predicted for that sequence. There is a wide variety of these programs available today, including GENSCAN (Burge and Karlin Citation1997), Genemark.hmm (Lukashin and Borodovsky Citation1998), GlimmerHMM (Majoros et al. Citation2004), FgeneSH (Salamov and Solovyev Citation2000), Augustus (Stanke and Waack Citation2003), geneid (Guigo Citation1998; Parra et al. Citation2000), SNAP (Korf Citation2004) and GeneMark.hmm-ES (Ter-Hovhannisyan et al. Citation2008). The caveat to using these tools is that most require training to find genes within a specific genome, and benefit from validation based on comparison to a reference (truth) gene set. Many of the gene prediction programs, including Augustus, GlimmerHMM, SNAP, and geneid, allow end users to both train and run the program. In these cases, a critical step is the construction of the training set composed of known gene structures within the corresponding genome, which is used to estimate the parameters for the splice junction signals, as well as length distribution and nucleotide composition of the exons, introns and intergenic regions.
A high-quality training set can be predicted for a new genome based on sequence homology. Given transcriptome sequences such those as derived from RNA-Seq, the PASA software can extract high-confidence gene models from full-length or near full-length reconstructed transcripts; these can serve as an excellent starting point for constructing a training set for ab initio gene predictors. In some cases, gene sequences from a closely related species can also be used to estimate the parameters for a gene predictor. One way to operate is to use an iterative approach (Brejova et al. Citation2009), whereby the initial gene sets are predicted with parameters estimated using a well-curated annotation from a distantly related species. Next, a subset of this initial prediction set is selected based on support from EST data and protein alignments and the parameters are then retrained. Yet another method for deriving a training set is CEGMA (Parra et al. Citation2007), which uses a set of 458 highly conserved eukaryotic proteins to search for orthlogous genes in the new genome, then uses these orthlogs to estimate the parameters of ab initio gene predictors.
Among the currently available ab initio gene prediction programs, GeneMark.hmm-ES is the only ab initio gene predictor we are aware of that does not require a user-generated and user-provided training set (Ter-Hovhannisyan et al. Citation2008). GeneMark.hmm-ES is the self-training version of the eukaryotic GeneMark.hmm software, which uses the genome sequence itself as input for an iterative process involving gene prediction and self-training.
2.3. Comparative gene prediction
It is clear that ab intio gene finding programs and sequence alignment utilities are both useful for finding genes. Transcript or protein alignments, or conserved regions found by related genome sequence comparisons, can be used to inform a hybrid breed of gene prediction tools that consider the intrinsic information corresponding to the DNA sequence composition together with extrinsic information derived from sequence homologies. In most cases, these programs are modified versions of the existing ab intio gene prediction programs (described above) that are enhanced to allow sequence homology information to augment the scores of supported gene structures. For example, GenomeScan (Yeh et al. Citation2001) is a version of GENSCAN that takes into account BLASTX matches of protein sequences to the genome and reports the most probable set of gene structures conditioned on the regions of sequence homology. SGP2 (Parra et al. Citation2003) uses TBLASTX alignments between genomes to augment scores of GENEID (Guigo Citation1998) predictions. TWINSCAN (Korf et al. Citation2001) couples a probabilistic model of sequence conservation, based on BLASTN matches between the informant and target genome, to the GHMM used by a reimplemented version of GENSCAN. A later version of the ab intio prediction program AUGUSTUS, called AUGUSTUS+ (Stanke et al. Citation2006), has the capacity to use externally supplied sequence homology data as a set of hints to guide improved gene finding accuracy. AUGUSTUS+ also accepts manually defined hints to force certain known parts of gene structures to be outputted when possible, which is particularly useful in cases where sources of evidence are conflicting and the user has advanced knowledge about a subset of genes or structural components, such as those supported by high quality transcript alignments.
ExonHunter (Brejova et al. Citation2005) employs a GHMM similar to that of GENSCAN and AUGUSTUS but has a mechanism to incorporate numerous sources of evidence including protein and EST matches, homologies resulting from pairwise genome sequence comparisons, and repeats, into gene predictions by using what are termed advisors specific to each evidence type. Each advisor yields a partial probabilistic statement that is summed into a single superadvisor probability using quadratic programming, and then integrated with the AUGUSTUS-like GHMM. The TWINSCAN algorithm can be implemented as a special case of ExonHunter where only a single evidence type is used as an informant of genome homology and a single corresponding advisor is employed (Brejova et al. Citation2005). Another more recent development in gene finding is to consider multiple genome homologies in a phylogenetic framework. A newer and improved version of TWINSCAN (N-SCAN a.k.a. TWINSCAN 3.0) considers homologies to a target genome from several other different related genomes and also their known phylogeny to accurately predict gene structures in the target genome (Gross and Brent Citation2006).
Another class of gene prediction tools compares two genome sequences to predict gene structures in both genomes by exploiting regions of conservation. The first attempt to utilize cross-species alignments to predict genes was performed by ROSETTA as applied to globally aligned pairs of orthologous mouse and human genes (Batzoglou et al. Citation2000). A similar approach was applied to re-annotate the well studied genome of Saccahromyces cerevisiae by comparison to three related sensu stricto species: Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces bayanus (Kellis et al. Citation2003). These four genomes were first aligned, and then the syntenic regions identified; reading frame conservation (RFC) was used to evaluate whether ORF predictions were biologically preserved or spurious. This analysis led to the revision of a large number of genes; conserved regions were further mined to identify known and novel regulatory motifs. This method was updated for the annotation of the Drosophila (Lin et al. Citation2007) and Candida (Butler et al. Citation2009) genomes, to include both the RFC test and a metric for conservative codon substitutions (CSF, for codon substitution frequencies).
Genome-wide conservation is also utilized by another program, SGP-1 (Wiehe et al. Citation2001), which uses nucleotide (BLASTN) or translated nucleotide (TBLASTX) alignments between two genomes to identify likely conserved exons. These candidate exons are then assembled into larger gene structures independently for both genomes. Soon thereafter, a theoretical framework was described for combining genome alignments and paired gene predictions in a single probabilistic model called a generalized pair HMM (GPHMM) (Pachter et al. Citation2002). The GPHMM combines the paired HMM that describes sequence alignment with the more traditional HMM that describes gene structures.
Gene prediction programs implementing the GPHMM include both SLAM (Alexandersson et al. Citation2003) and TWAIN (Majoros et al. Citation2005), the latter applied to the fungal genomes Aspergillus nidulans and Aspergillus fumigatus. One caveat in using these GPHMM-based software tools is that they emit gene structures that require identical numbers of introns and exons for the homologous gene pairs in the corresponding pair of genomes. This is a reasonable approximation for many closely related genomes such as between mouse and human. In theory, the GPHMM model could be extended to allow for different numbers of introns and exons, but this has practical ramifications in terms of increased memory usage and computational complexity (Majoros et al. Citation2005).
The most recent developments in computational gene prediction have leveraged the framework of conditional random fields (CRF) in place of the more traditional GHMMs, often leveraging cross-species genome alignments to inform gene predictions. Examples include Conrad (DeCaprio et al. Citation2007), demonstrating success in predicting genes in the fungal genomes of Crytpococcus neoformans and Aspergillus nidulans, outperforming competing ab initio or comparative approaches. Another comparative CRF-based gene predictor is CONTRAST (Gross et al. Citation2007), which demonstrated great success in predicting genes in the human genome by leveraging mouse alignments exclusively or in combination with additional vertebrate genome alignments.
Advances in the science of ab initio and homology-informed gene prediction over the last decade are apparent (Brent Citation2008). However, not all such advanced software tools are immediately at one's disposal. Although several advanced tools employing GPHMMs or CRFs have been published with demonstrated success in areas including fungal genomics and, although software and source code is often made publicly available by the authors, our personal experience is that achieving respectable performance when applying these tools to newly targeted genomes can be an enormous challenge. The strategy for best results in using such tools is to collaborate with the corresponding authors whenever possible. In contrast, we have achieved great success leveraging the more traditional GHMM-based gene finders as applied to diverse new fungal genomes, especially in the context of the flexible evidence combining strategies described below.
2.4. Automated gene modeling using evidence combiners
As a composite of gene prediction programs often produces the best gene set, additional algorithms are required to choose the best gene structure for a given locus. Such evidence-combining methods vary in complexity, from a simple majority-voting scheme to more complex stochastic methods as demonstrated by the linear and statistical Combiner software tools (Allen et al. Citation2004), respectively. Combiner was succeeded by JIGSAW (Allen and Salzberg Citation2005), software that combines diverse sources of evidence into gene structures using a GHMM-like algorithm. JIGSAW uses decision trees to weight the contribution each evidence type makes toward each possible label for each base (coding region, intron, start/stop codon, splice site, etc.) then selects the labeling with the highest overall probability. Another tool, GAZE (Howe et al. Citation2002), provides a general framework to assemble an optimal set of gene structures given a user-supplied feature set, scoring scheme, and model for how to build a gene structure.
The EVidenceModeler (EVM) (Haas et al. Citation2008) software generates a set of weighted consensus gene structures from ab initio gene predictions and protein and transcript alignments. EVM provides a flexible and intuitive framework for combining diverse evidence types into a single automated gene structure annotation system. Inputs to EVM include the genome sequence, sets of gene predictions produced by different gene-calling programs, protein and transcript sequence spliced alignments, and a list of numerical weight values to be applied to each type of evidence.
Maker (Cantarel et al. Citation2008) is another “combiner” annotation package. Maker combines ab initio gene predictions (SNAP, Augustus, FgeneSH and GeneMark.hmm), EST alignments (via Exonerate and Blastn), protein alignments (exonerate and BLASTX) and repeats from RepeatMasker and Maker's own internal RepeatRunner, synthesizes the input data into gene annotations, and tracks the evidence used in the process of gene model selection.
Evidence-combining methods to automate gene prediction have been shown to excel in comparison to both the ab initio and dual-genome gene finders (Guigo et al. Citation2006), (Haas et al. Citation2008). The ultimate goal is to reach a level of accuracy that meets or exceeds that of the human annotator, so that quality genome annotation can be generated at a rate that can keep pace with DNA sequencing. A flexible evidence combiner that is easily tuned to a wide array of evidence sources is an essential component of eukaryotic annotation efforts.
2.5. Manually modeling genes using a genome annotation editor
Manual evaluation of the data underlying a gene call is important in cases where data conflict or are otherwise sparse in support, and can be used to fix the more obvious errors. Genome annotation projects typically generate data such as cDNA sequences to assist in the gene finding effort, which almost always utilizes several different ab initio gene prediction programs, along with protein and transcript alignments. After an automatic gene set is built, the homologous protein and transcript alignments can help manual annotators to identify the regions where components of genes were predicted incorrectly, such as where the true gene structures were incorrectly split or merged as predicted, or in the simpler cases, identifying missed exons, wrong start or stop codons, or incorrect splice sites.
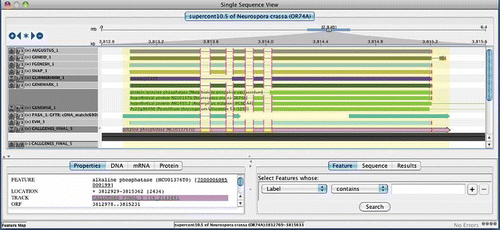
Although many genome browsers exist, such as the UCSC genome browser or Gbrowse, most focus on data viewing capabilities while comparatively few provide functionality that allows one to edit annotations. Genome browsers that also act as annotation editors include ARGO (Engels Citation2003–2011), GenomeView (Abeel Citation2006–2011), Apollo (Lewis et al. Citation2002) and Artemis (Rutherford et al. Citation2000). shows an example view provided by ARGO, which illustrates the many sources of evidence that can be used by an annotator in the process of manual gene structure curation. The editor allows the user to model new genes, delete unsupported genes, and modify intron and exon boundaries as needed. Such manual editing is hugely time-consuming and also imperfect, but still considered the most trusted mechanism to annotate a genome to the highest quality. Owing to the need for annotations of the highest quality, a method for manual inspection and approval has been applied to all fungal genomes previously sequenced and annotated at the Broad Institute. Similar approaches have been applied to other genomes considered to be of the greatest importance to biological and medical science, including human and other vertebrates (Ashurst et al. Citation2005), Arabidopsis (Haas et al. Citation2005; Wortman et al. Citation2003), C. elegans (Schwarz et al. Citation2006), and Drosophila (Misra et al. Citation2002).
Figure 4. ARGO genome annotation editor display. Shown is the evidence for the gene structure annotation of Neurospora crassa alkaline phosphatase (NCU01376) in the ARGO genome annotation editor. Evidence consists of, from top to bottom, Augustus, geneid, FgeneSH, SNAP, GLIMMERHMM, and GENEMARK.hmm ab initio predictions, followed by GENEWISE predictions based on top matching homologous proteins, PASA assemblies of EST alignments (ESTs not shown), EVidenceModeler consensus prediction, and the final annotated gene model for this locus. The intron boundaries that agree with the annotated gene model are highlighted as pink vertical bars. Positions of tart and stop codons are shown as green and red vertical bars, respectively. The ab initio predictors AUGUSTUS, FgeneSH, SNAP, and GENEMARK.hmm all perfectly agree on the structure of this gene, whereas GeneId and GLIMMERHMM propose different structures. The PASA assemblies of high quality EST alignments provide evidence for UTR annotations at both gene termini, extending upstream and downstream of the start and stop codons of the annotated gene model (pink model highlighted at bottom).
2.6. Comparative genomics as an annotation refinement tool
Subsequent to the initial gene structure annotations to a series of related complete genomes, more detailed comparisons between orthologous genes can yield insights that can significantly improve upon the quality of annotation after refinement, as was shown in the annotation evaluation and refinement of three Saccharomyces cerevisiae (Kellis et al. Citation2003) and Candida albicans genomes (Butler et al. Citation2009).
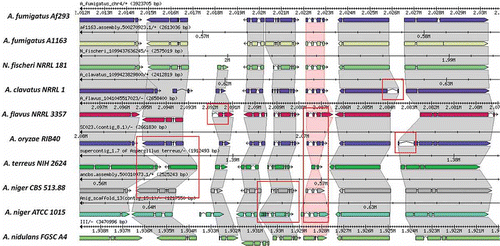
One strategy for examining orthologs as a focal point towards improved gene structure curations is exemplified by the Sybil web-based software (http://sybil.sf.net) for comparative genomics. Sybil's interface illustrates computed ortholog clusters in their genomic context; for more closely related genomes, orthologs are often found within large regions of synteny. Statistical summaries highlight ortholog clusters that are missing members, or with lower-than-expected alignment coverage or similarity, suggesting potential annotation inaccuracies that may need to be addressed. For example, closely related orthologs that vary substantially in their exon numbers, protein lengths, or intron lengths may indicate inaccurate, inconsistent gene predictions. An example Sybil comparative view of gene structure annotations across a syntenic region of Aspergillus species is provided in . Split or merged gene predictions are easily discerned in the display. ‘Missing’ genes manifest within syntenic regions as gaping holes arranged against orphaned (ortholog-lacking) genes. Upon more thorough inspection of the data in a genome annotation editor (as described above), the splits, merges, and recovery of such missed genes can be achieved.
Figure 5. The Sybil Comparative Genomics Interface. A short region of synteny among orthologous genes of Aspergillus and related genomes is shown within the Sybil interface. Similarities and differences among the annotated gene structures become readily apparent, and many differences are found to represent artifacts rather than true evolutionary differences among related genes. Examples of the most striking discrepancies among annotated gene structures, involving different numbers of exons, or intron and exon lengths are highlighted by red rectangles.
2.7. Annotation of fungal genomes with few spliced genes
Fungal genomes vary in the proportion of spliced genes, ranging from 0.7 to 97% (Ivashchenko et al. Citation2009). For example, only about 5% of genes are spliced in S. cerevisiae and the Candida species (Rossignol et al. Citation2008; Mitrovich et al. Citation2007), and <1% for E. cuniculi and other Microsporidia species (Katinka et al. Citation2001). For genomes with few spliced genes, the gene structures are more similar to those of the prokaryotes, except for the comparatively rare spliced genes. In these cases, we may leverage the prokaryotic ab initio gene predictors that target single-exon genes including Prodigal (Hyatt et al. Citation2010), GeneMark, and Glimmer. The remaining intron-containing genes can be identified by spliced protein and transcript alignments; some of these have a small initial exon that can be difficult to accurately predict. In such cases, transcript evidence is invaluable, and where high-quality transcript alignment evidence supports the existence of introns, PASA is often able to automatically incorporate the intron into the existing Prodigal or other single-exon prediction. Using this approach, manual refinement can be targeted to the more complex gene structure modifications required.
2.8. Annotation of non-coding RNA genes
Some RNA molecules do not encode proteins, but instead serve as the final functional products with enzymatic and/or structural roles in essential and ancient biomolecular processes (reviewed in Eddy (Citation2001) and Szymanski et al. (Citation2003)). Major classes of non-coding RNA (ncRNA) genes are involved in several essential biomolecular or biochemical processes including transcription, post-transcriptonal mRNA processing, and translation. Well known examples of ncRNAs include ribosomal RNAs (rRNA) involved as structural and functional components of ribosomes, transfer RNAs (tRNA) used to decode the mRNA to form proteins, the small nuclear RNAs (snRNA) involved in pre-mRNA splicing of introns, and the small nucleolar RNAs (snoRNA) that guide biochemical modifications to other RNA genes. Another class of seemingly ubiquitous RNA genes termed microRNAs (miRNA) has been more recently discovered. These microRNA genes encode very short products ∼22 nt in length, that generally act to downregulate the expression of specific gene targets (reviewed in He and Hannon (Citation2004) and Nelson et al. (Citation2003)). Similarly, small interfering RNA (siRNA) also play important roles in some fungal species. For example, siRNA in S. pombe are involved in heterochromatin assembly at centromeres (Grewal Citation2010; Lejeune et al. Citation2011). For a review of other short non-coding RNA genes, see Lee et al. (Citation2009).
Computational methods used to locate ncRNA genes are substantially different than those for finding protein-coding genes (reviewed in Eddy (Citation2002a)). Since protein-coding genes are encoded by non-random combinations of codons, the code can be recognized and deciphered as a translatable sequence with statistical properties consistent with known protein-coding genes. The information stored in ncRNA genes has no such codon structure, but instead can be recognized in the form of base-paired secondary structures consistent with the sequences and structures of known classes of structured ncRNA genes. The sequence and secondary structure for known classes of ncRNAs can be captured in a statistical models called profile stochastic context-free grammars (SCFGs) (Eddy Citation2002a, Eddy and Durbin Citation1994, Lowe and Eddy Citation1997). An essential ncRNA gene-finding resource is provided by Rfam (Griffiths-Jones et al. Citation2003, Citation2005), which includes profile SCFGs for classes of ncRNAs that span the tree of life and the INFERNAL software (Eddy Citation2002b) to search genome sequences with profile SCFGs to discover new members of known families. A search with a profile SCFG is computationally very expensive and thus slow, so in order to quicken the pace of the analysis, a blast heuristic is employed wherein the slower SCFG search is focused on a genomic region that first demonstrates BLAST-visible sequence similarity to a known member of the ncRNA family (Griffiths-Jones et al. Citation2003). Also, the computational complexity of the profile SCFG search imposes constraints on the size of the model such that modeling complete small subunit or large subunit rRNAs completely is impractical. Instead, a compromise was to model 5′ domains of the larger subunits (18S and 28S). Rfam and INFERNAL provide an essential resource for annotating tRNAs, snRNAs, snoRNAs, and other short ncRNAs with conserved sequences and secondary structures.
Larger rRNA sequences can often be annotated using BLAST against homologous rRNA genes. For example, the SILVA rRNA database project which has compiled a large collection of 5S, SSU and LSU rRNA sequences (Pruesse et al. Citation2007). However, in the case of newly sequenced fungi found to be distantly related from species represented by current rRNA sequence collections, low sequence similarity may prevent identification of rRNA sequences via BLAST. RNAmmer (Lagesen et al. Citation2007) identifies eukaryotic 5S, 18S and 28S ribosomal RNA genes with profile HMM models, and it has been used in the analysis of numerous fungal genomes. RNAmmer is generally more sensitive than BLAST, but has limitations; RNAmmer does not identify 5.8S rRNA sequences or rRNA genes in mitochondrial genomes. Furthermore, for draft genome assemblies, RNAmmer could fail to identify a rRNA gene if the 5′ end of the rRNA gene is missing from the assembled genome sequence, due to a heuristic employed within RNAmmer enabling runtime performance gains. Since searching large genome sequences with full-length 18S and 28S HMM profiles is computationally very expensive, RNAmmer first uses “spotter HMMs” constructed from the most highly conserved 18S and 28S rRNA segments to find the “seed” location of the rRNA genes, and only then uses the full-length HMMs to analyze the expanded regions around the seed location to define the boundary of the complete rRNA genes. In summary, RFAM is effective in identifying 5.8S rRNA genes, in addition to full-length 5S rRNA and the 5′ domain of the 18S and 28S rRNA. Thus, RNAmmer, RFAM and BLAST can complement each other, and the best rRNA gene predictions are achieved using a combined approach.
3. Repeats and transposable elements
Several attributes of repetitive sequences underscore their importance in eukaryotic genome annotation. Repeats are in many cases interesting sequences of great functional importance in their own right (reviewed in Shapiro and von Sternberg (Citation2005)). For example, the 5- to 7-mer tandem telomeric repeats protect the ends of linear chromosomes, and the ∼180-bp tandem repeat units found at plant and animal centromeres have been implicated in kinetochore formation. These tandem repeats form a class of repeats termed satellite sequences. Simple sequence repeats (SSRs) identified by tools such as MISA (http://pgrc.ipk-gatersleben.de/misa/download/misa.pl) are often used as genetic markers, and vary in abundance in different fungal genomes. Another class of repeats is formed by mobile DNA elements, including transposons, retrotransposons, MITEs, and SINEs (reviewed in Kazazian (Citation2004), and numerous references for MITEs provided in Yang and Hall (Citation2003)). Although these elements may sometimes cluster in specific regions of the genome, their distribution is often quite broad in large eukaryotic genomes, and they are sometimes found inserted within introns of genes and at gene termini. Unlike MITEs and SINEs, which are non-coding, transposons and retrotransposons encode genes for the proteins including transposases, integrases, and reverse transcriptases, which function to ensure their mobility. The DNA encoding these proteins is easily mistaken by ab initio gene prediction software as complete or fragmented host genes, falsely inflating the number of host gene predictions and occasionally resulting in gene predictions that are, in fact, chimeras between host genes and mobile elements. A solution to this problem is to first identify these repeat features and then conceal them from gene-finding software. A common mechanism used to mask these DNA regions is to replace the nucleotide characters of these sequences deemed repetitive with ‘N's.
Repeat sequence and transposable element content varies widely among different fungal species. For example, transposable elements account for 64% of the 103-Mb genome of barley powdery mildew, Blumeria graminis f.sp. hordei (Spanu et al. Citation2010), while other fungal genomes sequenced thus far have been found to harbor much smaller numbers but exhibiting considerable diversity of transposable elements. A wide variety of tools is available for the identification of sequence repeats and transposable elements (see review by Lerat (Citation2010)). In the simplest case, the repeat sequence content of a genome could be estimated by genome self-alignment via BLAST or CrossMatch (see protocol in Tarailo-Graovac and Chen (Citation2009)), as in examples of Coccidioides genomes (Sharpton et al. Citation2009), Rhizopus oryzae (Ma et al. Citation2009) and Fusarium oxysporum genomes (Ma et al. Citation2010).
To characterize the types of transposable elements in a fungal genome, additional tools and analyses are needed. For example, TransposonPSI (http://transposonpsi.sf.net) identifies and classifies repetitive protein or nucleic acid sequences based on homology to proteins encoded by diverse families of transposable elements. TransposonPSI uses PSI-Blast (Altschul et al. Citation1997) with a collection of (retro-) transposon ORF homology profiles to identify statistically significant alignments. This method can be used both to identify potential transposon ORFs within a set of predicted genes, and to identify regions of transposon homology within a larger genome sequence. TransposonPSI is particularly useful to identify degenerate transposon homologies within genome sequences that, due to their sequence divergence, successfully escape identification and masking by using RepeatMasker and an associated nucleotide library of repetitive elements. TransposonPSI has been routinely used to assist in the discovery of mobile elements across multiple fungal species and other eukaryotes including protozoa, plants and animals.
The RepeatMasker software (Smit 1996–2004) is used to identify regions of the genome with substantial sequence similarity to a repeat element in a library of known repeat sequences. Libraries of repeat family consensus sequences are provided by RepBase (Jurka et al. Citation2005, Jurka Citation2000) and these can be used with RepeatMasker to find and mask similar elements in raw genomic sequences. Once the repeat elements are identified, a BLAST (Altschul et al. Citation1990) search against RepBase could provide partial characterization of the repeat elements, as in the examples of Coccidioides genomes (Sharpton et al. 2009). The RepBase libraries are organism-specific and, although these are of fantastic utility if your genome of interest is represented, these libraries have limited application in the context of previously uncharacterized genomes, if these genomes are not sufficiently similar to the previously characterized genomes. Novel genome sequences must first be mined for repetitive elements in order to generate a corresponding repeat library containing such sequences. There are additional tools that provide for such de novo repeat library construction, including RepeatScout (Price et al. Citation2005), PILER (Edgar and Myers Citation2005), and RECON (Bao and Eddy Citation2002). A subset of the repeats reported by these programs may be found to be extraordinarily abundant, and to more likely represent mobile elements and noncoding repeats than they are to correspond to protein-coding host genes, and these can be used as a companion library to RepeatMasker to mask the genome in preparation for a more focused gene finding exercise. More recent efforts in repeat-finding attempt to combine and integrate multiple tools into a single package, since it has been noted that different repeat finders seem to complement each other. For example, the “RepeatModeler” package (Smit and Hubley) combines RepeatMasker, RECON, RepeatScout and TRF (Benson Citation1999) for repeat identification and classification. The “REPET” package (Flutre et al. Citation2010) combines the functionalities of two modules (TEdenovo and TEannot) and uses BLASTER for self-alignment, GROUPER and RECON for identification and comparison with RepBase for classification.
4. Pseudogenes
A persistent difficulty in genome annotation is distinguishing functional protein-coding genes from pseudogenes, their defunct counterparts. Pseudogenes arise in genome evolution via several mechanisms, the most predominant mechanisms being gene duplication followed by degeneration of one of the duplicated copies, and retrotranssposition – by which processed (intron-free) transcripts of functional genes are reverse transcribed and inserted into the genome, presumably accidentally by the machinery of active retrotransposons (reviewed in Mighell et al. (Citation2000); also visit http://www.pseudogene.org). Additionally, over the course of evolution and changing selective pressures, genes once important for some biological function may no longer be required and are free to accumulate mutations and degenerate over time. Today, we recognize the signatures of the remaining gene features as “genomic fossils”, remnants of the past retained, still recognizable in the sequence of the DNA, yet no longer coding for proteins.
Pseudogenes tend to have properties that facilitate their identification (nicely summarized in Zhang and Gerstein (Citation2004)); most tools rely on a comparison to a known functional homolog from which the candidate pseudogene was presumed to have been derived. The most recognizable property of a pseudogene is that its gene structure is interrupted by frameshifts and/or intervening in-frame stop codons. Degenerated pseudogenes will often have acquired numerous frameshifts and intervening stop codons, which in all likelihood preclude the translation of a functional polypeptide. In cases where a single DNA sequence aberration exists that would be suggestive of a pseudogene, it might be the result of a DNA sequencing error instead of a mutation. In certain cases, such suspicious regions of genome sequences are closely examined and potentially targeted for resequencing to confirm the sequence quality before a pseudogene is predicted.
Additional reasons for suspecting a pseudogene derive from other characteristics presumed to disable gene function. A truncated form a full-length gene elsewhere in the genome may be presumed nonfunctional. Similarly, a gene missing a start codon or required regulatory sequence might be flagged as a candidate pseudogene. Retroposed pseudogenes are perhaps the most readily identified; these genes are reminiscent of the parental gene's fully processed mRNA, lacking introns and having a string of adenines at the 3′ end. They also may have characteristics of mobile element-mediated genome insertion, such as having short direct repeats in the flanking sequence.
The identification and annotation of pseudogenes currently relies heavily on homology to known genes. Most DNA sequence-based ab initio gene prediction tools model coding and noncoding sequences, but do not model degenerate pseudogenes. Since pseudogenes have detectable remnants of their earlier protein-coding potential, which makes their identification possible, this remaining coding signal may be detected by gene prediction software, and bizarre and entirely bogus predictions are often the result. Gene models are sometimes predicted with an inflated number of introns that are introduced to sidestep the frameshifts and intervening stop codons that preclude any proper gene modeling.
Many pseudogenes are so degenerate that their identification and deduction as a pseudogene is obvious to the annotator. Other cases may not be so obvious. For example, disruption of an important regulatory sequence (promoter, transcription factor binding site, splicing enhancer, etc.) by a recent transposon insertion or non-consensus splice sites may render a gene nonfunctional. Given comparative genomic sequence, a statistical test can be used to determine if the gene remains under selective pressure (positive or negative), or if it appears to be evolving at a neutral rate (randomly). Pseudogenes are mostly expected to be defunct and not under selective pressure, so in most cases we would expect them to be evolving neutrally. The Ka/Ks test describes the evolution of the coding sequence by measuring the rate of substitution at synonymous and non-synonymous codon positions (Li Citation1997). A neutrally evolving sequence such as a pseudogene (which by definition is not evolving under any selective pressure at all) would acquire synonymous substitutions at the same rate as nonsynonymous substitutions, and hence have a Ka/Ks value that approximates the value of one (Li et al. Citation1981).
PPFINDER (van Baren and Brent Citation2006) is a tool that exploits two characteristics of processed pseudogenes to first identify and then exclude them from subsequent gene-finding efforts using the N-SCAN gene prediction program. The characteristics of retroposed pseudogenes sought by PPFINDER include the loss of introns and the lack of detectable homology within a significantly diverged related genome. The system works as follows. An intron location method is employed that involves finding sequence homology among gene predictions output from an initial N-SCAN execution. The genome sequence of the candidate pseudogene is aligned to the genome sequence of the tentative parental gene and, if the candidate pseudogene is aligned with gaps that coincide with predicted introns, then that model is flagged. A second method based on conserved synteny involves analyzing the syntenic region of a related genome for homology to the candidate pseudogene; recently occurring processed pseudogenes are expected to lack conserved synteny as compared to the parental genes from which the processed pseudogenes were derived.
The processed pseudogenes found by PPFINDER are restricted to only those genes that are initially predicted by N-SCAN; hence the caveat, genes that are not predicted by N-SCAN remain undetected by this process. Even so, PPFINDER represents the first publicly available software tool that effectively predicts pseudogenes, and is the first effort to include pseudogene detection and exclusion as a direct component of the automated gene finding process. A more general, standalone pipeline for pseudogene detection is provided by PseudoPipe (Zhang et al. Citation2006), which relies on BLAST to correlate candidate pseudogenes with their homologous parental genes and, subsequently, classifies pseudogenes as retroposed, duplicated or fragments. Over time, we expect to see more of these types of software tools and methods being developed, and integrated at various stages of the genome annotation pipeline, either as a component of the initial gene finding strategy as in PPFINDER, or as a subsequent quality-control step to interrogate gene models output from a larger annotation pipeline, flagging candidate pseudogenes for further evaluation.
5. Annotation of mitochondrial genomes
Fungal mitochondrial genomes show great diversity in the genome size, number of genes and genome architecture. For example, the S. pombe mitochondrion contains only 19,431 bases with 10 protein-coding genes, while Podospora anserina mitochondrion is about 100 kb with 50 protein-coding genes (Cummings et al. Citation1990) and Moniliophthora perniciosa genome is even larger at 109 kb, but only has 14 protein-coding genes (Formighieri et al. Citation2008). This is largely consistent with observations in eukaryote mitochondrial genomes, where the number of protein-coding genes range from 3 to 67 (Adams and Palmer Citation2003). In addition, fungal mitochondrial genome architectures are also diverse. Most fungal mitochondrial genomes form a single circle, as in Stagonospora nodorum (Hane et al. Citation2007), Aspergillus niger (Juhasz et al. Citation2008), several dermatophytes (Wu et al. Citation2009), and Paracoccidioides brasiliensis (Cardoso et al. Citation2007). However, exceptions exist, such as the Spizellomyces punctatus mitochondrial genome, which consists of three circular chromosomes (58.8, 1.4 and 1.1 kb) with 31, 1, and 0 protein-coding genes (Forget et al. Citation2002). A comparative study of multiple Candida mitochondrial genomes suggests that these are all linear genomes with telomeres, sometimes with multiple chromosomes (Valach et al. Citation2011). Incidentally, in the extreme case of Diplonema papillatum (which is not a fungus), the mitochondrion consists of multipartite (“segmented”) genomes with numerous circular chromosomes, where 10 of the 11 recognizable protein-coding genes are split among 3–12 chromosomes (Vlcek et al. Citation2011).
Annotation of mitochondrial genomes has unique challenges, since the mitochondrial and nuclear genomes have very different nucleotide composition and use different codon translation tables (NCBI Translation Table 4, see http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi ). For example, the nuclear stop-codon UGA is translated as a tryptophan or leucine. In a subset of Candida species, all cytoplasmic CUG codons are translated as a serine instead of leucine (Massey et al. Citation2003). Mitochondrial genomes can use alternative start codons, such as AUU, AUA and UUA. Due to the small genome size, most ab initio gene predictors cannot be readily trained to work on the mitochondrion. As a result, most gene prediction programs developed for the annotation of the nuclear genome cannot be used effectively for the annotation of mitochondrial genomes.
Fungal mitochondrial genomes generally contain a standard set of 14 or 15 conserved protein-coding genes. These include the ATP synthase subunits (atp6, atp8, atp9), apocytochrome b (cob), cytochrome oxidase subunits (cox1, cox2, cox3), NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5 and nad6), and the rps5 ribosomal protein in some fungal species (Cardoso, Tambor and Nobrega Citation2007, Woo et al. Citation2003; Torriani et al. Citation2008; Valach et al. Citation2011, Vlcek et al. Citation2011). Most of these protein-coding genes are transcribed from the same DNA strand. For single-circle mitochondrial genomes, the standard is to break the circle at a consistent location, generally after cox2 (Cardoso et al. Citation2007, Woo et al. Citation2003). This provides to compare gene content and gene order among mitochondrial genomes from different fungal species. For closely related species, gene order can be very similar, as was the case with Penicillium marneffei and Aspergillus nidulans, where the order of protein-coding genes is conserved for all protein-coding genes except atp9 (Woo et al. Citation2003). However, gene order and orientation in Candida species appears less well conserved (Valach et al. Citation2011). The high conservation of mitochondrial protein-coding genes greatly facilitates mitochondrial genome annotation, most of which can be readily identified using a combination of BLAST, GeneWise, and Pfam domain analysis. Additional gene candidates can be identified by exploring long ORFs using the proper codon translation table. Finally, gene annotations are manually refined using a genome annotation editor.
Fungal mitochondrions also contain non-coding RNA genes. The mitochondrial rRNA genes are single copy genes for the large and small ribosomal RNAs (rrl and rrs). These are significantly shorter than their counterpart in the nuclear genome and cannot be identified by RNAmmer (Lagesen et al. Citation2007). Instead, they are identified by a combination of RFAM and BLASTN against a rRNA database. This approach has been used for the annotation of multiple fungal mitochondrial genomes, such as Cryptococcus gattii R265 (D'Souza et al. Citation2011). Most fungal species contain tRNAs for all 20 amino acids. In some fungal mitochondrial genomes that appear be deficient in tRNA content, it is likely that these “absent” tRNAs are nuclear encoded and later imported into the mitochondrion (Forget et al. Citation2002). tRNA genes can be identified with tRNAScan using the organelle option and the sensitive mode (Lowe and Eddy Citation1997). The fungal mitochondrial tRNA genes tend to form clusters, often around the rRNA genes (rnl and rns), but the clusters can also be dispersed throughout the mitochondrial genome. The order of tRNA genes is largely conserved among closely related fungal species (Cardoso et al. Citation2007, Woo et al. Citation2003, Torriani et al. Citation2008).
6. Projecting reference genome annotations
There are often instances where reference genome annotations must be propagated from one genome assembly to another. For example, many fungal species have multiple sequentially improved assemblies generated over a period of time, sometimes involving several years, as in the case of Neurospora crassa (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html). Gene structures are projected from earlier assemblies to the latest assemblies, keeping track of the gene identifiers (“locus tags”) and other annotation attributes across each new assembly release. Another example involves leveraging a reference genome annotation for annotating a newly sequenced genome of a related species, e.g. from the highly curated Schizosaccharomyces pombe to the newly sequenced Schizosaccharomyces octosporus genome (Rhind et al. Citation2011), or between strains of C. gattii (D'Souza et al. Citation2011). For this purpose, we developed an alignment-based gene mapping strategy at the Broad Institute and used this strategy for mapping genes in all updated genome assemblies since 2005. In our gene mapping strategy, we first align the two genomes using NUCmer (Delcher et al. Citation2002) to establish base-to-base mapping between the two assemblies, and then use this info to project the gene coordinates from the reference genome to the target genome. A similar strategy is used in the RATT tool published recently (Otto et al. Citation2011).
7. Functional annotation
Once the gene structural annotation phase is completed for a fungal genome, the focus shifts to the functional annotation of the gene set. The purpose is to assign gene product names largely based on in silico functional characterization of the genes.
For gene product naming, we follow the GenBank naming guidelines (http://www.ncbi.nlm.nih.gov/genbank/genomesubmit_annotation.html#CDS), and assign gene product names according the SwissProt naming conventions (http://www.uniprot.org/docs/proknameprot). Specifically, we first assign gene product names via BLAST hits to SwissProt database with manually curated gene product names, using stringent criteria (≥70% protein sequence identity, ≥70% coverage of both the query and the database hit sequence, and length difference ≤10%). For the remaining genes with unassigned product names, we then use HMMER equivalogs (related proteins with presumed equivalent functions) from TIGRfam (Haft et al. Citation2003) and Pfam (Finn et al. Citation2010) hits to assign the name based on the HMMER hits, if the hit score is above the trusted cutoff value. This usually results in name assignment for 10–30% of genes. Since our naming standards require high identity, genomes corresponding to newly sequenced clades of the fungal phylogeny often have fewer genes that are assigned meaningful names based on sequence homology to known proteins.
Additional functional characterizations include assigning Gene Ontology identifiers (via pfam2go (http://www.geneontology.org/external2go/pfam2go) and blast2go (Conesa et al. Citation2005)), enzyme commission codes (EC numbers), KEGG-pathway membership, KOG-homology (Tatusov et al. Citation2003), protein domains (Pfam and TIGRfam), secretion signals (Choi, 2010), and transmembrane domains (Krogh et al. Citation2001). Of particular interest to fungal genomes is the analysis of specialized function categories, including protein kinases (Manning et al. Citation2002, Stajich et al. Citation2010), histidine kinases (Nemecek et al. Citation2006, Citation2007), carbohydrate-activating enzymes (CAZy) (Cantarel et al. Citation2009), GPI-anchored proteins (PredGPI) (Pierleoni et al. Citation2008), transporters (Coleman and Mylonakis Citation2009), GPCR (Xue et al. Citation2008), secreted proteins (signalP) (Emanuelsson et al. Citation2007) and effectors (Stergiopoulos and de Wit Citation2009), other candidate pathogenicity factors (PHI-base), secondary metabolite gene clusters (SMURF) (including PKS, NRPS, etc.) (Khaldi et al. Citation2010), proteases (Merops peptidase database; Rawlings et al. (Citation2004)), and transcription factors (via SUPERFAMILY, (Shelest (Citation2008)). Taken together, the general function profiles and the specialized function profiles provide a comprehensive overview of the biochemical characteristics of a genome, which can be correlated with the biological phenotypes of a fungal species.
8. The Broad Institute fungal/eukaryotic genome annotation pipeline
A production genome annotation system ties many of the above genome and gene annotation tasks into a set of components of a larger pipeline. The general annotation pipeline applied to most eukaryotic genomes annotated by the Broad Institute is shown in . The tools and processes we describe are those that we have found to be most reliable and effective in a production genome annotation environment and represent a subset of the tools described within the earlier sections.
Figure 6. The Broad Institute Eukaryotic Genome Annotation Pipeline. Genome sequences are annotated by leveraging multiple sources of evidence for genes, including ab initio gene predictions, protein and transcript alignments, all of which are distilled into a consensus gene set. Gene products are named based on homology to proteins or domains of known function, manually refined, and ultimately released to public databases.
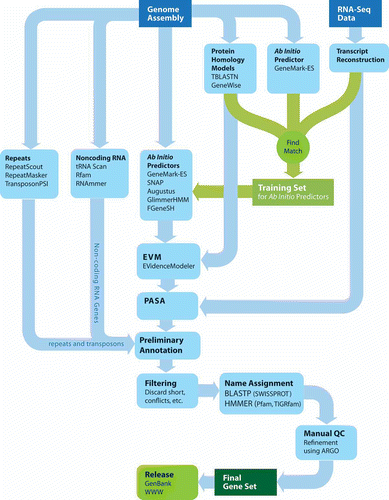
The input to the pipeline is a set of sequences provided as a multi-FASTA file. The first stage of the pipeline involves decorating the genome with primary evidence to be leveraged for downstream annotation efforts, collecting data based on analysis of the genome sequence alone or in combination with general sequence resources. The self-training GeneMark-ES is run to identify an initial set of ab initio predictions, since no prior training is required. Regions of the genome with homology to known protein sequences are identified by TBLASTN of the genome sequences against the UniRef90 non-redundant protein dataset. Regions shown to have homology to known proteins are subject to subsequent gene modeling using GeneWise. Any available RNA-Seq data is processed using our hybrid genome-guided transcript reconstruction method described earlier (and illustrated in ).
Repeats are identified using several methods. A genome-specific repeat library is generated using RepeatScout, which is then searched against the genome using RepeatMasker. In addition, we leverage TransposonPSI to identify genomic regions with detectable sequence homology to known families of transposable element proteins. We also search the genome assembly against a custom fungal repeat protein database of sequences collected from repbase and fungal genome projects to identify regions with homology to known repeat proteins.
This initial round of data collection can identify a set of high quality gene structures to be used for training additional ab initio gene predictors, including Augustus, GlimmerHMM, and SNAP. High quality reference gene structures are extracted from two sources: genes reconstructed from transcript data (e.g. RNA-Seq), and those complete GeneMark-ES predictions that appear to be of high quality and supported by transcript and protein alignments. This process is detailed below.
ESTs and FL-cDNAs, and now RNA-Seq data, are the single best resource available at the earliest stage of characterizing a new genome, particularly when no other highly similar genome sequence exists. Complete and partial gene structures based on spliced transcript sequence alignment data are used as inputs to train gene-finding software. This is mediated in part by components of the PASA software (Haas et al. Citation2003). To automatically generate a training set, the longest ORF is located within each PASA transcript alignment assembly, and those complete and partial ORFs that exceed a specified length (e.g. ≥300 nt) and that appear to have strong coding potential are output for gene-finder training. We find that by aligning transcripts to the unmasked genome instead of a repeat-masked genome, we obtain more robust transcript alignments; since many UTR regions tend to extend into the beginnings of repetitive regions, it is important to retain the repeats to allow the alignments to extend completely. Furthermore, GMAP has an option to report only the single best alignment for each transcript, so we need not examine copious amounts of output that would normally be associated with repeat-matching transcripts, which makes the unmasked genome a feasible choice of alignment target.
Second, the subset of GeneMark-ES predictions that have structures consistent with transcript alignments and GeneWise predictions are extracted to further supplement the training set. Predicted proteins found to exhibit homology to known repeats (ascertained by using TransposonPSI) are excluded from this gene collection. With this set of trusted genes, we train the ab initio gene prediction programs including Augustus (Stanke and Waack Citation2003), geneid (Parra et al. Citation2000), Fgenesh (Salamov and Solovyev Citation2000), GlimmerHMM (Majoros et al. Citation2004), SNAP (Korf Citation2004), in addition to re-training GeneMarkES (Lukashin and Borodovsky Citation1998) that was initially run in the self-training mode.
In the next phase, the ab initio gene predictions, the high-quality reference gene models, PASA alignment assemblies, protein and cross-species transcript alignments, and GeneWise predictions are then combined into consensus gene structure annotations using EVidenceModeler (Haas et al. Citation2008). PASA is then used to further update these annotations based on the high-quality transcript alignments, primarily adding UTRs and modeling alternative splicing isoforms.
The product of the pipeline thus far corresponds to the final output of an automated protein-coding gene discovery pipeline. Our noncoding RNA gene-finding relies almost exclusively on INFERNAL (Eddy Citation2002b) and Rfam (Griffiths-Jones et al. Citation2003, Griffiths-Jones et al. Citation2005), RNAmmer (Lagesen et al. Citation2007) and tRNAScan (Lowe and Eddy Citation1997). The small and large subunit rDNAs are located and annotated by aligning representative sequence entries.
We next filter the candidate gene set to remove spurious genes from repeat sequences and transposable elements. Specifically, we filter out genes with substantial overlap to RepeatScout, blast hits to known fungal transposon proteins, Pfam domains corresponding to regions of known transposable element proteins, and Transposon-PSI matches. We also inspect those coding sequences with multiple hits (≥10) to different parts of the assembly at ≥90% identity, which may represent previously undiscovered repetitive elements rather than host coding sequences. We also check for genes with similarity to other repeats based on assigned gene product names and remove short proteins with low complexity and having insufficient supporting evidence (e.g. non-repeat Pfam domains, ESTs, RNA-Seq). After filtering, the resulting feature set is then ready for targeted manual review.
Manual review of genome annotations at the Broad Institute consists of a team of bioinformaticists examining genes flagged as suspicious and most likely to benefit from manual inspection. Such targeted genes include those with long introns, very short ORFs, those ORFs that have only partial sequence homology to known proteins and perhaps represent gene fragments, and those genes that potentially represent merged or split gene products based on the range of protein sequence homologies across the lengths of the genes. Annotators inspect these genes using our ARGO genome browser (shown in ), which brings all evidence and features into a single view, to allow review of the gene structures and manual edits if warranted.
Once the gene set is finalized, the genes are numbered using the locus tags assigned to the genome (a unique locus prefix is provided by NCBI for each genome). The gene product names are assigned by BLAST against SwissProt or by HMMER against TIGRfam equivalogs. The rest of the genes are assigned the name “hypothetical protein” according to the current GenBank guidelines. Finally, genome annotations are released on the Broad website and submitted to GenBank.
9. Access to fungal genome annotations
The worldwide destinations for all biological sequence data include GenBank (Benson et al. Citation2005) in the United States of America, the European Molecular Biology Laboratory (EMBL) (Kanz et al. Citation2005) in the United Kingdom, and the DNA Databank of Japan (DDBJ) (Tateno et al. Citation2005). The sequence data can be searched based on keyword terms or based on computationally determined similarity to a query sequence. The depth of annotation associated with genome sequence data can be very rich, especially for model organisms in which many genes have been well characterized. To gain additional insight into gene function, gene annotations are linked with gene expression data, to key pathways in metabolic maps, and to the most recent literature that further elucidates knowledge about a particular gene function from a fungal genome. It is mostly beyond the mission of the sequence archives to maintain these types of specialized data, and so specialty databases have arisen over the years to better cater towards serving the fungal scientific community. These include:
-
• Aspergillus Genome Database (AspGD) at http://www.aspgd.org/
-
• Fungal Genome Initiative site at
-
• JGI MycoCosm site at
-
• Saccharomyces Genome Database (SGD) (Christie et al. Citation2004) at
-
• Candida Genome Database (CGD) (Costanzo et al. Citation2006) at
-
• CandidaDB (Rossignol et al. Citation2008) at
-
• Comparative Fungal Genomics Platform (Park et al. Citation2008) at
-
• Ensembl Fungi athttp://fungi.ensembl.org/
10. Summary
Accurate eukaryotic gene structure annotation is a complex task that couples homology-based methods with prediction algorithms to reveal introns and exons of genes otherwise hidden within the string of bases that comprise the DNA sequence. Repeat sequences increase the difficulty of gene finding by diluting the gene content and confusing ab initio gene predictors. Recent developments in repeat element identification have greatly alleviated this problem, but repeats still undermine gene discovery in previously uncharacterized genomes. Since the late 1990s, eukaryotic ab initio gene prediction tools have evolved from using the genome sequence exclusively, to incorporating external data sources indicating genome homologies to further improve gene-finding accuracy. Flexible evidence combiners are well suited to yield consensus gene structures given a wide array of evidence types. High quality databases of expressed transcript sequences provide an invaluable resource to genome annotation efforts; at the earliest stage, they can be leveraged to model gene structures for training ab initio gene prediction tools and, in the final stage, they can be used to add the finishing touches to gene annotations by yielding UTR structures, alternative splicing variations, and identification of polyadenylation sites; all tasks mediated by our PASA software (Haas et al. Citation2003). All automated methods are less than perfect, so it is incumbent upon interested scientists to manually repair errant annotations using genome annotation editing software such as ARGO (Engels Citation2003–2011), Apollo (Lewis et al. Citation2002), or Artemis (Rutherford et al. Citation2000).
The Broad Institute fungal genome annotation pipeline exemplifies how bioinformatics tools are coupled together to find and model genes in any newly sequenced genome. The set of automatically generated gene structures produced by the annotation pipeline is, in many ways, the end of the beginning. As knowledge accrues from further studying the genome sequence coupled with downstream experimentation, the genome annotations and related resources will need to be continually refined.
Acknowledgements
We thank Christian Stolte and Leslie Gaffney for help in figure illustration; Zehua Chen and Sharvari Gujja for comments on the manuscript; Li-Jun Ma for inviting us to submit to this special issue; and the National Human Genome Research Institute for initial support of the fungal genome initiative (FGI) at the Broad Institute.
References
- Abeel T. 2006–2011. GenomeView http://genomevieworg/ (http://genomevieworg/)
- Adams KL, Palmer JD. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 29(3):380–395. doi nrg2220 [pii] 10.1038/nrg2220 http://www.ncbi.nlm.nih.gov/pubmed/14615181 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18087260 (http://www.ncbi.nlm.nih.gov/pubmed/14615181 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18087260)
- Alexandersson M, Cawley S, Pachter L. 2003. SLAM: cross-species gene finding and alignment with a generalized pair hidden Markov model. Genome Res. 13(3):496–502. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12618381 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12618381)
- Allen JE, Pertea M, Salzberg SL. 2004. Computational gene prediction using multiple sources of evidence. Genome Res. 14(1):142–148. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14707176 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14707176)
- Allen JE, Salzberg SL. 2005. JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics. 21(18):3596–3603. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16076884 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16076884)
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi 10.1006/jmbi.1990.9999 http://www.ncbi.nlm.nih.gov/pubmed/2231712 (http://www.ncbi.nlm.nih.gov/pubmed/2231712)
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. http://www.ncbi.nlm.nih.gov/pubmed/9254694 (http://www.ncbi.nlm.nih.gov/pubmed/9254694)
- Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. 2008. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Func Genom Proteom. 7(1):50–62. doi 10.1093/bfgp/eln010 http://www.ncbi.nlm.nih.gov/pubmed/18334489 (http://www.ncbi.nlm.nih.gov/pubmed/18334489)
- Ashurst JL, Chen CK, Gilbert JG, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, Trevanion S, et al. 2005. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 33(Database issue):D459–465. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608237 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608237)
- Bao Z, Eddy SR. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12(8):1269–1276. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12176934 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12176934)
- Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. 2000. Human and mouse gene structure: comparative analysis and application to exon prediction. Genome Res. 10(7):950–958. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10899144 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10899144)
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res. 33(Database issue):D34–38. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608212 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608212)
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. doi gkc131 [pii] http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9862982 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9862982)
- Birney E, Clamp M, Durbin R. 2004a. GeneWise and Genomewise. Genome Res. 14(5):988–995. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15123596 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15123596)
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, et al. 2004b. An overview of Ensembl. Genome Res. 14(5):925–928. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15078858 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15078858)
- Bitton DA, Wood V, Scutt PJ, Grallert A, Yates T, Smith DL, Hagan IM, Miller CJ. 2011. Augmented Annotation of the Schizosaccharomyces pombe Genome Reveals Additional Genes Required for Growth and Viability. Genetics. doi genetics.110.123497 [pii]10.1534/ genetics.110.123497 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21270388 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21270388)
- Brejova B, Brown DG, Li M, Vinar T. 2005. ExonHunter: a comprehensive approach to gene finding. Bioinformatics 21 Suppl 1:i57–65. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961499 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961499)
- Brejova B, Vinar T, Chen Y, Wang S, Zhao G, Brown DG, Li M, Zhou Y. 2009. Finding genes in Schistosoma japonicum: annotating novel genomes with help of extrinsic evidence. Nucleic Acids Res. 37(7):e52. doi gkp052 [pii]10.1093/nar/gkp052 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19264800 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19264800)
- Brent , MR. 2008 . Steady progress and recent breakthroughs in the accuracy of automated genome annotation . Nat Rev Genet. , 9 ( 1 ) : 62 – 73 . Available from
- Brent MR, Guigo R. 2004. Recent advances in gene structure prediction. Curr Opin Struct Biol. 14(3):264–272. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15193305 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15193305)
- Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268(1):78–94. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9149143 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9149143)
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459(7247):657–662. Available from doi nature08064 [pii] 10.1038/nature08064 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19465905 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19465905)
- Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. 2006. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7:327. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17194304 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17194304)
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. doi gr.6743907 [pii]10.1101/gr.6743907 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18025269 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18025269)
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37(Database issue):D233–238. doi gkn663 [pii]10.1093/nar/gkn663 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18838391 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18838391)
- Cardoso MA, Tambor JH, Nobrega FG. 2007. The mitochondrial genome from the thermal dimorphic fungus Paracoccidioides brasiliensis. Yeast. 24(7):607–616. doi 10.1002/yea.1500 http://www.ncbi.nlm.nih.gov/pubmed/17492801 (http://www.ncbi.nlm.nih.gov/pubmed/17492801)
- Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. 2004. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32(Database issue):D311–314. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14681421 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14681421)
- Coleman JJ, Mylonakis E. 2009. Efflux in fungi: la piece de resistance. PLoS Pathog. 5(6):e1000486. doi 10.1371/journal.ppat.1000486 http://www.ncbi.nlm.nih.gov/entrez/query.cmd=fcgi?Retrieve&db=PubMed&dopt=Citation&list_uids=19557154 (http://www.ncbi.nlm.nih.gov/entrez/query.cmd=fcgi?Retrieve&db=PubMed&dopt=Citation&list_uids=19557154)
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. doi 10.1093/bioinformatics/bti610 http://www.ncbi.nlm.nih.gov/pubmed/16081474 (http://www.ncbi.nlm.nih.gov/pubmed/16081474)
- Costanzo MC, Arnaud MB, Skrzypek MS, Binkley G, Lane C, Miyasato SR, Sherlock G. 2006. The Candida Genome Database: facilitating research on Candida albicans molecular biology. FEMS Yeast Res. 6(5):671–684. doi 10.1111/j.1567-1364.2006.00074.x http://www.ncbi.nlm.nih.gov/pubmed/16879419 (http://www.ncbi.nlm.nih.gov/pubmed/16879419)
- Cummings DJ, McNally KL, Domenico JM, Matsuura ET. 1990. The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr Genet. 17(5):375–402. http://www.ncbi.nlm.nih.gov/pubmed/2357736 (http://www.ncbi.nlm.nih.gov/pubmed/2357736)
- Cuomo CA, Birren BW. 2010. The fungal genome initiative and lessons learned from genome sequencing. Methods Enzymol. 470:833–855. doi 10.1016/S0076-6879(10)70034-3 http://www.ncbi.nlm.nih.gov/pubmed/20946837 (http://www.ncbi.nlm.nih.gov/pubmed/20946837)
- DeCaprio D, Vinson JP, Pearson MD, Montgomery P, Doherty M, Galagan JE. 2007. Conrad: gene prediction using conditional random fields. Genome Res. 17(9):1389–1398. doi 10.1101/gr.6558107 http://www.ncbi.nlm.nih.gov/pubmed/17690204 (http://www.ncbi.nlm.nih.gov/pubmed/17690204)
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30(11):2478–2483. http://www.ncbi.nlm.nih.gov/pubmed/12034836 (http://www.ncbi.nlm.nih.gov/pubmed/12034836)
- Do JH, Choi DK. 2006. Computational approaches to gene prediction. J Microbiol. 44(2):137–144. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16728949 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16728949)
- D'Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, et al. 2011. Genome Variation in Cryptococcus gattii, an Emerging Pathogen of Immunocompetent Hosts. MBio. 2(1). doi mBio.00342-10 [pii] 10.1128/mBio. 00342-10 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21304167 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21304167)
- Eddy SR. 2001. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2(12):919–929. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11733745 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11733745)
- Eddy SR. 2002a. Computational genomics of noncoding RNA genes. Cell. 109(2):137–140. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12007398 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12007398)
- Eddy SR. 2002b. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 3:18. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12095421 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12095421)
- Eddy SR, Durbin R. 1994. RNA sequence analysis using covariance models. Nucleic Acids Res. 22(11):2079–2088. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8029015 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8029015)
- Edgar RC, Myers EW. 2005. PILER: identification and classification of genomic repeats. Bioinformatics 21 Suppl 1:i152–158. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961452 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961452)
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2(4):953–971. doi 10.1038/nprot.2007.131 http://www.ncbi.nlm.nih.gov/pubmed/17446895 (http://www.ncbi.nlm.nih.gov/pubmed/17446895)
- Engels R. 2003–2011. ARGO Genome Browser http://wwwbroadinstituteorg/annotation/argo/ (http://wwwbroadinstituteorg/annotation/argo/)
- Eyras E, Caccamo M, Curwen V, Clamp M. 2004. ESTGenes: alternative splicing from ESTs in Ensembl. Genome Res. 14(5):976–987. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15123595 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15123595)
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38(Database issue):D211–222. doi 10.1093/nar/gkp985 http://www.ncbi.nlm.nih.gov/pubmed/19920124 (http://www.ncbi.nlm.nih.gov/pubmed/19920124)
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8(9):967–974. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9750195 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9750195)
- Florea L, Di Francesco V, Miller J, Turner R, Yao A, Harris M, Walenz B, Mobarry C, Merkulov GV, Charlab R, et al. 2005. Gene and alternative splicing annotation with AIR. Genome Res. 15(1):54–66. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15632090 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15632090)
- Flutre , T , Inizan , O , Hoede , C and Quesneville , H. REPET: pipelines for the identification and annotation of transposable elements in genomic sequences . Plant & Animal Genome (PAG) XVIII Conference . January 9–13 2010 .
- Forget L, Ustinova J, Wang Z, Huss VA, Lang BF. 2002. Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol Biol Evol. 19(3):310–319. http://www.ncbi.nlm.nih.gov/pubmed/11861890 (http://www.ncbi.nlm.nih.gov/pubmed/11861890)
- Formighieri EF, Tiburcio RA, Armas ED, Medrano FJ, Shimo H, Carels N, Goes-Neto A, Cotomacci C, Carazzolle MF, Sardinha-Pinto N, et al. 2008. The mitochondrial genome of the phytopathogenic basidiomycete Moniliophthora perniciosa is 109 kb in size and contains a stable integrated plasmid. Mycol Res. 112(Pt 10):1136–1152. doi 10.1016/j.mycres.2008.04.014 http://www.ncbi.nlm.nih.gov/pubmed/18786820 (http://www.ncbi.nlm.nih.gov/pubmed/18786820)
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. 1996. Life with 6000 genes. Science. 274(5287):546, 563–547. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8849441 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8849441)
- Grewal SI. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 20(2):134–141. doi S0959-437X(10)00028-6 [pii]10.1016/j.gde.2010.02.003 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20207534 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20207534)
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31(1):439–441. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12520045 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12520045)
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33(Database issue):D121–124. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608160 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608160)
- Gross SS, Brent MR. 2006. Using multiple alignments to improve gene prediction. J Comput Biol. 13(2):379–393. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16597247 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16597247)
- Gross SS, Do CB, Sirota M, Batzoglou S. 2007. CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction. Genome Biol. 8(12):R269. doi 10.1186/gb-2007-8-12-r269 http://www.ncbi.nlm.nih.gov/pubmed/18096039 (http://www.ncbi.nlm.nih.gov/pubmed/18096039)
- Guigo R. 1998. Assembling genes from predicted exons in linear time with dynamic programming. J Comput Biol. 5(4):681–702. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10072084 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10072084)
- Guigo R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E, et al. 2006. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 7 Suppl 1:S2 1–31. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16925836 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16925836)
- Haas BJ, Zody MC. 2010. Advancing RNA-Seq analysis. Nat Biotechnol. 28(5):421–423. doi 10.1038/nbt0510-421 http://www.ncbi.nlm.nih.gov/pubmed/20458303 (http://www.ncbi.nlm.nih.gov/pubmed/20458303)
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL. 2002. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 3(6): RESEARCH0029. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12093376 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12093376)
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Jr., Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14500829 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14500829)
- Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith RK, Jr., Maiti R, Chan AP, Yu C, Farzad M, Wu D, et al. 2005. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 3:7. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15784138 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15784138)
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9(1):R7. doi gb-2008-9-1-r7 [pii] 10. 1186/gb-2008-9-1-r7 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18190707 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18190707)
- Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31(1):371–373. http://www.ncbi.nlm.nih.gov/pubmed/12520025 (http://www.ncbi.nlm.nih.gov/pubmed/12520025)
- Hane JK, Lowe RG, Solomon PS, Tan KC, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Galagan JE, et al. 2007. Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 19(11):3347–3368. doi 10.1105/tpc.107.052829 http://www.ncbi.nlm.nih.gov/pubmed/18024570 (http://www.ncbi.nlm.nih.gov/pubmed/18024570)
- He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 5(7):522–531. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15211354 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15211354)
- Howe KL, Chothia T, Durbin R. 2002. GAZE: a generic framework for the integration of gene-prediction data by dynamic programming. Genome Res. 12(9):1418–1427. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12213779 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12213779)
- Huang X, Adams MD, Zhou H, Kerlavage AR. 1997. A tool for analyzing and annotating genomic sequences. Genomics 46(1):37–45. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9403056 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9403056)
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi 1471-2105-11-119 [pii] 10.1186/1471-2105-11-119 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20211023 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20211023)
- Ivashchenko AT, Tauasarova MK, Atambaeva Sh A. 2009. [Exon-intron structure of genes of fungi genomes]. Mol Biol (Mosk). 43(1):28–35. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19334523 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19334523)
- Jurka J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16(9):418–420. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10973072 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10973072)
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1–4):462–467. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16093699 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16093699)
- Juhasz A, Pfeiffer I, Keszthelyi A, Kucsera J, Vagvolgyi C, Hamari Z. 2008. Comparative analysis of the complete mitochondrial genomes of Aspergillus niger mtDNA type 1a and Aspergillus tubingensis mtDNA type 2b. FEMS Microbiol Lett. 281(1):51–57. doi 10.1111/j.1574-6968.2008.01077.x http://www.ncbi.nlm.nih.gov/pubmed/18318841 (http://www.ncbi.nlm.nih.gov/pubmed/18318841)
- Kanz C, Aldebert P, Althorpe N, Baker W, Baldwin A, Bates K, Browne P, van den Broek A, Castro M, Cochrane G, et al. 2005. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 33(Database issue):D29–33. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608199 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608199)
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414(6862):450–453. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11719806 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11719806)
- Kazazian HH, Jr. 2004. Mobile elements: drivers of genome evolution. Science 303(5664):1626–1632. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15016989 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15016989)
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423(6937):241–254. doi 10.1038/nature01644nature01644 [pii] http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12748633 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12748633)
- Kent WJ. 2002. BLAT – the BLAST-like alignment tool. Genome Res. 12(4):656–664. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11932250 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11932250)
- Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 11(5):345–355. doi 10.1038/nrg2776 http://www.ncbi.nlm.nih.gov/pubmed/20376054 (http://www.ncbi.nlm.nih.gov/pubmed/20376054)
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 47(9):736–741. doi S1087- 1845(10)00105-2 [pii]10.1016/j.fgb.2010.06.003 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20554054 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20554054)
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15144565 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15144565)
- Korf I, Flicek P, Duan D, Brent MR. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17 Suppl 1:S140–148. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11473003 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11473003)
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. doi 10.1006/jmbi.2000.4315 http://www.ncbi.nlm.nih.gov/pubmed/11152613 (http://www.ncbi.nlm.nih.gov/pubmed/11152613)
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. doi gkm160 [pii]10.1093/nar/gkm160 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17452365 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17452365)
- Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23(22):2639–2649. doi 23/22/2639 [pii]10.1101/gad.1837609 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19933153 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19933153)
- Lejeune E, Bayne EH, Allshire RC. 2011. On the Connection between RNAi and Heterochromatin at Centromeres. Cold Spring Harb Symp Quant Biol. doi sqb.2010.75.024 [pii]10.1101/sqb.2010.75.024 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21289046 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21289046)
- Lerat E. 2010. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity 104(6):520–533. doi hdy2009165 [pii]10.1038/hdy.2009.165 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19935826 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19935826)
- Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al. 2002. Apollo: a sequence annotation editor. Genome Biol. 3(12):RESEARCH0082. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12537571 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12537571)
- Li , W-H. 1997 . Molecular evolution , Sunderland, Mass : Sinauer Associates. Wen-Hsiung Li. ill .
- Li W-H, Gojobori T, Nei M. 1981. Pseudogenes as a paradigm of neutral evolution. Nature. 292(5820):237–239. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7254315 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7254315)
- Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. 2007. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17(12):1823–1836. doi 10.1101/gr.6679507 http://www.ncbi.nlm.nih.gov/pubmed/17989253 (http://www.ncbi.nlm.nih.gov/pubmed/17989253)
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9023104 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9023104)
- Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26(4):1107–1115. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9461475 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9461475)
- Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5(7):e1000549. doi 10.1371/journal.pgen.1000549 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19578406 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19578406)
- Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 464(7287):367–373. doi nature08850 [pii]10.1038/nature08850 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20237561 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20237561)
- Manning G, Plowman GD, Hunter T, Sudarsanam S. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 27(10):514–520. doi S0968000402021795 [pii] http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12368087 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12368087)
- Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA. 2003. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 13(4):544–557. doi 10.1101/gr.811003 http://www.ncbi.nlm.nih.gov/pubmed/12670996 (http://www.ncbi.nlm.nih.gov/pubmed/12670996)
- Majoros WH, Pertea M, Salzberg SL. 2004. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20(16):2878–2879. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15145805 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15145805)
- Majoros WH, Pertea M, Salzberg SL. 2005. Efficient implementation of a generalized pair hidden Markov model for comparative gene finding. Bioinformatics. 21(9):1782–1788. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15691859 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15691859)
- McGuire AM, Pearson MD, Neafsey DE, Galagan JE. 2008. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 9(3):R50. doi 10.1186/gb-2008-9-3-r50 http://www.ncbi.nlm.nih.gov/pubmed/18321378 (http://www.ncbi.nlm.nih.gov/pubmed/18321378)
- Mighell AJ, Smith NR, Robinson PA, Markham AF. 2000. Vertebrate pseudogenes. FEBS Lett. 468(2–3):109–114. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10692568 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10692568)
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3(12):RESEARCH0083. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12537572 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12537572)
- Mitrovich QM, Tuch BB, Guthrie C, Johnson AD. 2007. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 17(4):492–502. doi 10.1101/gr.6111907 http://www.ncbi.nlm.nih.gov/pubmed/17351132 (http://www.ncbi.nlm.nih.gov/pubmed/17351132)
- Mott R. 1997. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 13(4):477–478. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9283765 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9283765)
- Nemecek JC, Wuthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312(5773):583–588. doi 312/5773/583 [pii] 10.1126/science.1124105 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16645097 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16645097)
- Nemecek JC, Wuthrich M, Klein BS. 2007. Detection and measurement of two-component systems that control dimorphism and virulence in fungi. Methods Enzymol. 422:465–487. doi S0076-6879(06)22024-X [pii] 10.1016/S0076-6879(06)22024-X http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17628155 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17628155)
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. 2003. The microRNA world: small is mighty. Trends Biochem Sci. 28(10):534–540. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14559182 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14559182)
- Nicholson P, Muhlemann O. 2010. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochemical Soc Trans. 38(6):1615–1620. doi 10.1042/BST0381615 http://www.ncbi.nlm.nih.gov/pubmed/21118136 (http://www.ncbi.nlm.nih.gov/pubmed/21118136)
- Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. doi gkq1268 [pii] 10.1093/nar/gkq1268 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21306991 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21306991)
- Pachter L, Alexandersson M, Cawley S. 2002. Applications of generalized pair hidden Markov models to alignment and gene finding problems. J Comput Biol. 9(2):389–399. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12015888 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12015888)
- Park J, Park B, Jung K, Jang S, Yu K, Choi J, Kong S, Kim S, Kim H, Kim JF, et al. 2008. CFGP: a web-based, comparative fungal genomics platform. Nucleic Acids Res. 36(Database issue):D562–571. doi gkm758 [pii] 10.1093/nar/gkm758 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17947331 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17947331)
- Parra G, Blanco E, Guigo R. 2000. GeneID in Drosophila. Genome Res. 10(4):511–515. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10779490 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10779490)
- Parra G, Agarwal P, Abril JF, Wiehe T, Fickett JW, Guigo R. 2003. Comparative gene prediction in human and mouse. Genome Res. 13(1):108–117. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12529313 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12529313)
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. doi btm071 [pii] 10.1093/bioinformatics/btm071 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17332020 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17332020)
- Pierleoni A, Martelli PL, Casadio R. 2008. PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9:392. doi 1471-2105-9-392 [pii] 10.1186/1471-2105-9-392 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18811934 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18811934)
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21 Suppl 1:i351–358. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961478 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15961478)
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35(21):7188–7196. doi gkm864 [pii] 10.1093/nar/gkm864 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17947321 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17947321)
- Rawlings ND, Tolle DP, Barrett AJ. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32(Database issue):D160–164. doi 10.1093/nar/gkh071 32/suppl_1/D160 [pii] http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14681384 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14681384)
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332(6032):930–936. doi 10.1126/science.1203357 http://www.ncbi.nlm.nih.gov/pubmed/21511999 (http://www.ncbi.nlm.nih.gov/pubmed/21511999)
- Rossignol T, Lechat P, Cuomo C, Zeng Q, Moszer I, d'Enfert C. 2008. CandidaDB: a multi-genome database for Candida species and related Saccharomycotina. Nucleic Acids Res. 36(Database issue):D557–561. doi gkm1010 [pii] 10.1093/nar/gkm1010 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18039716 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18039716)
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11120685 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11120685)
- Salamov AA, Solovyev VV. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10(4):516–522. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10779491 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10779491)
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101(6):671–684. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10892653 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10892653)
- Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. 2006. WormBase: better software, richer content. Nucleic Acids Res. 34(Database issue):D475–478. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16381915 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16381915)
- Shapiro JA, von Sternberg R. 2005. Why repetitive DNA is essential to genome function. Biol Rev Camb Philos Soc. 80(2):227–250. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15921050 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15921050)
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19(10):1722–1731. Available fromDoi gr.087551.108 [pii]10.1101/gr.087551.108 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19717792. (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19717792.)
- Shelest E. 2008. Transcription factors in fungi. FEMS Microbiol Lett. 286(2):145–151. doi FML1293 [pii] 10.1111/j.1574-6968.2008.01293.x http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18789126 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18789126)
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15713233 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15713233)
- Smit A, Hubley R. RepeatModeler http://wwwrepeatmaskerorg/RepeatModelerhtml (http://wwwrepeatmaskerorg/RepeatModelerhtml)
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stuber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ, et al. 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330(6010):1543–1546. doi 330/6010/1543 [pii] 10.1126/science.1194573 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21148392 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21148392)
- Stajich JE. http://fungalgenomes.org/. http://fungalgenomesorg/. Available
- Stajich JE, Wilke SK, Ahren D, Au CH, Birren BW, Borodovsky M, Burns C, Canback B, Casselton LA, Cheng CK, et al. 2010. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA 107(26):11889–11894. doi 1003391107 [pii] 10.1073/pnas.1003391107 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20547848 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20547848)
- Stalder L, Muhlemann O. 2008. The meaning of nonsense. Trends Cell Biol. 18(7):315–321. doi 10.1016/j.tcb.2008.04.005 http://www.ncbi.nlm.nih.gov/pubmed/18524595 (http://www.ncbi.nlm.nih.gov/pubmed/18524595)
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. 2005. Function of alternative splicing. Gene 344:1–20. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15656968 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15656968)
- Stanke M, Waack S. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 Suppl 2:II215-II225. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14534192 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14534192)
- Stanke M, Schoffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16469098 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16469098)
- Stergiopoulos I, de Wit PJ. 2009. Fungal effector proteins. Annu Rev Phytopathol. 47:233–263. doi 10.1146/annurev.phyto.112408.132637 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19400631 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19400631)
- Szymanski M, Barciszewska MZ, Zywicki M, Barciszewski J. 2003. Noncoding RNA transcripts. J Appl Genet. 44(1):1–19. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12590177 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12590177)
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter 4:Unit 4 10. doi 10.1002/0471250953.bi0410s25 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19274634 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19274634)
- Tateno Y, Saitou N, Okubo K, Sugawara H, Gojobori T. 2005. DDBJ in collaboration with mass-sequencing teams on annotation. Nucleic Acids Res. 33(Database issue):D25–28. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608189 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15608189)
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi 10.1186/1471-2105-4-41 http://www.ncbi.nlm.nih.gov/pubmed/12969510 (http://www.ncbi.nlm.nih.gov/pubmed/12969510)
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18(12):1979–1990. doi gr.081612.108 [pii] 10.1101/gr.081612.108 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18757608 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18757608)
- Torriani SF, Goodwin SB, Kema GH, Pangilinan JL, McDonald BA. 2008. Intraspecific comparison and annotation of two complete mitochondrial genome sequences from the plant pathogenic fungus Mycosphaerella graminicola. Fungal Genet Biol. 45(5):628–637. doi 10.1016/j.fgb.2007.12.005 http://www.ncbi.nlm.nih.gov/pubmed/18226935 (http://www.ncbi.nlm.nih.gov/pubmed/18226935)
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. doi 10.1093/bioinformatics/btp120 http://www.ncbi.nlm.nih.gov/pubmed/19289445 (http://www.ncbi.nlm.nih.gov/pubmed/19289445)
- Usuka J, Zhu W, Brendel V. 2000. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics 16(3):203–211. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10869013 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10869013)
- Valach M, Farkas Z, Fricova D, Kovac J, Brejova B, Vinar T, Pfeiffer I, Kucsera J, Tomaska L, Lang BF, et al. 2011. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. doi 10.1093/nar/gkq1345 http://www.ncbi.nlm.nih.gov/pubmed/21266473 (http://www.ncbi.nlm.nih.gov/pubmed/21266473)
- van Baren MJ, Brent MR. 2006. Iterative gene prediction and pseudogene removal improves genome annotation. Genome Res. 16(5):678–685. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16651666 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16651666)
- Vlcek C, Marande W, Teijeiro S, Lukes J, Burger G. 2011. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 39(3):979–988. doi 10.1093/nar/gkq883 http://www.ncbi.nlm.nih.gov/pubmed/20935050 (http://www.ncbi.nlm.nih.gov/pubmed/20935050)
- Wang BB, Brendel V. 2006. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103(18):7175–7180. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16632598 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16632598)
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456(7221):470–476. doi 10.1038/nature07509 http://www.ncbi.nlm.nih.gov/pubmed/18978772 (http://www.ncbi.nlm.nih.gov/pubmed/18978772)
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. 2010. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 38(18):e178. doi 10.1093/nar/gkq622 http://www.ncbi.nlm.nih.gov/pubmed/20802226 (http://www.ncbi.nlm.nih.gov/pubmed/20802226)
- Wheelan SJ, Church DM, Ostell JM. 2001. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 11(11):1952–1957. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11691860 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11691860)
- Wiehe T, Gebauer-Jung S, Mitchell-Olds T, Guigo R. 2001. SGP-1: prediction and validation of homologous genes based on sequence alignments. Genome Res. 11(9):1574–1583. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11544202 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11544202)
- Woo PC, Zhen H, Cai JJ, Yu J, Lau SK, Wang J, Teng JL, Wong SS, Tse RH, Chen R, et al. 2003. The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett. 555(3):469–477. http://www.ncbi.nlm.nih.gov/pubmed/14675758 (http://www.ncbi.nlm.nih.gov/pubmed/14675758)
- Wortman JR, Haas BJ, Hannick LI, Smith RK, Jr., Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al. 2003. Annotation of the Arabidopsis genome. Plant Physiol. 132(2):461–468. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12805579 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12805579)
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. 2006. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34(Database issue):D187–191. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16381842 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16381842)
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9):1859–1875. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15728110 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15728110)
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881. doi 10.1093/bioinformatics/btq057 http://www.ncbi.nlm.nih.gov/pubmed/20147302 (http://www.ncbi.nlm.nih.gov/pubmed/20147302)
- Wu Y, Yang J, Yang F, Liu T, Leng W, Chu Y, Jin Q. 2009. Recent dermatophyte divergence revealed by comparative and phylogenetic analysis of mitochondrial genomes. BMC Genomics 10:238. doi 10.1186/1471-2164-10-238 http://www.ncbi.nlm.nih.gov/pubmed/19457268 (http://www.ncbi.nlm.nih.gov/pubmed/19457268)
- Xue C, Hsueh YP, Heitman J. 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 32(6):1010–1032. doi FMR131 [pii] 10.1111/j.1574-6976.2008.00131.x http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18811658 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18811658)
- Yang G, Hall TC. 2003. MAK, a computational tool kit for automated MITE analysis. Nucleic Acids Res. 31(13):3659–3665. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12824388 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12824388)
- Yeh RF, Lim LP, Burge CB. 2001. Computational inference of homologous gene structures in the human genome. Genome Res. 11(5):803–816. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11337476 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11337476)
- Zhang MQ. 2002. Computational prediction of eukaryotic protein-coding genes. Nat Rev Genet. 3(9):698–709. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12209144 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12209144)
- Zhang Z, Gerstein M. 2004. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev. 14(4):328–335. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15261647 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15261647)
- Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. 2006. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22(12):1437–1439. doi 10.1093/bioinformatics/btl116 http://www.ncbi.nlm.nih.gov/pubmed/16574694 (http://www.ncbi.nlm.nih.gov/pubmed/16574694)