Abstract
Muscodor kashayum (MycoBank no.: MB 803800; GenBank no.: KC481680) is a newly described endophytic fungus of a medicinal plant Aegle marmelos (Bael tree), growing in the tropical conserved rainforest in the Western Ghats of India. Muscodor kashayum possesses distinct morphological, molecular and physiological features from the earlier reported Muscodor species. The fungus forms characteristic rings of the ropy mycelium on potato dextrose agar medium. This sterile fungus is characterised by the presence of a pungent smell which is attributable to a blend of more than 23 volatile organic constituents, predominantly 3-cyclohexen-1-ol,1-(1,5-dimethyl-4-hexenyl)-4-methyl; 1,6-dioxacyclododecane-7,12-dione; 2,6-bis(1,1-dimethylethyl)-4-(1-oxopropyl) phenol; 2,4-di-tert-butylthiophenol and 4-octadecylmorpholine. In the in vitro anti-microbial assay using M. kashayum, growth of 75% of test fungi/yeasts and 72% of the test bacteria were completely inhibited. Therefore, M. Kashayum holds potential for future application to be used as a myco-fumigation agent.
Introduction
The Western Ghats of India is endowed with a rich biodiversity of more than 4700 different plant species (Rajeshkumar and Singh Citation2012). These plants harbour a suite of micro-organisms known as endophytes which do not exhibit any visible signs of their existence in the internal plant tissues (Bacon and White Citation2000; Strobel and Daisy Citation2003). Today it is well established that every plant possesses its own endophytic fungal community and this existence is ubiquitous (Firakova et al. Citation2007). These endophytic fungi produce an array of metabolites which may defend the host plant against different stress conditions such as pathogenic invasions and drought (Suwannarach et al. Citation2013). Muscodor is a genus of sterile endophytic fungi that has been first isolated from cinnamon tree in Honduras which has been phylogenetically placed in Xylariaceae based on its internal transcribed spacer (ITS) rRNA gene sequence. The genus is expanding with the discovery of more species which have been reported from different hosts, geographical regions and possessing distinct features based on scanning electron microscope (SEM) and phylogenetic studies. Morphologically, these endophytic fungi are differentiated on the basis of colony and mycelial characteristics due to lack of reproductive structures in any medium. However, each species exhibits a particular hyphal arrangement which has been used for establishing the cultural uniqueness (Strobel et al. Citation2001; Zhang et al. Citation2010). To date 12 species have been added to this genus based on ITS sequencing, volatile gas composition analysis and hyphal structures using SEM (Strobel et al. Citation2001; Suwannarach et al. Citation2013). The volatiles produced by Muscodor albus and other species synergistically induce lethal effects against plant and human pathogens (Strobel et al. Citation2001; Strobel and Daisy Citation2003). Thus, Muscodor species holds enormous potential to be used as a biocontrol agent with possible replacement of hazardous chemical fumigants such as methyl bromide, and therefore considered to be a promising source of anti-microbial compounds with eco- friendly mycofumigation potential (Strobel Citation2006; Mercier et al. Citation2007).
Aegle marmelos (Bael Tree) is an Indian plant with medicinal and religious importance. It grows in India, Sri Lanka, Nepal, South Africa, Pakistan, Myanmar, etc. Various parts of this plant have been used in the Ayurvedic as well as Unani medicinal preparations to treat diarrhoea, dysentery and dyspeptic symptoms. The plant parts are also reported to possess anti-diabetic, anti-fungal and anti-bacterial properties (Dhankar et al. Citation2011). Several endophytic fungi have been reported from plant parts of Aegle marmelos (Gond et al. Citation2007). However, this is the first report of Muscodor species #16AMLWLS obtained from the leaf tissue of Bael tree growing in Wayanad Wildlife Sanctuary, Kerala, India. The isolated fungus produced a unique set of volatile compounds which possess antagonistic activity and can further be used to identify the particular Muscodor sp. The strain, #16AMLWLS exhibits cultural, morphological, molecular and chemical characteristics that differ from previously reported Muscodor sp. namely M. albus, M. roseus, M. fengyangensis, M. sutura, etc. Based on its unique features, we conclude that #16 AMLWLS represents a new species of Muscodor for which the name Muscodor kashayum is proposed.
Materials and methods
Fungal isolation and preservation
Leafs and twigs of the medicinal plant, Aegle marmelos, were collected from the rainforest areas of Muthanga region of Wayanad Wildlife Sanctuary, Kerala, India, during July 2009. Plant samples were kept in sterile packets and stored at 4°C till further use. The fungal isolation was accomplished using M. albus CZ620 Woropong, Strobel and Hess, as screening tool as reported by Ezra et al. (Citation2004). Briefly, Potato Dextrose Agar (PDA) was poured into one quadrant of the four sectioned commercially available petri dishes. Subsequently, into that quadrant on each plate a 3 mm agar plug of actively growing M. albus CZ620 was placed while other quadrants of the petri dishes contained water agar (WA). The plates were then incubated at 24°C for 4 days for the production of VOCs (volatile organic compounds) production by M. albus. The plant samples were cut into segments (leaf, 5 mm ×5 mm; stem, 5 mm long) and surfaces were sterilised with 75% ethanol for 30 s, 2% sodium hypochlorite for 3 min and 95% ethanol under a laminar flow hood. The sterile plant segments were placed in other quadrants containing WA, thereby exposing them to the VOCs of M. albus arising in the plates. The fungi emerging out of the host tissue was aseptically subcultured onto a fresh PDA plate so as to obtain pure isolates, which were maintained on PDA slants supplemented with 10% glycerol. The fungus was preserved on sterilised grain (rye and wheat) and Whatmann paper stored at –70°C till further use. The fungus remained viable for 12–14 months (Ezra et al. Citation2004; Strobel et al. Citation2007; Mitchell et al. Citation2008).
Morphotaxonomy
Morphotaxonomic studies of the endophytic fungal isolate were done by mounting the culture in lactophenol cotton blue and then observing under Nikon Stereozoom microscope (Nikon SMZ 745T, Tokyo, Japan) coupled with Nikon Instruments Software (NIS) element D 3.2 software (Nikon) and Nikon Eclipse Compound microscope (E100, Kawasaki, Japan). Micrometry was carried out using Image J software (National Institutes of Health, Bethesda, MD, USA) with at least 30 observations per structure. Morphological and microscopic characters were studied on six different media comprising of PDA, MEA (Malt Extract Agar), CMA (Corn Meal Agar), CDA (Czapek Dox Agar), WA and BLA (Banana Leaf Agar). The colony texture, colour, colony growth rate, pigment and VOCs production along with its microscopic structures like hyphal characteristics and other cellular bodies were minutely observed and recorded (Guo et al. Citation1998; Ezra et al. Citation2004; Mitchell et al. Citation2008).
ITS-based phylogenetic analysis
For the genomic DNA extraction, about 0.1–0.2 g of cultured mycelia was scrapped off from the 3–4-day-old culture of #16 AMLWLS with a sterile inoculation loop and crushed to very fine powder in pestle and mortar using liquid nitrogen. Further, DNA extraction was done by using the Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA) following the manufacturer instructions.
Amplification of the ITS region of #16 AMLWLS was carried out using Muscodor-specific primers, M. albus F (5′GGGAGGCTACCCTATAGGGGATAC3′) and M. albus R (5′CAGGGGCCGGAACCACTACAGAGG3′) using a My Cycler TM thermal cycler (Bio Rad, Hercules, CA, USA ). The Polymerase Chain Reaction (PCR) mixture comprised 25 ng of extracted genomic DNA, 25 mM MgCl2, 2.5 mM dNTP, 10 pmol/µl of each primer, 1.5 U of Taq DNA polymerase in 10X Taq buffer while the PCR amplification conditions used have been previously described by Ezra et al. (Citation2009). The ITS rDNA PCR amplicon of approximately 500–600 bp was resolved onto 1.5% agarose gel and further purified by Wizard® SV Gel and PCR clean-up system kit (Promega). The purified ITS rDNA amplicons were sent for direct PCR sequencing to Xcleris Genomics, Xcleris labs, Gujarat, India. The final sequence was obtained by assembling the obtained sequences using Sequencher ver. 5 (www.genecodes.com) and was submitted in the GenBank under the accession No. KC481680.
The obtained sequence was then subjected to sequence similarity search by using BLAST software accessed form National Center for Biotechnology Information (NCBI) server. Sequences with highest similarity were aligned with the obtained ITS sequence of #16AMLWLS using Clustal W option in Molecular Evolutionary Genetics Analysis (MEGA) 5.0 (Tamura et al. Citation2011). The alignment was made uniform by manually correcting the aligned sequences. The alignment was exported to the MEGA format. The evolutionary distances were inferred by using maximum likelihood method based upon the Kimura-2-parameter model. Bootstrap replicates (1000) were taken into account to infer the bootstrap consensus tree for the representation of evolutionary history. Branches corresponding to partitions reproduced in less than 26% bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained by applying the neighbour joining method to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach. Gaps were treated as missing data.
Computation of genetic distances
The genetic relatedness of # 16AMLWLS (M. kashayum) with already reported species of Muscodor was deduced by the determination of pairwise distances implemented in MEGA 5.2 by using the p-distance. p-Distance is the proportion (p) of nucleotides sites at which the two sequences being compared are different. This value is obtained by dividing the number of nucleotide differences by the total number of nucleotides compared. Gaps were treated as missing characters and maximum composite likelihood was chosen as a model for the analysis (Tajima Citation1989).
The levels of DNA polymorphism such as number of variable sites (η), haplotypes, haplotype diversity, nucleotide diversity (π), evolutionary models were deduced with DNASp5 (Librado and Rozas Citation2009).
Scanning electron microscopy
Agar plugs of 10-day-old fungal samples were placed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) overnight at 4°C. Next day, they were washed twice with 0.1 M phosphate buffer at a 10 min interval. Thereafter, the samples were slowly dehydrated using acetone-graded series (10 min each at 30%, 40%, 50%, 60%, 70%, 80%, 95% and 100%). The samples were then brought to critical point drying and coated with gold palladium using a sputter coater. The images were then recorded in high vacuum mode using Zeiss Evo40 (Carl-Zeiss, Oberkochen, Germany) SEM with a magnification range in between 307 X and 2.02 KX at 15-kV extra high tension (Ezra et al. Citation2004; Kudalkar et al. Citation2012).
Volatile analysis of M. kashayum
The VOC mixture produced by a 10-day-old culture of isolate #16 AMLWLS was entrapped using a solid-phase micro-extraction (SPME) syringe having a stable flex fibre made up of 50/30 divinylbenzene/carboxen on polydimethylsiloxane (Supelco, Sigma Aldrich, St. Louis, MO, USA) following the method of Ezra et al. (Citation2004). The fibre was exposed for 45 min by placing the SPME syringe through a small bore made of using sterile needle over the headspace of culture in the petri dish. Subsequently, the fibre was injected for 30 s in the Shimadzu QP 2010 (Columbia, MD, USA) plus Gas chromatograph with TD-20 (Shimadzu, Columbia, Maryland, USA) thermal desorption system. An Restek column (diphenyl (95%) and dimethyl polysiloxane (5%)) with 30 m × 0.25 mm ID and 0.25 mm film diameter was used for separating the fungal volatiles. The column was programmed at 100°C for 2 min and the temperature was then raised to 250°C for 2 min and then finally to 300°C for 13 min. The carrier gas was helium and the initial column head pressure was 94.4 KPa. Data acquisition and processing was done on GCMS (Shimadzu, Columbia, Maryland, USA) solution software. The compounds obtained after gas chromatography/mass spectroscopy (GC/MS) analysis were then subtracted from the control plate consisting only PDA medium. The obtained compounds were then tentatively identified based on their high-quality matching (above 70% similarity) with database of National Institute of Standard and Technology for compounds (NIST05) and compared with all reported species of Muscodor to date (Ezra et al. Citation2004; Kudalkar et al. Citation2012).
Bioassay of VOCs produced by M. kashayum
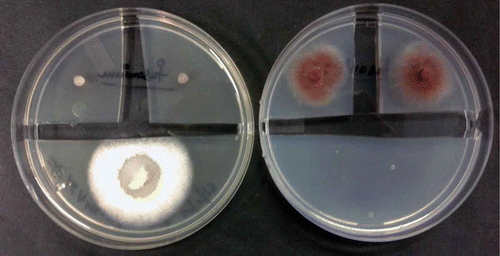
Anti-microbial activity of the volatiles produced by M. kashayum was tested by using a bioassay method employing 90-mm petri dishes with PDA (Ezra et al. Citation2004; Mitchell et al. Citation2008). Agar strips were removed to create quadrants as well as to prohibit movement of any diffusible inhibitory compound from the Muscodor sp. to the test micro-organism(s). Into one quadrant, an agar plug of actively growing M. kashayum was placed. The plates were sealed and incubated at 24 ± 2°C for 5 days for VOC production. Thereafter, individual test fungi were inoculated by placing a 3 mm plug of 7-day-old culture on the rest of the quadrants. Bacteria and yeasts were tested by individual streaking in other quadrants. Correspondingly, the control plates comprised only inoculated test bacteria or fungi were devoid of isolate #16 AMLWLS, allowing it to grow normally. Anti-microbial action of VOC was determined by monitoring the difference in the growth of micro-organisms in test and control plates. To check the inhibitory or killing effect of the VOCs after 3-day exposure, the culture plugs were transferred on fresh solid media (PDA, Yeast Extract Peptone Dextrose Agar or Muller Hinton Agar) to assess their viability (Mitchell et al. Citation2010). All the tests were performed in triplicates and values calculated as mean ± SD.
Results and discussion
Taxonomy
Muscodor kashayum Meshram, N. Kapoor & S. Saxena, sp. nov.
()
MycoBank no.: MB 803800 GenBank no.: KC481680
Etymology
The epithet ‘kashayum’ signifies a Hindi word ‘Kashay’, literally meaning pungent smell.
Diagnosis
Differ from the M. yucantanesis and M. equiseti by the absence of swollen hyphae. Differ from M. albus and M. vitigenus by the presence of coiling structures. Vary from M. cinnamomi, M. crispans and M. sutura by the absence of cauliflower-like or non-descript structures. Muscodor kashayum does not produce hairy aerial mycelium and wavy margins-like M. musae. Furthermore, it does not produce any pigment such as M. roseus, M. suthepensis, M. oryzae and M. sutura.
Type
India, Muthanga region of Wayanad Wildlife Sanctuary, Kerala, India, with its geographical coordinates from 11° 35′–11° 51′ N to 76° 02′–76° 27′ E as an endophytic fungi from leaves of Aegle marmelos, 12th July 2009, Sanjai Saxena (Holotype: AMLWLS #16; ex-type culture: National Fungal Culture Collection of India (NFCCI), NFCCI-2947)
Teleomorph
Unknown.
rDNA sequence ex-holotype: KC481680
Description
The fungus in nature is associated with Aegle marmelos and an Ascomycete with sterile mycelium. Fungal colonies are milky white and form a floccose mycelium pattern on PDA when grown in a 12-h photoperiod for 10 days at 26 ± 2°C (). Hyphae (2.14–5.41-µm thick) (see ) with coils (47.38–50.38 µm diameter) appear like a fused rope-like hyphal strands with branching at right angle (2.33–11.25 µm). Mycelium on PDA produced a growth rate of 4.5–4.7 ± 1.41 mm day−1 having a pungent smell. Spores and fruiting bodies did not develop under any tested conditions.
Figure 1. Morphological traits of Muscodor kashayum (#16 AMLWLS) after 10 days of growth in different medium. (a) Macromorphological culture in PDA medium ((b)–(d)); compound microscope micrographs of hyphae in PDA, MEA and CDA medium stained with lactophenol cotton blue ((e)–(g)); light microscope micrographs of coiling formation of fungal hyphae over CMA, WA and BLA medium; and (h) Scanning electron micrograph of the mycelial arrangement and morphology (Bar 10 µm).
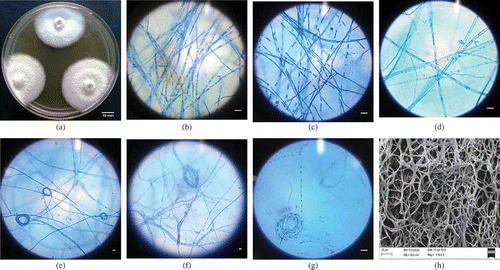
Fungus in MEA medium attains a colony of diameter of 64–67 mm. The isolate produces white coloured colonies with interwoven mycelium that intertwine to form long rope like structures which are (2.45-) 4.07 ± 1.07 (–6.37) μm wide (see ).
The fungus exhibits variation in morphology when grown in different media. On CMA, CDA, WA and BLA, the culture forms hyaline colonies which are slow growing with a mean colony diameter of 23, 36, 39 and 43 mm, respectively, after 10-day incubation. Microscopic studies reveal that hyphae are septate, branched at right angle, forming coiling structures either centrally or terminally. The average width of the mycelium in CMA, CDA, WA and BLA medium was 4.67 ± 1.23, 4.49 ± 0.97, 3.01 ± 0.68 and 3.03 ± 0.92 µm, whereas mean diameter of the coils are 40.82, 24.68, 15.05 and 44.89 µm, respectively (see –). The fungal culture produces VOCs with a strong pungent odour only in PDA, MEA and CDA medium. Volatile production was comparably higher in PDA than to MEA and CDA. Spores and fruiting bodies did not develop under any of the tested conditions.
Detailed morphological studies using SEM
The micrographs of M. kashayum prepared using SEM displayed typical characters of Muscodor species forming long, thick and branched mycelium similar to roots of a Banyan tree (see ). The mycelium was interconnected forming rings of different sizes. No fruiting bodies were observed.
Muscodor kashayum shows distinct morphological features as compared to other type strains of Muscodor namely M. crispans and M. cinnamomi, which exhibit a cauliflower-like sterile structure and possess a ropy-coiled mycelia. Muscodor yucatanensis has a ropy structure with swollen hyphae whereas M. albus only exhibits a ropy mycelium without any fruiting bodies, spores or sterile structures hence different forms of Muscodor kashayum. Muscodor sutura forms knitting pattern mycelia over the PDA plate making it remarkably dissimilar from M. kashayum.
ITS-based phylogenetic analysis of M. kashayum
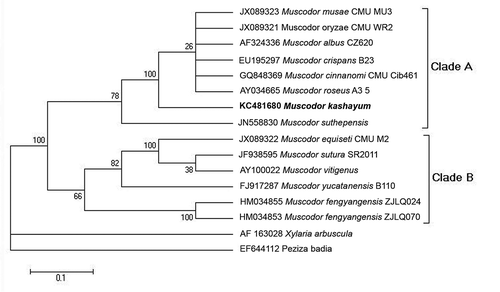
To ascertain the evolutionary relationship of the putative taxon within the Muscodor lineage, ITS1-5.8S-ITS2 sequence of #16 AMLWLS was subjected to BLAST similarity search. The result showed the 99% sequence identity with M. albus CZ620, M. oryzae, M. musae, M. cinnamomi followed by 95% sequence similarity with M. vitigenus and M. sutura. It exhibited 88% sequence identity with M. yucatanensis. The phylogenetic map was constructed by using all 12 reported species of Muscodor. Xylaria arbuscula and Peziza badia were taken as outgroup.
Maximum likelihood analysis of ITS1-5.8S-ITS2 region generated a well-resolved phylogram which shows two highly supported Clades A and B (see ). Clade A clustered M. musae, M. oryzae, M. cinnanomi, M. albus CZ620, M. crispans, M. roseus with M. kashayum which confirmed its placement basal to this clade with a significantly high bootstrap support value. M. suthepensis was further emerged as basal sister to this clade. The second Clade B comprised M. sutura, M. vitigenus, M. equiseti, M. fengyanensis and M. yucatanensis.
Figure 2. Phylogenetic relationships among Muscodor spp. Phylogenetic reconstruction based on the ITS rDNA gene sequences, applying maximum likelihood as optimality criterion.
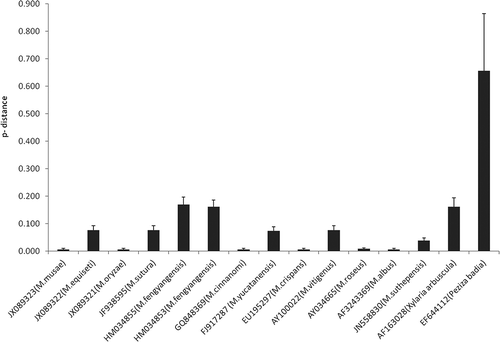
Pairwise distance proportion of nucleotide sites of the ITS region sequence comparisons between all the known species of Muscodor sp. and M. kashayum indicates that M. kashayum is different from other species of Muscodor (see and ).
Table 1. p-Distance of nucleotide sites among the ITS sequences compared between Muscodor Kashayum and 12 species of Muscodor.
Figure 3. Phylogenetic distance of Muscodor kashayum to congeneric species. Shown are p-distance values calculated by MEGA5.0.
DNA analysis can provide insights into the significant evolutionary factors acting on the population and species and can be employed to compare alternative evolutionary scenarios. The comparative analysis of within-species DNA polymorphism and between-species variation is noticeably an effective approach to understand the evolutionary process and obtain insights into the functional significance of genomic regions.
The number of polymorphic or segregating sites (η), nucleotide diversity (π) and number of haplotypes of ITS region are shown in . Fu & Li’s (Citation1993) D* and F* statistics, and Tajimas test (Tajima Citation1989) revealed that ITS region locus evolved neutral and were thus suitable for the phylogenetic analysis. Muscodor species were grouped into eight haplotypes based on ITS region locus. Furthermore, a 12% nucleotide variation was exhibited within this locus which is far greater than 2.7% as observed in the case of five strains of M. fengyangensis (Zhang et al. Citation2010). This variation was greater than 3% which is considered as a benchmark for intra-specific variation in fungal systems (Nilsson et al. Citation2008) (see ), thereby indicating M. kashayum to be a new species of the genus Muscodor. The above phylogenetic analyses prove that M. kashayum is a new member of monophyletic lineage of Muscodor.
Table 2. Nucleotide properties of the ITS region of the Muscodor kashayum and other species.
Volatile analysis of M. kashayum
Muscodor kashayum produces a mixture of 23 volatile compounds which were identified by comparing the GC/MS spectra with the NIST database (see ). Of all the volatile compounds produced, 3-cyclohexen-1-ol, 1-(1,5-dimethyl-4-hexenyl)-4-methyl- (synonymn : beta-Bisabolol) is the most abundant with percentage peak area of 20.67. Other major VOC produced by M. kashayum are 2, 6-bis-(1,1-dimethylethyl)-4-(1-oxopropyl) phenol, 1,6-dioxacyclododecane-7,12-dione, 2, 3-dihydro-1,1-dimethyl-6-tert-butyl-1H-indene-4-acetic acid, 2,4-di-tert-butylthiophenol and 4-octadecylmorpholine. It also produces some unknown volatile moieties that cannot be identified on the basis of NIST data. Muscodor kashayum possesses entirely different gas chemistry from the earlier reported Muscodor species which predominantly produces esters of 2-methylpropanoic acid, azuelene, naphthalene derivatives and thujopsene (Kudalkar et al. Citation2012). The volatiles produced by M. kashayum are unique and belonging to ketones, amines, phenols and alcohols, and have not been previously reported by any other Muscodor species.
Table 3. Composition of the volatiles produced by Muscodor kashayum after 10-day incubation at 24 ± 2°C on Potato Dextrose Agar (PDA) entrapped using a solid-phase micro-extraction (SPME) fibre and GC/MS analysis.
Bioassay of VOCs produced by M. kashayum
The preliminary evaluation of the VOCs produced by M. kashayum exhibited a potent and broad anti-microbial activity against a test panel of micro-organisms comprising bacteria, yeast and fungi. After the exposure of 5-day-old culture for 72 h, the VOC produced by M. kashayum completely inhibited 26 out of 36 tested pathogens. A total of 15 among the 20 tested fungi were completely killed whereas the other pathogenic fungi such as Aspergillus flavus and Alternaria alternata showed a 20–60% inhibition. Muscodor albus exhibited 100% survival against volatiles produced by M. kashayum. All the isolates of Candida albicans were susceptible while Saccharomyces cerevisiae was resistant to volatiles. The VOC exhibited a broad-spectrum anti-microbial activity against human bacterial pathogens namely Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa and completely inhibited their growth after the exposure for 48–72 h. However, Bacillus subtilis, Staphylococcus epidermidis and Salmonella typhi remained unaffected by the VOC (see and ).
Table 4. Growth inhibition of reference organisms by volatile compounds produced by Muscodor kashayum after 3-day exposure.
Conclusion
Tropical ecosystems possess highest diversity of the fungal species than any other environment. The Western Ghats of India is a repository of nature’s hidden wealth. It shelters numerous fungal species that live in a symbiotic relationship with the host plant which are commonly known as endophytes. Muscodor is basically a genus of sterile endophytic fungi which are associated with different plant families worldwide producing characteristic VOCs, which are generally used as signatures of their identification apart from anti-microbial potential. Thus various ecological niches round the globe have been explored over the last two decades to unveil whether other species or biotypes of the same organism exists in nature or not. The present study is the first to report a novel Muscodor species from Aegle marmelos. It differed from the previously reported Muscodor species on the basis of its morphological characters, ITS-rDNA sequence and VOC profile. The striking feature of this fungus was its unique gas chemistry and its broad spectrum of anti-microbial activity. Thus, further studies are warranted to determine its potential as a biological control agent or as a preservative for enhancing the shelf life of harvested fruits and vegetables. Apart from this, studies on plant–microbe interaction will add to our current knowledge of micro-organisms associated with plants. The life cycle of Muscodor still remains an untold story.
Acknowledgements
We thank Department of Biotechnology, National Biodiversity Development Board, for sponsoring the project no. BT/PR/10083/NDB/52/95/2007. We also acknowledge Dr. Gary Strobel, Montana State University, Bozeman, USA, for providing Muscodor albus CZ620-type strain and plant pathogenic fungi and for constructive discussions. We are also thankful to Dr. Marc-Andre Lachance, University of Western Ontario, Canada, for constructive discussions on phylogenetic analysis. We also gratefully acknowledge Dr. Naresh M. Meshram and Ms. Salam Rita Devi from Division of Entomology, IARI, PUSA, New Delhi, and Dr. Ruchita Paland Shri Ajay Kumar from ARIF, JNU, New Delhi, for SEM analysis and GC/MS analysis, respectively.
References
- Bacon CW, White JF. 2000. Microbial endophytes. New York (NY): Marcel Dekker.
- Dhankar S, Ruhil S, Balhara M, Dhankar S, Chhillar AK. 2011. Aegle marmelos (Linn.) Correa: a potential source of phytomedicine. J Med Plants Res. 5(9):1497–1507.
- Ezra D, Hess WM, Strobel GA. 2004. New endophytic isolates of Muscodor albus, a volatile – antibiotic-producing fungus. Microbiology. 12:4023–4031.
- Ezra D, Skovorodnikova J, Kroitor-Keren T, Denisov Y, Liarzi O. 2009. Development of methods for detection and Agrobacterium-mediated transformation of the sterile, endophytic fungus Muscodor albus. Biocontrol Sci Technol. 20(1):83–97.
- Firakova S, Sturdikova M, Muckova M. 2007. Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia. 62:251–257.
- Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics. 133:693–709.
- Gond SK, Verma VC, Kumar A, Kumar V, Kharwar RN. 2007. Study of endophytic fungal community from different parts of Aegle marmelos Correa (Rutaceae) from Varanasi (India). World J Microbiol Biol. 23(10):1371–1375.
- Guo LD, Hyde KD, Liew ECY. 1998. A method to promote sporulation in palm endophytic fungi. Fung Divers. 1:109–113.
- Kudalkar P, Strobel G, Hasan SRU, Geary G, Sears J. 2012. Muscodor sutura, a novel endophytic fungus with volatile antibiotic activities. Mycoscience. 53:319–325.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Mercier J, Santamaria JIJ, Guerra PT. 2007. Development of the volatile producing fungus Muscodor albus Worapong, Strobel, and Hess as a novel antimicrobial bio-fumigant. Rev Mex Fitopatol. 25:173–179.
- Mitchell A, Strobel G, Hess W, Vargas P, Ezra D. 2008. Muscodor crispans, a novel endophyte from Ananas ananassoides in the Bolivian Amazon. Fung Divers. 31: 37–43.
- Mitchell AM, Strobel GA, Moore E, Robinson R, Sears J. 2010. Volatile antimicrobials from Muscodor crispans, a novel endophytic fungus. Microbiology. 156:270–277.
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. 2008. Intraspecific ITS Variability in the Kingdom Fungias expressed in the International Sequence Databases and its implications for molecular species identification. Evol Bioinform Online. 4:193–201.
- Rajeshkumar KC, Singh SK. 2012. Manoharachariella indica sp. nov. from the Western Ghats, India. Mycotaxon. 120:43–48.
- Strobel G. 2006. Muscodor albus and its biological promise. J Ind Microbiol Biotechnol. 33:514–522.
- Strobel G, Daisy B. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 4:491–502.
- Strobel GA, Dirske E, Sears J, Markworth C. 2001. Volatile antimicrobials from Muscodor albus, a novel endophytic fungus. Microbiology. 147:2943–295.
- Strobel GA, Katreena K, Hess WM, Sears J, Ezra D, Vargas PN. 2007. Muscodor albus E-6, an endophyte of Guazuma ulmifolia making volatile antibiotics: isolation, characterization and experimental establishment in host plant. Microbiology. 153:2613–2620.
- Suwannarach N, Kumla J, Bussaban B, Hyde KD, Matsui K, Lumyong S. 2013. Molecular and morphological evidence support four new species in the genus Muscodor from northern Thailand. Ann Microbiol. doi:10.1007/s13213-012-0593-6
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–59.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Zhang CL, Wang GP, Mao LJ, Komon- Zelazowska M, Yuan ZL, Lin FC, Druzhinina IS, Kubicek CP. 2010. Muscodor fengyangensis sp. nov. from south east China: morphology, physiology and production of volatile compounds. Fungal Biol. 114:797–808.