ABSTRACT
Oligotrophs are microorganisms that can grow in environments where concentrations of nutrients are low or even absent. Caves are typical oligotrophic environments distinctly characterised by constant low temperature, high humidity, scarcity of organic matter and darkness, which encompass a high diversity of fungi. In our investigation of microorganisms from carbonate caves in China, 169 strains belonging to at least 84 taxa were isolated using oligotrophic carbon free silica gel medium (SGM). Cephalotrichum appeared to be one of the dominant genera. Further morphological comparisons and molecular phylogenetic analyses using DNA sequences of four loci (LSU, ITS, TUB2 and EF-1α) revealed that the 30 strains of Cephalotrichum represent three new species, which are described and named C. guizhouense, C. laeve and C. oligotriphicum. This study also significantly improved our understanding on fungi being able to grow on carbon free medium, with the known species increased from 18 to 99.
Introduction
Oligotrophs are microorganisms that can grow in environments where concentrations of nutrients are low or even absent (Wainwright Citation1993). Soil of extreme environments, ocean and polar environments have been universally regarded as low nutrient habitats, from which a variety of oligotrophic fungi have been isolated (Gundersen et al. Citation1976; Hiroyuki and Tsutomu Citation1983; Bergero et al. Citation1999; Godinho et al. Citation2015). Oligotrophic habitats are generally regarded with a nutrient flux from 1–15 mg of carbon per litre (Poindexter Citation1981), and medium containing a carbon concentration of 10 mg per litre was widely suggested to cultivate oligotrophic fungi (Martin and MacLeod Citation1984; Parkinson et al. Citation1989; Wainwright et al. Citation1997). Since agar contains available carbon, silica gel is more preferable for the isolation of oligocarbotrophic fungi for laboratory studies (Payton et al. Citation1976; Wainwright and Grayston Citation1988). Up to now, a wide range of fungi have been isolated from oligotrophic habitats, while only a few of them were confirmed possessing the ability for growth on carbon free Silica Gel Medium (SGM) (Mirocha and DeVay Citation1971; Wainwright and Grayston Citation1988; Parkinson et al. Citation1989, Citation1990; Wainwright Citation1993; Wainwright and Al-Talhi Citation1999; Connell and Staudigel Citation2013; Liu et al. Citation2013; Godinho et al. Citation2015) ().
Table 1. Species able to grow on carbon free silica gel medium (SGM).
Caves have been generally regarded as typical oligotrophic environments (Bastian et al. Citation2009), distinctly characterised by constantly low temperature, high humidity, scarcity of organic matter and darkness (Gabriel and Northup Citation2013). However, studies on oligotrophic fungi from caves are rare. The objective of this study was to verify oligotrophic fungi in a cave with ability of growing on carbon free SGM and describe several new oligotrophic Cephalotrichum species by means of morphological examinations and multi-locus phylogenetic analysis.
Materials and methods
Fungal isolation and growth on carbon free medium
Fifteen air samples, 25 limestone samples, 6 water samples, and 14 soil samples were collected from a carbonate cave (28°13ʹ819ʹʹ N, 107°18ʹ041ʹʹ E; about 750 m deep; 19°C) located at the Shuanghe National Geographic Park, Guizhou Province, China. From the entrance of the cave, each sampling site was approximately 200 m distant from the next. Collections of air samples followed the Koch sedimentation method described by Borda et al. (Citation2004) and Kuzmina et al. (Citation2012). Three Petri dishes that contained 2% potato-dextrose agar (PDA, Difco) were exposed to the atmosphere in the cave for 15 min at each sampling site, then sealed with parafilm and placed in zip-locked plastic bags. Limestones of the cave wall were collected following the method of Ruibal et al. (Citation2005). Five pieces of limestone in different orientations were collected at each site. Seeping, stream and pool water was collected (10 ml) and kept in 15 ml sterile centrifuge tubes. Ten grams of soil samples were collected at shallow depth (1–5 cm) after removing surface layer (ca. 1 cm). Water and soil samples were placed onto PDA (potato dextrose agar) medium following the dilution plate method (a series of concentrations, i.e. 10–1, 10–2, 10–3, 10–4, 10–5 and 10–6) (Zhang et al. Citation2015).
All isolates were cultured on 1/2000 strength PDA (Liu et al. Citation2013) and then inoculated on carbon free silica gel medium (SGM) to screen for oligotrophic strains at room temperature (approximately 20–25°C). SGM was prepared from the following three solutions: (a) mineral salts solution consisting of KH2PO4 1.0 g, KCl 0.5 g, MgSO4.7H2O 0.5 g and FeSO4.7H2O 0.01 g in 1 L ultrapure water (UPW); (b) orthophosphoric acid, 20 mL in 100 mL of UPW; (c) silicic acid 10 g and KOH 7 g added to 100 mL of UPW. Gels were prepared by mixing 10 mL of solution (a) with 10 mL of solution (c) and 2 mL of solution (b) into an autoclaved plastic Petri dish (9 cm) (Wainwright and Al-Talhi Citation1999). All glassware used in the preparation of silica gel medium were washed with chromic acid and then rinsed with ultrapure water (Parkinson et al. Citation1989).
Type specimens of the novel species were deposited in Mycological Herbarium of Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), with the ex-type living cultures deposited in China General Microbiological Culture Collection Center (CGMCC) and LC culture collection (personal collection of Lei Cai housed in the Institute of Microbiology, Chinese Academy of Sciences).
Morphological observation of cephalotrichum isolates
Isolates of Cephalotrichum were incubated on SGM, oatmeal agar (OA, BD, France), malt extract agar (MEA, BD, France) and potato dextrose agar (PDA, BD, France) at different temperatures (0–40°C at intervals of 5°C, as well as 37°C) and examined at 7, 14 and 28 d to determine colony growth rates (Sandoval-Denis et al. Citation2016b). Measurements and photographs of colony and micromorphological descriptions were made according to methods described by Sandoval-Denis et al. (Citation2016b), with colours assessed according to the colour chart of Rayner (Citation1970). Observations were performed with a Leica M125 dissecting microscope and a Zeiss Axio Imager A2 compound microscope under differential interference contrast (DIC) illumination.
DNA extraction and PCR
Genomic DNA was extracted from mycelia taken from PDA cultures after 7 days following the protocol of Cubero et al. (Citation1999). Primer ITS1/ITS4 (White et al. Citation1990) was used to amplify internal transcribed spacer regions 1 and 2 including 5.8S nrDNA (ITS) for all oligotrophic fungi. In addition, for Cephalotrichum, the primer pairs LR0R/LR5 (Vilgalys and Hester Citation1990; Rehner and Samuels Citation1994), Bt2a/Bt2b (Glass and Donaldson Citation1995) and EF 1-983F/EF 1-2218R (Rehner and Buckley Citation2005) were used to amplify partial regions of 28S large subunit of the nrRNA (LSU), β-tubulin (TUB2) and translation elongation factor 1α (EF-1α), respectively. The PCR amplifications were performed following the methods of Sandoval-Denis et al. (Citation2016a). DNA sequencing was conducted by the Biomed Genetics Company (Beijing, China).
Molecular identification and phylogenetic analysis
Sequences from forward and reverse primers were assembled to obtain a consensus sequence with MEGA v. 6.0 (Tamura et al. Citation2013). All strains were megablast searched in NCBI and assigned to potential genera and species. For Cephalotrichum, the obtained sequences and related sequences downloaded from GenBank () were aligned with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html; Katoh and Standley Citation2013). Ambiguous regions were excluded from the analyses and gaps were treated as missing data. A 70% neighbour-joining (NJ) reciprocal bootstrap method with maximum-likelihood distance was applied to check the congruence of the individual loci in the multi-locus dataset (Mason-Gamer and Kellogg Citation1996). Phylogenetic analysis of four genes combined datasets was conducted.
Table 2. Isolates used in this study and their GenBank accession numbers.
The jModelTest v. 2.1.4 (Posada Citation2008) was used to determine the best nucleotide substitution model settings for each locus. The Bayesian analyses of the combined four-locus dataset and individual locus data were performed with MrBayes v. 3.2.1 (Ronquist et al. Citation2012). The Markov Chain Monte Carlo sampling (MCMC) analysis of four chains started in parallel from a random tree topology. The number of generations was set at 10 million and the run was stopped automatically when the average standard deviation of split frequencies fell below 0.01. Trees were saved each 1 000 generations. Burn-in was set at 25% after which the likelihood values were stationary and the remaining trees were used to calculate posterior probabilities. Maximum-likelihood analyses including 1 000 bootstrap replicates were conducted using RAxML v. 7.2.6 (Stamatakis and Alachiotis Citation2010). A general time reversible model (GTR) was applied with a gamma-distributed rate variation.
Results
A total of 510 strains were isolated from air, limestone, water and soil using PDA medium. Among them, only 169 strains were determined as oligocarbotrophic fungi through their cultivation on SGM. A preliminary identification based on ITS megablast searches in GenBank assigned these isolates to 84 taxa in 51 genera, 28 families and 20 orders. Species able to grow on carbon free medium, including those previously known and those determined in this study, are summarised in . The result demonstrated that oligotrophic fungi are of multiple evolutionarily origins. The most common genera obtained in this study include Cephalotrichum (3 species, 30 strains), Plectosphaerella (2 species, 16 strains), Clonostachys (1 species, 14 strains), Cladosporium (8 species, 13 strains) and Fusarium (6 species, 10 strains) (). The most common species include Plectosphaerella cucumerina, Clonostachys rosea, Cephalotrichum oligotriphicum and C. guizhouense (). For the substrates of isolation, 85 oligotrophic strains from limestones belong to 41 species; 53 strains from air belong to 41 species; 25 strains from soil belong to 17 species; and 6 strains from water belong to 6 species ().
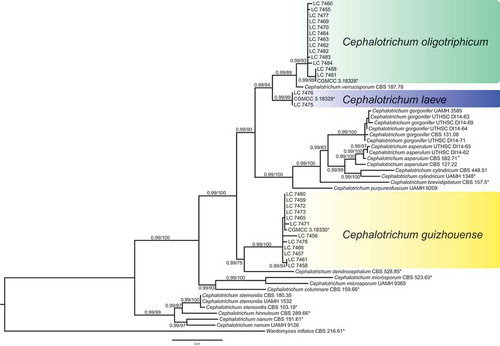
The multi-locus dataset of Cephalotrichum used for phylogenetic analysis included 55 in-group strains, including 30 oligotrophic strains obtained from the present study, and Wardomyces inflatus CBS 216.61 as an out-group. The dataset comprises 2708 characters including gaps (805 characters for LSU, 498 for ITS, 498 for TUB2 and 895 for EF-1α). TIM3 + I + G was selected as the best-fit model for the ITS dataset, while TrN+I for LSU, TrN+G for TUB2 and TIM2 + I + G for EF-1α were also selected. The phylogenetic trees obtained from Bayesian inference and RAxML were similar in topology. The ML consensus tree with Bayesian posterior probabilities (BPP) and RAxML bootstrap support (MLBS) values is shown in . Thirty strains of Cephalotrichum from caves clustered in three distinct clades with high bootstrap supports.
Figure 1. Phylogenetic tree inferred from a Maximum likelihood analysis based on a concatenated alignment of LSU, ITS, TUB2 and EF-1α sequences. The BPP and MLBS are given at the nodes (BPP/MLBS). Ex-type strains are marked by asterisks (*). The tree is rooted with Wardomyces inflatus (CBS 216.61).
Taxonomy
Cephalotrichum guizhouense J.R. Jiang, L. Cai & F. Liu, sp. nov.
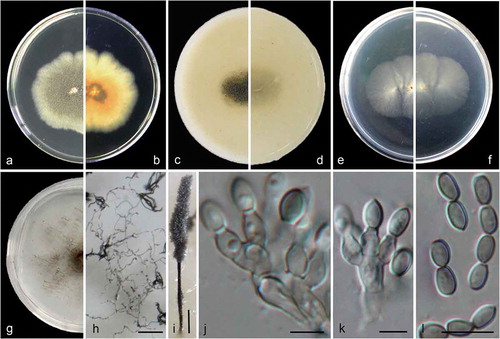
Figure 2. Cephalotrichum guizhouense (ex-holotype CGMCC 3.18330). (a–b). Colony on PDA (front and reverse). (c–d). Colony on OA (front and reverse). (e–f). Colony on MEA (front and reverse). (g). Colony on SGM (front). (h). Hyphae and conidia chains on the surface of SGM. (i). Synnema. (j–k). Conidiophores and conidiogenous cells. (l). Conidia. Scale bars: H = 100 μm; I = 200 μm; J–K = 5 μm; L = 10 μm.
Fungal names: FN 570479.
Etymology: named after its distribution: China, Guizhou province.
Culture characteristics
Colonies on PDA 40–45 mm diam in 14 d at 25°C, flat, lobate, felty, grey-olivaceous to greenish grey, with pale gray regular margin; reverse pale brown to faint yellow from centre to margin. Colonies on OA 35–40 mm diam in 14 d at 25°C, margin regular, flat, velvety with scarce white aerial mycelia, olive-grey, synnemata abundant, more or less powdery; reverse buff, pale brown near the centre. Colonies on MEA 30–35 mm diam in 14 d at 25°C, flat, velvety with scarce aerial mycelia, front and reverse white. Colonies on carbon free SGM growing more slowly compared to that on PDA, OA and MEA, aerial mycelia scarce but forming hyphae networks, with brown and sparse synnemata in one month.
Hyphae subhyaline, septate, pale brown, smooth- and thin-walled, 1.5–2.5 μm wide. Conidiophores unbranched or slightly branched, often in groups of 2–3 annellides on basal cells, pale brown, smooth- and thin-walled. Synnemata 800–1500 μm high, stipes dark brown, 15–28 μm wide, conidial heads olive-grey, ellipsoidal to clavate; setae absent. Annellides ampulliform to doliiform, 4.5–7.5 × 2.5–4 μm, subhyaline, pale brown, smooth- and thin-walled. Conidia ellipsoidal, ovoid, obovoid, 4.5–6.5 × 3–4 μm(), with truncate base and rounded or slightly acute apex, pale brown to brown, smooth- and thin-walled, arranged in long chains.
Cardinal temperatures for growth: optimum 25–30°C, maximum 35°C, minimum 5°C.
Specimens examined
China, Guizhou Province, Suiyang, the Shuanghe National Geographic Park, from cave air, 8 May 2015, Zhi-Feng Zhang, HMAS 247177 (holotype designated here), ex-holotype living culture CGMCC 3.18330 (= LC 7485); from cave limestones, 8 May 2015, Jia-Rui Jiang and Xin Zhou, LC 7456; ibid. LC 7457; ibid. LC 7458; ibid. LC 7459; ibid. LC 7461; ibid. LC 7465; ibid. LC 7466; ibid. LC 7471; ibid. LC 7472; ibid. LC 7473; ibid. LC 7478; ibid. LC 7480.
Notes
Isolates of C. guizhouense formed a well-supported clade distinct from its most closely related species C. dendrocephalum (). Morphologically C. guizhouense is distinct from the latter in the absence of undulating setae and the number of annellides (2–3 vs. 4(–6)) (Udagawa et al. Citation1985).
Cephalotrichum laeve J.R. Jiang, L. Cai & F Liu, sp. nov.
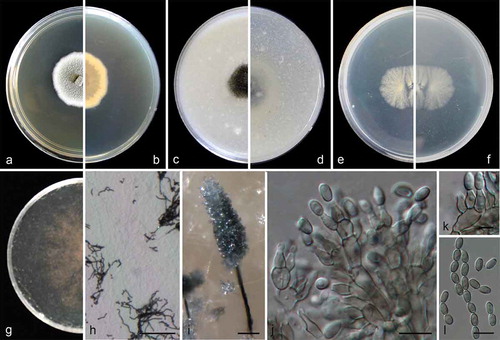
Figure 3. Cephalotrichum laeve (ex-holotype CGMCC 3.18329). (a–b). Colony on PDA (front and reverse). (c–d). Colony on OA (front and reverse). (e–f). Colony on MEA (front and reverse). (g). Colony on SGM (front). (h). Hyphae and conidia chains on the surface of SGM. (i). Synnema. (j). Detail of the apical portion of synnema. (k). Conidiogenous cells. (l). Conidia. Scale bars: H–J = 100 μm; K = 5 μm; L = 10 μm.
Fungal names: FN 570480.
Etymology: named after its smooth conidia.
Culture characteristics
Colonies on PDA 40–45 mm diam in 14 d at 25°C, flat, felty to floccose, pale olivaceous near the centre, with white regular margin; reverse pale brown near the centre, white near the margin. Colonies on OA 45–50 mm diam in 14 d at 25°C, margin regular, flat, velvety with scarce white aerial mycelia, leaden, synnemata abundant; reverse pale yellow, buff near the margin. Colonies on MEA 25–30 mm diam in 14 d at 25°C, flat, velvety with scarce aerial mycelia, front and reverse white. Colonies on carbon free SGM growing more slowly compared to that on PDA, OA and MEA, aerial mycelia scarce but forming hyphae networks, with pale brown and sparse synnemata in one month. Hyphae septate, hyaline to pale brown, smooth- and thin-walled, 2–3.5 μm wide. Conidiophores unbranched or branched, in groups of 3–5 annellides on basal cells, pale brown, smooth- and thin-walled, usually forming synnemata. Synnemata 600–1000 μm high, stipes brown, 10–28 μm wide, conidial heads olive-grey, ellipsoidal; setae absent. Annellides ampulliform, 5.5–8 × 3–4 μm, subhyaline, pale brown, smooth- and thin-walled. Conidia ellipsoidal to ovoid, 5.5–7 × 3–4 μm (), with truncate base and rounded or slightly acute apex, pale brown, smooth- and thin-walled, arranged in long chains.
Cardinal temperatures for growth: optimum 15–25°C, maximum 37°C, minimum 5°C.
Specimens examined
China, Guizhou, Suiyang, the Shuanghe National Geographic Park, from cave limestone, 8 May 2015, Zhi-Feng Zhang, Jia-Rui Jiang and Xin Zhou, HMAS 247178 (holotype designated here), ex-holotype living culture CGMCC 3.18329 (= LC 7474); ibid. LC 7475; ibid. LC 7476.
Notes
Cephalotrichum laeve showed close phylogenetic relationships with C. oligotriphicum (2 bp differences in LSU; 7 bp in ITS; 19 bp in EF-1α; 1 bp in TUB2) and C. verrucisporum (2 bp differences in LSU; 7 bp in ITS; 14 bp in EF-1α; 7 bp in TUB2) (). In morphology, C. laeve differs from C. oligotriphicum in producing broadly ellipsoidal (vs. cylindrical) conidiophores and in groups of 3–5 (vs. 2–4) annellides; from C. verrucisporum in producing smooth (vs. verrucouse) conidia (Jiang and Zhang Citation2008). The colony of C. laeve is felty to floccose with white regular margin on PDA, while that of C. oligotriphicum is velvety to felty with white crenate to fimbriate margin. The maximum growth temperature of C. laeve is 37°C, whereas that for C. oligotriphicum and C. verrucisporum are 30°C (Sandoval-Denis et al. Citation2016b).
Cephalotrichum oligotriphicum J.R. Jiang, L. Cai & F. Liu, sp. nov.
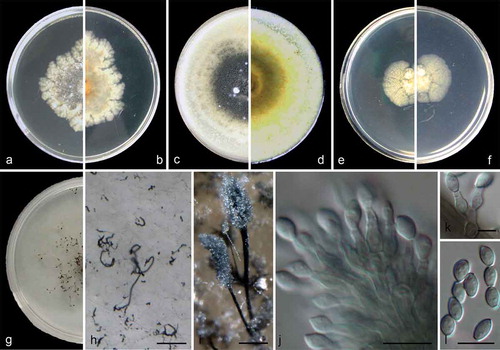
Figure 4. Cephalotrichum oligotriphicum (ex-holotype CGMCC 3.18328). (a–b). Colony on PDA (front and reverse). (c–d). Colony on OA (front and reverse). (e–f). Colony on MEA (front and reverse). (g). Colony on SGM (front). (h). Hyphae and conidia chains on the surface of SGM. (i). Synnemata. (j–k). Conidiophores and conidiogenous cells. (l). Conidia. Scale bars: H–I = 100 μm; J, L = 10 μm; K = 5 μm.
Fungal names: FN 570481.
Etymology: referred to the oligotrophic substrate of the fungus.
Culture characteristics
Colonies on PDA 35–40 mm diam in 14 d at 25°C, flat, crenate to fimbriate, velvety to felty, pale grey near the centre, with white crenate margin; reverse pale amber to white from centre to margin. Colonies on OA 45–50 mm diam in 14 d at 25°C, margin regular, flat, velvety with scarce white aerial mycelia, iron grey, synnemata abundant; reverse light olivaceous, buff near the margin. Colonies on MEA 20–25 mm diam in 14 d at 25°C, flat, radially striate with lobate edge, velvety with scarce aerial mycelia, front and reverse pale yellow. Colonies on carbon free SGM growing more slowly compared to that on PDA, OA and MEA, aerial mycelia scarce but forming hyphae networks, with dark brown and sparse synnemata in one month.Hyphae septate, hyaline to pale brown, smooth- and thin-walled, 1.5–2.5 μm wide. Conidiophores unbranched or sparingly branched, often consisting of single annellides borne sessile on the aerial hyphae or in groups of 2–4 annellides on short basal cells, pale brown, smooth- and thin-walled, usually forming synnemata. Synnemata 400–1000 μm high, stipes dark brown to black, 15–25 μm wide, conidial heads iron grey, ellipsoidal; setae absent. Annellides ampulliform, 5.5–9.5 × 2.5–4 μm, subhyaline, pale brown, smooth- and thin-walled. Conidia ellipsoidal to ovoid, 5.5–7.5 × 3–4.5 μm (), with truncate base and rounded or slightly acute apex, pale brown, smooth- and thin-walled, arranged in long chains.
Cardinal temperatures for growth: optimum 15–25°C, maximum 30°C, minimum 5°C.
Specimens examined
China, Guizhou Province, Suiyang, the Shuanghe National Geographic Park, from cave limestone, 8 May 2015, Zhi-Feng Zhang, Jia-Rui Jiang and Xin Zhou, HMAS 247176 (holotype designated here), ex-holotype living culture CGMCC 3.18328 (= LC 7467); ibid. LC 7455; ibid. LC 7460; ibid. LC 7462; ibid. LC 7463; ibid. LC 7464; ibid. LC 7468; ibid. LC 7469; ibid. LC 7470; ibid. LC 7477; from cave soil, 8 May 2015, Zhi-Feng Zhang, Jia-Rui Jiang and Xin Zhou, LC 7481; ibid. LC 7482; ibid. LC 7483; ibid. LC 7484.
Notes
Cephalotrichum oligotriphicum showed a close phylogenetic relationship with C. verrucisporum (2 bp differences in ITS; 5 bp in EF-1α; 10 bp in TUB2) (). However, the former species morphologically differed from the latter in producing smooth (vs. verrucouse) and slightly smaller (5.5–7.5 × 3–4.5 μm vs. 6–9 × 3–5.5 μm) conidia (Jiang and Zhang Citation2008). In addition, the colony margin of C. oligotriphicum is white crenate on PDA, but white velvety in C. verrucisporum (Jiang and Zhang Citation2008).
Discussion
Caves are typical oligotrophic environments, with total organic carbon (TOC) < 0.5 mg/kg, two orders lower than an average terrestrial environment (Barton and Jurado Citation2007). Among the oligotrophic fungi tested with the carbon free medium SGM in this study, air and limestone harbour the highest fungal diversity (41 spp. respectively), followed by soil (17 spp.) and water (6 spp.). It is premature to speculate why these different substrates harboured different number of oligotrophic species, as the isolation protocols are probably more determinate for the obtained fungal communities.
Although numerous fungi have been reported from various oligotrophic habitats, most are likely facultative rather than obligate oligotrophic fungi (Parkinson et al. Citation1989, Citation1990; Wainwright Citation1993; Wainwright and Al-Talhi Citation1999; Connell and Staudigel Citation2013; Liu et al. Citation2013; Godinho et al. Citation2015; Jiang et al. Citation2017). Hitherto only 18 species have been proven for being able to grow on SGM (). The data presented here increased the known oligotrophic species to 99 (). Within the 84 species from the preliminary identification, up to 31 (exclude three new species in this study) are considered potential new species, and they will be examined in future studies.
Limestone is predominantly composed of calcium carbonate (Li et al. Citation2009), and the carbonate minerals play important roles in global carbon cycling. In the present study, 83% of 30 Cephalotrichum strains were obtained from limestone in caves. These fungi may utilise energy gained from inorganic oxidation (Wainwright and Grayston Citation1988), as well as play important roles in the limestone weathering and dissolution (Sterflinger Citation2000; Northup and Lavoie Citation2001; Burford et al. Citation2003; Gadd Citation2007). In addition to the three species of Cephalotrichum described in this study, C. stemonitis from Slovakia and Spain, and C. verrucisporum from Japan were also previously reported from caves environments (wall and ceiling, rodent feces, and rhizomorphs) (Nováková Citation2009; Kuzmina et al. Citation2012; Kiyuna et al. Citation2017).
Three novel species of Cephalotrichum, C. guizhouense, C. laeve, and C. oligotriphicum, detailed in this study produce little typical aerial mycelia under oligotrophic conditions, but their hyphae stick on the surface of the silica gel, forming fine hyphal networks (i.e. gossamers) ((h); (h); (h)), which provided a large surface area aiding nutrient scavenging from the gel and atmosphere (Parkinson et al. Citation1989; Wainwright Citation1993). In contrast, they produced abundant synnemata and dry and light airborne conidia on the carbon-free silica gel, which might be a survival strategy to promote the spore dispersal in a nutrient-less environment (Wainwright Citation1993; Sandoval-Denis et al. Citation2016b). The genus Cephalotrichum (Microascaceae) was established by Link (Citation1809) and its type species is C. stemonitis. Synnemata is the crucial morphological characters distinguishing Cephalotrichum from other genera in Microascaceae, i.e. Microascus, Scopulariopsis and Fuscoannellis (Sandoval-Denis et al. Citation2016b). Recent phylogenetic analyses revealed that Doratomyces (Sturm Citation1829) and Trichurus (Clements Citation1896) were conspecific to Cephalotrichum and they have been synonymised with Cephalotrichum (Sandoval-Denis et al. Citation2016b). Together with the current study, a total of 78 species have been described in Cephalotrichum, although only 15 of them have reliable molecular data and 43 names are of uncertain application (Sandoval-Denis et al. Citation2016b). An identification key for the 15 species is provided.
Key to Cephalotrichum species
1a Setae present on the upper part of the synnemata 2
1b Setae absent 4
2a Setae straight, branched or unbranched C. cylindricum
2b Setae curved or flexuous 3
3a Setae flexuous, coiled, unbranched C. gorgonifer
3b Setae undulate and dichotomously branched C. dendrocephalum
4a Echinobotryum-like synasexual morph present 5
4b Echinobotryum-like synasexual morph absent 6
5a Synnemata up to 3000 μm tall; annelloconidia ellipsoidal to cylindrical, 5–9 × 4–5 μm with rounded apices C. stemonitis
5b Synnemata up to 1600 μm tall; annelloconidia subglobose to ellipsoidal, 6–7.5 × 2.5–4 μm with slightly pointed apices C. hinnuleum
6a Conidia distinctively rough 7
6b Conidia smooth or finely ornamented 9
7a Synnemata up to 1000 μm tall; conidia oval to ellipsoidal C. asperulum
7b Synnemata often higher; conidia globose to ovoid 8
8a Conidia 4.5–7.5 μm wide, coarsely warted, grey-brown C. nanum
8b Conidia 3–5.5 μm wide, spirally sculpted, dark brown C. verrucisporum
9a Growth at 37°C 10
9b No growth at 37°C 11
10a Synnemata up to 500 μm tall, conidia dark brown C. columnare
10b Synnemata up to 1000 μm tall, conidia pale brown C. laeve
11a No growth at 35°C 12
11b Growth at 35°C 13
12a Conidia green-brown, 3.5–5 × 2–3 μm C. microsporum
12b Conidia pale brown, 5.5–7.5 × 3–4.5 μm C. oligotriphicum
13a Conidia smooth, 4.5–6.5 × 3–4 μm C. guizhouense
13b Conidia smooth to slightly roughened, larger than C. guizhouense 14
14a Synnemata up to 500 μm tall, conidia pale brown C. brevistipitatum
14b Synnemata up to 1600 μm tall, conidia green-brown C. purpureofuscum
Acknowledgements
This study was financially supported by Project for Fundamental Research on Science and Technology, Ministry of Science and Technology of China (2014FY120100). Jia-Rui Jiang acknowledges CAS grant (153211KYSB20160029) for providing her postgraduate studentship. Zhi-Feng Zhang and Xin Zhou are thanked for the help on sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barton HA, Juardo V. 2007. What’s up down there? Microbe-Am Soc Microbiol. 2:132–138.
- Bastian F, Alabouvette C, Saiz-Jimenez C. 2009. The impact of arthropods on fungal community structure in Lascaux Cave. J Appl Microbiol. 106:1456–1462.
- Bergero R, Girlanda M, Varese GC, Intili D, Luppi AM. 1999. Psychrooligotrophic fungi from arctic soils of Franz Joseph Land. Polar Biol. 21:361–368.
- Borda D, Borda C, Tămaş T. 2004. Bats, climate, and air microorganisms in a Romanian cave. Mammalia. 68:337–343.
- Burford EP, Kierans M, Gadd GM. 2003. Geomycology: fungi in mineral substrata. Mycologist. 17:98–107.
- Clements FE. 1896. Report on collections made in 1894–95. Bot Surv Nebraska. 4:1–48.
- Connell L, Staudigel H. 2013. Fungal diversity in a dark oligotrophic volcanic ecosystem (DOVE) on Mount Erebus, Antarctica. Biology. 2:798–809.
- Cubero OF, Crespo ANA, Fatehi J, Bridge PD. 1999. DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant Syst Evol. 216:243–249.
- Gabriel CR, Northup DE. 2013. Microbial ecology: caves as an extreme habitat. Cave Microbiomes: A novel resource for drug discovery. Springer New York, p.85–108
- Gadd GM. 2007. Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol Res. 111:3–49.
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb. 61:1323–1330.
- Godinho VM, Gonçalves VN, Santiago IF, Figueredo HM, Vitoreli GA, Schaefer CE, Barbosa EC, Oliveira JG, Alves TM, Zani CL, et al. 2015. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles. 19:585–596.
- Gundersen KR, Corbin JS, Hanson CL, Hanson ML, Hanson RB, Russell DJ, Stollar A, Yamada O. 1976. Structure and biological dynamics of the oligotrophic ocean photic zone off the Hawaiian Islands. Pac Sci. 30:45–68.
- Hiroyuki O, Tsutomu H. 1983. Oligotrophic bacteria on organic debris and plant roots in a paddy field soil. Soil Biol Biochem. 15:1–8.
- Jiang JR, Chen Q, Cai L. 2017. Polyphasic characterisation of three novel species of Paraboeremia. Mycol Prog. 16:285–295.
- Jiang YL, Zhang TY. 2008. Two new species of Doratomyces from soil. Mycotaxon. 104:131–134.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kiyuna T, An K, Kigawa R, Sano C, Sugiyama J. 2017. Noteworthy anamorphic fungi, Cephalotrichum verrucisporum, Sagenomella striatispora, and Sagenomella griseoviridis, isolated from biodeteriorated samples in the Takamatsuzuka and Kitora Tumuli, Nara, Japan. Mycoscience. doi:10.1016/j.myc.2017.02.003
- Kuzmina L, Galimzianova N, Abdullin S, Ryabova A. 2012. Microbiota of the Kinderlinskaya cave (South Urals, Russia). Microbiology. 81(2):251–258.
- Li W, Zhou PP, Jia LP, Yu LJ, Li XL, Zhu M. 2009. Limestone dissolution induced by fungal mycelia, acidic materials, and carbonic anhydrase from fungi. Mycopathologia. 167:37–46.
- Link HF. 1809. Observationes in Ordines plantarum naturales [ Dissertatio 1ma. (Berlin Ges. NatKde 3:1–42)]. Germany: Berlin.
- Liu TT, Hu DM, Liu F, Cai L. 2013. Polyphasic characterization of Plectosphaerella oligotrophica, a new oligotrophic species from China. Mycoscience. 54:387–393.
- Martin P, MacLeod RA. 1984. Observations on the distinction between oligotrophic and eutrophic marine bacteria. Appl Environ Microb. 47:1017–1022.
- Mason-Gamer RJ, Kellogg EA. 1996. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol. 45:524–545.
- Mirocha CJ, DeVay JE. 1971. Growth of fungi on an inorganic medium. Can J Microbiol. 17:1373–1378.
- Northup DE, Lavoie KH. 2001. Geomicrobiology of caves: a review. Geomicrobiol J. 18:199–222.
- Nováková A. 2009. Microscopic fungi isolated from the Domica Cave system (Slovak Karst National Park, Slovakia). A review. Int J Speleol. 38(1):71–82.
- Parkinson SM, Killham K, Wainwright M. 1990. Assimilation of 14CO2 by Fusarium oxysporum grown under oligotrophic conditions. Mycol Res. 94:959–964.
- Parkinson SM, Wainwright M, Killham K. 1989. Observations on oligotrophic growth of fungi on silica gel. Mycol Res. 93:529–534.
- Payton M, McCullough W, Roberts CF. 1976. Agar as a carbon source and its effect on the utilization of other carbon sources by acetate non-utilizing (acu) mutants of Aspergillus nidulans. J Gen Microbiol. 94:228–233.
- Poindexter JS. 1981. Oligotrophy. Adv Microb Ecol. 5:63–89.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Rayner RW. 1970. A mycological colour chart. UK: Commonwealth Mycological Institute and British Mycological Society.
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 97:84–98.
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 98:625–663.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Ruibal C, Platas G, Bills GF. 2005. Isolation and characterization of melanized fungi from limestone formations in Mallorca. Mycol Prog. 4:23–38.
- Sandoval-Denis M, Gené J, Sutton DA, Cano-Lira JF, de Hoog GS, Decock CA, Wiederhold NP, Guarro J. 2016a. Redefining Microascus, Scopulariopsis and allied genera. Persoonia. 36:1–36.
- Sandoval-Denis M, Guarro J, Cano-Lira JF, Sutton DA, Wiederhold NP, de Hoog GS, Abbott SP, Decock C, Sigler L, Gené J. 2016b. Phylogeny and taxonomic revision of Microascaceae with emphasis on synnematous fungi. Stud Mycol. 83:193–233.
- Stamatakis A, Alachiotis N. 2010. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics. 26:i132–i139.
- Sterflinger K. 2000. Fungi as geologic agents. Geomicrobiol J. 17:97–124.
- Sturm J. 1829. Deutschlands Flora, Abt. III. Die Pilze Deutschlands. 2:1–136.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Udagawa S, Horie Y, Abdullah SK. 1985. Trichurus dendrocephalus sp. nov., from Iraqui soil. Mycotaxon. 23:253–259.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172:4238–4246.
- Wainwright M, Al-Talhi A. 1999. Selective isolation and oligotrophic growth of Candida on nutrient-free silica gel medium. J Med Microbiol. 48:1130.
- Wainwright M, Al-Wajeeh K, Grayston SJ. 1997. Effect of silicic acid and other silicon compounds on fungal growth in oligotrophic and nutrient-rich media. Mycol Res. 101:933–938.
- Wainwright M, Grayston SJ. 1988. Fungal growth and stimulation by thiosulphate under oligocarbotrophic conditions. Tr Brit Mycol Soc. 91:149–156.
- Wainwright M. 1993. Oligotrophic growth of fungi: stress or natural state? In: Jennings DH, editor. Stress tolerance of fungi. New York: Marcel Dekker; p. 127–144.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; p. 315–322.
- Zhang Y, Liu F, Wu W, Cai L. 2015. A phylogenetic assessment and taxonomic revision of the thermotolerant hyphomycete genera Acrophialophora and Taifanglania. Mycologia. 107:768–779.
