ABSTRACT
A large polyporoid mushroom from the West Usambara Mountains in North-eastern Tanzania produces dark brown, up to 60-cm large fruiting bodies that at maturity may weigh more than 10 kg. It has a high rate of mycelial growth and regeneration and was found growing on both dry and green leaves of shrubs; attached to the base of living trees, and it was also observed to degrade dead snakes and insects accidentally coming into contact with it. Phylogenetic analyses based on individual and concatenated data sets of nrLSU, nrSSU and the RPB2 and TEF1 genes showed it, together with Laetiporus, Phaeolus, Pycnoporellus and Wolfiporia, to form a monophyletic group in Polyporales. Based on morphological features and molecular data, it is described as Kusaghiporia usambarensis.
Introduction
Polyporales is an order of fungi in Basidiomycota containing more than 1800 species in 216 genera and 13 families (Kirk et al. Citation2008). However, Justo et al. (Citation2017) recognised 37 families in Polyporales. Seven clades have been recognised in Polyporales: the “antrodia”; “core polyporoid”; “residual polyporoid”; “phlebioid”; “tyromyces”; “gelatoporia” and “fragiliporia” clades (Binder et al. Citation2013; Zhao et al. Citation2015).
The “antrodia clade” was first identified by Hibbett and Donoghue (Citation2001) and currently more than 26 genera are recognised in this clade (Ortiz-Santana et al. Citation2013). Members in the “antrodia clade” are of economic importance as a source of food, and also of pharmaceutical and biotechnological products. However, it also contains species that are plant pathogens detrimental to forests and forest plantations (Dai et al. Citation2007; Banik et al. Citation2010). The “antodia clade” is morphologically diverse and includes species that have resupinate, stipitate or pileatebasidiomata that are either annual or perennial; the hyphal system is monomitic, dimitic or trimitic; the basidiospores are hyaline thin- to thick-walled, subglobose to cylindricaland they cause brown rots (Ryvarden and Melo Citation2014).
The “antrodia clade” has been widely studied and additional genera have been suggested to belong there. Recent taxonomic and phylogenetic studies, including that of Lindner and Banik (Citation2008), have presented molecular phylogenies of the clade. In a study of Laetiporus and other polypores, Banik et al. (Citation2010) inferred relationships among North American and Japanese Laetiporus isolates; Ortiz-Santana et al. (Citation2013) presented a phylogenetic overview of the “antrodia clade” and Binder et al. (Citation2013) used genomic data and a six-gene data set for evaluating phylogenetic relationships in Polyporales. Further studies include those of Han et al. (Citation2014) in which two new species of Fomitopsis from China were described, and Zhao et al. (Citation2015) used a multi-gene dataset to support the recognition of Fragiliporiaceae, a new family of Polyporales. Han et al. (Citation2016) offered a study of the phylogeny of the brown-rot fungi, including Fomitopsis and related genera, while Justo et al. (Citation2017) revised the phylogeny of Polyporales at family-level.
A mushroom locally known as “Kusaghizi” has a long tradition of being used as food by local communities in the Usambara mountains as first reported by Powell et al. (Citation2013). In a study by Juma et al. (Citation2016), which assessed antioxidant activities of saprobic mushrooms from Tanzania, “Kusaghizi” was included, but neither the study by Powell et al. nor that of Juma et al. reported a scientific name for “Kusaghizi”. Here we aim to describe this species and infer its phylogenetic position.
Material and methods
Material
Material of the fungus locally named “Kusaghizi” was collected during the rainy seasons in February 2016 and March 2017 close to the villages Bungu, Buti and Makuri in the Usambara Mountains. These villages are located in the Korogwe District of the Tanga region, Tanzania. The west Usambara Mountains are part of the “Eastern Arc” of ranges in eastern Tanzania, from the Taita Hills in Kenya to the Udzungwa Mountains in southern Tanzania. The samples were examined in a fresh condition for macro-morphological features including colour changes upon cut, bruising and exposure to air. A fruit body was divided into two parts; one was sun dried for 5 days while the remaining part was stored in a freezer at −20°C for further investigations.
Morphological characterisation
Basidioma colours of the holotype were indicated according to Kornerup and Wanschern (Citation1967). Photographs of the fruit body were taken before and after removing it from its substrate. Microscopic characterisation was done from preparations of a rehydrated specimen sectioned with a freezing microtome and stained with Lactic Blue, or treated with 10% KOH and Melzer’s reagent.
A total of 40 mature basidiospores were randomly selected and measured. Statistical averages were used to estimate the observed features as follows: AL = mean spore length (arithmetic mean of the length of spores); AW = mean spore width (arithmetic mean of the width of spores); Q = AL/AW ratio; n (a/b) = number of spores (a) measured from given number (b) of specimen. Melzer’s reagent was used where IKI+ = Melzer’s reagent positive; IKI− = both inamyloid and indextrinoid.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from both fresh and dried material and kept at −20°C following the protocol of the plant Genomic DNA extraction kit (E.Z.N.A. Fungal DNA Mini Kit Protocols). Diluted samples (10–1) of DNA were used for PCR amplification of the nrLSU, nrSSU, RPB2 and TEF1. Primers LR0R, LR7, LR5 were used for nrLSU (Vilgalys and Hester Citation1990), and PNS1 and NS41 for nrSSU (Hibbett Citation1996). PCR conditions for nrLSU and nrSSU were: initial denaturation for 4 min at 95°C, followed by 35 cycles of 1 min at 94°C, 1 min at 54°C, 45 s at 72°C, and a final elongation for 5 min at 72°C. The RPB2 region was amplified using degenerated primers fRPB2-5f and RPB2-7.1R (Matheny Citation2005). For amplification of TEF1the EF1-983Fand EF1-1567R primers were used (Rehner and Buckley Citation2005). Touchdown PCR was used with an initial annealing temperature of 66°C following the protocol of Rehner and Buckley (Citation2005). The PCR products were visualised by electrophoresis on 1.5% agarose gels. Products were purified using Illustra™ ExoStar buffer diluted 10×, following the manufacturer’s protocol. Sequencing was carried out by Macrogen.
Data analyses
Sequences from GenBank were selected based on their quality and with an intention of wide coverage of Polyporales and the “antrodia clade” as in Zhao et al. (Citation2015) and Han et al. (Citation2016) respectively. The sequences produced in this study were aligned along with those downloaded from GenBank () using MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/) and manually adjusted using AliView (Larsson Citation2014). Ambiguously aligned regions were excluded from the analyses. For RPB2 and TEF1 only coding parts of the sequences were used for the analyses. The concatenated data matrix of Polyporalesand the “antrodia clade” contained 4940 and 3760 unambiguously aligned sites respectively. All alignments were based on the nucleotide sequences with each gene analysed separately.
Table 1. Species, collection and GenBank accession number of sequences used in this study. New sequences in bold
Single-gene analyses were performed to detect significant conflicts among datasets. A conflict among single-locus datasets (nrLSU, nrSSU, RPB1, TEF1) was considered significant if a well-supported monophyletic group, for example posterior probability (PP) ≥ 0.95, was found to be well supported as non-monophyletic when different loci were used. No significant incongruence among the single-gene trees was detected (Supplementary Figures S1A, S1B S1C and S1D), hence the four matrices were concatenated.
Further analyses were carried out after concatenation using Sequence Matrix (Vaidya et al. Citation2011).
The best-fit model of DNA evolution for the analyses, for both individual codon positions and genes, was obtained using the Akaike Information Criterion as implemented in MrModeltest 2.3 (Nylander Citation2004). For the Polyporales dataset the GTR+I + G model was employed across sites for nrLSU, nrSSU, and for the 1st and 2nd codon for RPB2. For the 1st and the 2nd codon for TEF1 the model F81 + I + G was applied, while GTR + G was implemented for both the 3rd codon of RPB2 and TEF1. For the “antrodia clade” dataset the GTR+I + G model was employed across sites for nrLSU, nrSSU, for all three codons for RPB2, and the 2nd codon for TEF1. For the 1st codon for TEF1 the F81 + I model was applied while the HKY + I + G model was implemented for the 3rd codon for TEF1. Bayesian Inference was conducted with MrBayes 3.2.6, and branch support was estimated by PP (Ronquist and Huelsenbeck Citation2003). Four Markov chains were run for 2 runs from random starting trees for 10 million generations, trees were sampled every 100 generations and 25% were discarded as burn-in.
Maximum likelihood estimates were carried out by RAxML v.8.2.10 using the GTR +G + I model of site substitution (Stamatakis Citation2014). The branch support was obtained by maximum likelihood bootstrapping (MLbs) of 1000 replicates (Hillis and Bull Citation1993).
Bayesian PPs ≥ 0.95 (Alfaro et al. Citation2003), and MLb ≥ 70% were considered to be significant. Sequence alignments and phylogenetic trees were deposited in TreeBase, submission ID: (http://purl.org/phylo/treebase/phylows/study/TB2:S223838).
Results
Phylogenetic analyses
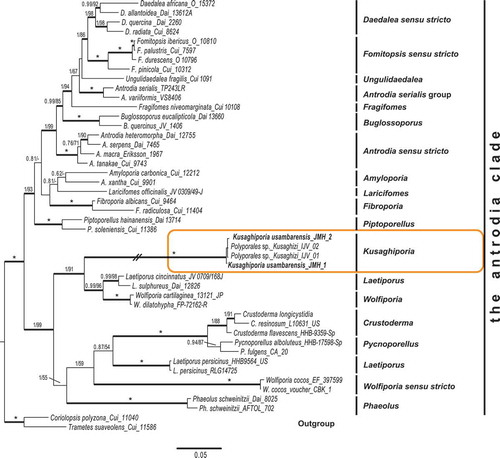
Analyses were based on a total of 209 sequences representing 201 species of Polyporales, with two russuloid species as out-group. The phylogeny of the Polyporales and the position of the “Kusaghizi” was inferred from four datasets: 36 nrLSU sequences, 25 nrSSU sequences, 26 RPB2 sequences and 22 TEF1 sequences. The Polyporales concatenated dataset () contained 100 sequences of 34 nrLSU, 21 nrSSU, 23 RPB2 and 22 TEF1. Further analyses included members of the “antrodia clade” () containing 145 concatenated sequences of 46 nrLSU, 33 nrSSU, 32 RPB2 and 35 TEF1. Maximum likelihood and Bayesian analyses of these datasets were undertaken, first separately and then also of the concatenated dataset. The analysis of the concatenated Polyporales dataset retrieved a phylogeny with five distinct clades () in addition to Stereum hirsutum and Heterobasidion annosum, as outgroup. Clade annotations follow Zhao et al. (Citation2015). Kusaghiporia usambarensis was found to belong in the “antrodia clade”. The annotation of the concatenated phylogeny of the “antrodia clade” () follows Han et al. (Citation2016).
Figure 1. Phylogenetic relationships among Kusaghiporia usambarensis and allied taxa in Polyporales, based on Bayesian and ML analyses of concatenated nrLSU, nrSSU, RPB1 and TEF1 datasets. The tree was rooted using two species from Russulales (Heterobasidion annosum and Stereum hirsutum). The two support values associated with each internal branch correspond to PPs and MLbs proportions, respectively. Branches in bold indicate a support of PP ≥ 0.95 and MLbs ≥ 70%. An asterisk on a bold branch indicates that this node has a support of PP = 1.0 and MLbs = 100. The branch with double-slash is shortened. Clade names follow Zhao et al. (Citation2015)
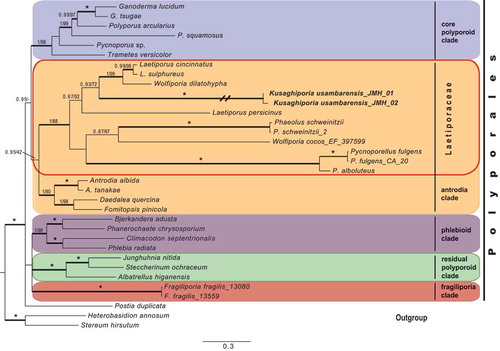
Figure 2. Phylogenetic relationships among Kusaghiporia usambarensis and allied taxa in the “antrodia clade”, based on Bayesian and ML analyses of concatenated nrLSU, nrSSU, RPB1 and TEF1 datasets. The tree was rooted using two species from the “core polyporoid clade” (Coriolopsis polyzona and Trametes suaveolens). The two support values associated with each internal branch correspond to PPs and MLbs proportions, respectively. Branches in bold indicate a support of PP ≥ 0.95 and MLbs ≥ 70%. An asterisk on a bold branch indicates that this node has a support of PP = 1.0 and MLbs = 100. The branch with double-slash is shortened. Clade names follow Han et al. (Citation2016)
Taxonomy
Kusaghiporia usambarensis Hussein J., Tibell S. & Tibuhwa, gen. et sp. nov. MycoBank no.: MB824538 [, , .]
Figure 3. (a) Basidiocarp of Kusaghiziporia usambarensis(holotype). (b) Vertical section of basidiocarp. (c) Lower part of basidiocarp. (d) Bruise reaction, the creamy pores (5A2) turned brown (5D6)

Figure 4. Microscopic structures of Kusaghiziporia usambarensis (holotype). (a) Skeletal hyphae (sk) with Y-shaped branches; gloeplerous hyphae (gl). (b) Septate generative hyphae (gen). (c) Basidia. (d) Basidia with spores attached to sterigmata. (e) Globular to subglobular basidiospores
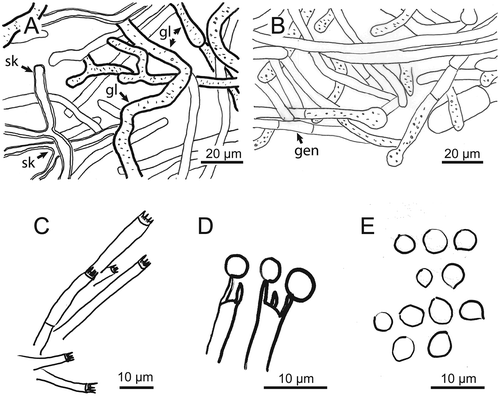
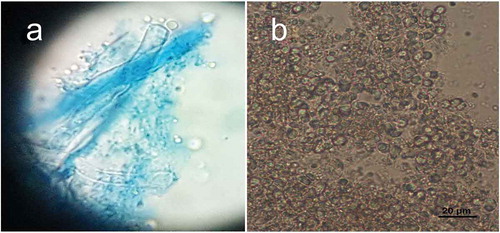
Figure 5. (a) Picture of basidia with spores attached to sterigmata. (b) Globular to subglobular basidiospores
Basidioma annual, spathulate, viscid when young, at maturity saucer-shaped, bumpy, and with a spongy surface. Upper surface mottled dark brown with creamy patches. Hyphal system dimitic, with generative and skeletal hyphae. Gloeplerous hyphae present.
Holotype TANZANIA
Korogwe district, Tanga, Bungu, 18 February 2016, J. Hussein 01/16 (UPS); GenBank MH010044, MH010046, MH048871, MH048870.
Additional material examined
TANZANIA, Korogwe district, Tanga, Buti, 21 March 2017, J. Hussein 01/17 (UPS); GenBank MH010045, MH048869; Tanga, Makuri, 21 March 2017, J. Hussein 02/17 (UPS).
Etymology
Kusaghiporia refers to the sambaa name of the mushroom “Kusaghizi”, which means the collector or accumulator, and –poria (Lat.): with pores; usambarensis (Lat.): referring to the Usambara mountain range.
Fruitbody
Basidioma annual, spathulate, when young viscid, when mature depressed saucer-shaped, up to 60 cm in diameter with an uneven, velvety surface, surface dark brown at the centre (5E8), eroded (wrinkled), dark brown (5E8) to creamy (5A2); basidioma margin fleshy, up to 6-cm thick, dark brown (5E6) with pale brown (5C3) stripes. Stipe central, c. 12-cm high, c. 10 cm in diameter at the base, clavate, with creamy small dots (5A2), tough/woody, dark brown (5F8) in the inner part, without ring. The pores are creamy (5A2), turning brown (5D6) upon bruising. Cap in section dark brown (5E8) with creamy stripes (5A2), not changing upon exposure to air. Spore print whitish to creamy (5A2).
Hyphal structure
Hyphal system dimitic; generative hyphae with simple septa, hyaline, thin-walled 2.7–10.9 µm in diam ()); skeletal hyphae hyaline, thick-walled, with Y-shaped branches 2.7–6.3 µm in diam ()); gloeplerous hyphae present 3.6–11.8 µm in diam ()).
Basidia and basidiospores
Basidia thin-walled, hyaline, tetrasterigmatic, 2.7–5.4 µm in diam (, ) and (a)). Basidiospores hyaline, globose to subglobose, thin-walled, smooth, IKI-, 2.7–8.1 × 2.7–7.2 µm, AL = 5.9 µm, AW = 5.7 µm, Q = 1.04 (n = 40/1) (, ) and )).
Rot type
Brown rot.
Host
Found growing at the base of the trees Maesopsis eminii and Ficus natalensis.
Discussion
In earlier studies (Binder et al. Citation2013; Zhao et al. Citation2015), seven clades were found in Polyporales: the “core polyporoid clade”; the “residual polyporoid clade”; the “antrodia clade”; the “gelatoporia clade”; the “phlebioid clade”; the “tyromyces clade” and the “fragiliporia clade”. Justo et al. (Citation2017) recognised six clades, excluding the “fragiliporia clade” reported by Zhao et al. (Citation2015). We found Kusaghiporia to be nested within the “antrodia clade” in all analyses; concatenated dataset (), and nrLSU, nrSSU, TEF1, RPB2 (Fig S1A, S1B, S1C; S1D). Analyses of Kusaghiporia and related taxa in the “antrodia clade” grouped Kusaghiporia with Laetiporus and Wolfiporia, a clade receiving strong support (; 1 PP, 91% MLbs). Despite the strong support of Laetiporaceae Jülich () K. usambarensis, L. persicinus, W. cocos, Phaoelus, and Crustoderma together with Pycnoporellus displayed long branches indicating a high genetic divergence. A high genetic divergence of L. persicinus has previously been reported (Binder et al. Citation2013; Ortiz-Santana et al. Citation2013; Han et al. Citation2016; Justo et al. Citation2017). Lindner and Banik (Citation2008) suggested placing L. persicinus in a separate genus due to its genetic remoteness as compared to other species of Laetiporus. Justo et al. (Citation2017) suggested further studies to be needed for the delimitation of Wolfiporia and Laetiporus. However, a detailed discussion of L. persicinus and W. cocos is beyond the scope of this study.
Kusaghiporia usambarensis is morphologically similar to Crustoderma, Pycnoporellus, Phaeolus, Wolfiporia and Laetiporus, insofar that they all have hyphae with simple septa, produce annual polyporoid fruiting bodies with hyaline spores and cause brown rots (Lindner and Banik Citation2008). Crustoderma (Eriksson and Ryvarden Citation1975) differs from K. usambarensis in having resupinate basidio carps and a monomitic hyphal system. Pycnoporellus (Ryvarden and Melo Citation2014) differs from K. usambarensis in having yellow to orange basidiocarps and a monomitic hyphal system. Phaeolus (Lindner and Banik Citation2008) differs from K. usambarensis in having a monomitic hyphal system and producing hymenial cystidia. Wolfiporia and Laetiporus, like K. usambarensis, have dimitic hyphal systems. Wolfiporia, however, has resupinate basidiocarps and oblong-ellipsoid basidiospores (Zmitrovich et al. Citation2006). With the exception of L. persicinus, other Laetiporus species produce brightly coloured basidiocarps (Lindner and Banik Citation2008). Kusaghiporia usambarensis is different from L. persicinus in basidioma morphology (up to 60 cm) and the basiodiospores being globose to subglobose, while broadly ovoid in L. persicinus.
The BLAST results from GeneBank (NCBI, from 2017-10-16), using blastn with the program “discontiguous megablast” (for cross-species comparison, searching with coding sequences) with Kusaghiporia sequences, showed a highest sequence similarity for all four genes with Laetiporus sulphureus. Based on RPB2 and TEF1 they were: Query cover 99% and Ident. 87%, and Query cover 99%, Ident. 84% respectively. For nrSSU the highest similarity has a Query cover of 91% and Ident. 87%; while for nrLSU the Query cover was 100% and the Ident. 86%. Among the five top scores “Polyporales sp. Kusaghizi”, voucher IJV40-2 was found: Query cover only 60% and Ident. 100%. In our opinion the genetic isolation of K. usambarensis as compared to Laetiporus justifies the proposal of a new genus to accommodate the species investigated. The monophyly and strong support of the clade containing K. usambarensis, Laetiporus, Wolfiporia, Crustoderma, Pycnoporellus, and Phaeolus as shown in our phylogeny (), also justifies the incorporation of K. usambarensis in Laetiporaceae.
Conclusion
The new genus Kusaghiporia was described based on morphological characters and phylogenetic analyses based on concatenated sequence data from four genes. Kusaghiporia produces large fruit bodies. Together with Laetiporus, Pycnoporellus, Phaeolus, and Wolfiporia it formed a strongly supported clade () belonging in Laetiporaceae, which is nested in the “antrodia clade”. Kusaghiporia is a resource in the local communities of the Usambaras, where it is collected and eaten.
Supplemental Material
Download Zip (726.4 KB)Acknowledgements
Special thanks to Frank Mbago for his help in identifying host plants, local field guides, traditional healers, and the local community for their cooperation throughout the study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental data can be accessed here.
Additional information
Funding
References
- Alfaro ME, Zoller SLutzoni F. 2003. Bayes or bootstrap? a simulation study comparing the performance of bayesian markov chain monte carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol and Evol. 20(2):255-266.
- Banik MT, Lindner DL, Ota Y, Hattori T. 2010. Relationships among North American and Japanese Laetiporus isolates inferred from molecular phylogenetics and single-spore incompatibility reactions. Mycologia. 102(4):911–917.
- Binder M, Justo A, Riley R, Salamov A, Lopez-Giraldez F, Sjökvist E, Copeland A, Foster B, Sun H, Larsson E. 2013. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia. 105(6):1350–1373.
- Chen J, Cui B, He S, Cooper JA, Barrett MD, Chen J, Dai Y. 2016. Molecular phylogeny and global diversity of the remarkable genus Bondarzewia (Basidiomycota, Russulales). Mycologia. 108(4):697–708.
- Chen YY,Cui BK. 2016. Phylogenetic analysis and taxonomy of the antrodia heteromorpha complex in china. Mycoscience. 57(1):1-10.
- Dai YC, Cui BK, Yuan HS, Li BD. 2007. Pathogenic wood decaying fungi in China. Forest Pathol. 37(2):105–120.
- Eriksson J, Ryvarden L. 1975. The Corticiaceae of north Europe 3. Norway: Fungiflora; p. 288–546.
- Floudas D, Hibbett DS. 2015. Revisiting the taxonomy of Phanerochaete (Polyporales, Basidiomycota) using a four gene dataset and extensive ITS sampling. Fungal Biol. 119(8):679–719.
- Han ML, Chen YY, Shen LL, Song J, Vlasák J, Dai YC, Cui BK. 2016. Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 80(1):343–373.
- Han ML, Song J, Cui BK. 2014. Morphology and molecular phylogeny for two new species of Fomitopsis (Basidiomycota) from South China. Mycol Prog. 13:905–914.
- Han ML, Vlasák J, Cui BK. 2015. Daedalea americana sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Phytotaxa. 204(4):277–286.
- Hibbett DS. 1996. Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroom-forming fungi. Mol Biol Evol. 13(7):903–917.
- Hibbett DS, Donoghue MJ. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol. 50(2):215–242.
- Hibbett DS, Gilbert LB, Donoghue MJ. 2000. Evolutionary instability of ectomycorrhizal symbioses in basidiomycetes. Nature. 407(6803):506–508.
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 42(2):182–192. http://mafft.cbrc.jp/alignment/server/
- Juma I, Mshandete A, Tibuhwa D, Kivaisi A. 2016. Assessment of antioxidant potentials of the wild and domesticated saprophytic edible mushrooms from Tanzania. Curr Res Environ App Mycol. 6(2016):1–10.
- Justo A, Hibbett DS. 2011. Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset. Taxon. 60(6):1567–1583.
- Justo A, Miettinen O, Floudas D, Ortiz-Santana B, Sjökvist E, Lindner D, Nakasone K, Niemelä T, Larsson KH, Ryvarden L, et al. 2017. A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 121:798–824.
- Kim KM, Lee JS, Jung HS. 2007. Fomitopsis incarnatus sp. nov. based on generic evaluation of Fomitopsis and Rhodofomes. Mycologia. 99(6):833–841.
- Kirk P, Cannon P, Minter D, Stalpers J. 2008. Dictionary of the Fungi. Wallingford (UK): CABI.
- Kornerup A, Wanscher JH. 1967. Methuen handbook of colour. ( 2nd English edition, revised.). London: Methuen & Co.
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30(22):3276–3278.
- Larsson KH. 2007. Re-thinking the classification of corticioid fungi. Mycol Res. 111(9):1040–1063.
- Lindner DL, Banik MT. 2008. Molecular phylogeny of Laetiporus and other brown rot polypore genera in North America. Mycologia. 100(3):417–430.
- Matheny PB. 2005. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol. 35(1):1–20.
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Langer E. 2007. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Phyl Evol. 43(2):430–451.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://github.com/nylander/MrModeltest2
- Ortiz-Santana B, Lindner DL, Miettinen O, Justo A, Hibbett DS. 2013. A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales). Mycologia. 105(6):1391–1411.
- Powell B, Maundu P, Kuhnlein HV, Johns T. 2013. Wild foods from farm and forest in the East Usambara Mountains, Tanzania. Ecol Food Nutr. 52(6):451–478.
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 97(1):84–98.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ryvarden L, Melo I. 2014. Poroid fungi of Europe. Oslo: Fungiflora.
- Seelan JSS, Justo A, Nagy LG, Grand EA, Redhead SA, Hibbett D. 2015. Phylogenetic relationships and morphological evolution in Lentinus, Polyporellus and Neofavolus, emphasizing southeastern Asian taxa. Mycologia. 107(3):460–474.
- Shen LL, Cui BK. 2014. Morphological and molecular evidence for a new species of Postia (Basidiomycota) from China. Cryptogamie, Mycologie. 35(2):199–207.
- Song J, Chen YY, Cui BK, Liu HG, Wang YZ. 2014. Morphological and molecular evidence for two new species of Laetiporus (Basidiomycota, Polyporales) from southwestern China. Mycologia. 106(5):1039–1050.
- Song J, Cui BK. 2017. Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evol Biol. 17(1):102.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172(8):4238–4246.
- Wu SH, Hibbett DS, Binder M. 2001. Phylogenetic analyses of Aleurodiscus sl and allied genera. Mycologia, 93(4):720–731.
- Wu SH, Nilsson HR, Chen CT, Yu SY, Hallenberg N. 2010. The white-rotting genus Phanerochaete is polyphyletic and distributed throughout the phleboid clade of the Polyporales (Basidiomycota). Fungal Divers. 42(1):107–118.
- Zhao CL, Cui BK, Song J, Dai YC. 2015. Fragiliporiaceae, a new family of Polyporales (Basidiomycota). Fungal Divers. 70(1):115–126.
- Zmitrovich IV, Malysheva VF, Spirin WA. 2006. A new morphological arrangement of the Polyporales. I. Phanerochaetineae. Mycena. 6:4–56.
