?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Sponge-associated fungi are the least explored marine fungal groups. It is only in recent years that fungal symbionts of marine sponges have received attention mainly due to the isolation of bioactive metabolites while not much attention was given to their specificity, biogeography and exact roles in marine sponges. The diversity of fungi associated with mangrove sponges (Axinella sp., Halichondria cf. panicea, Haliclona sp., Tedania sp.) collected from New Washington, Aklan, Philippines were investigated using morphological observation. A total of 110 species of sponge-associated fungi belonging to 22 genera of ascomycetes with 18 genera of asexual morphs whose sexual stage is unknown, 2 genera of basidiomycetes, 21 morphospecies of Mycelia sterilia, 1 unidentified yeast species and 11 unidentified hyphomycetes were isolated from four species of mangrove sponges. This is the first study that explored the diversity and ecology of sponge-associated fungi in mangrove habitats from the Philippines. The results of the study suggest host-preference by various fungal taxa and the development of fungi on these hosts appeared to be strongly influenced by the characteristics or nature of the immediate environment.
Introduction
Fungi have long been known to exist in the marine environment but considered to be the underexplored group in the oceans compared to bacteria, plants and animals (Jones and Pang Citation2012). Marine fungi live as saprophytes, parasites and symbionts on various matrices such as sediments, logs, water, as well as algae, vascular plants, invertebrates and fishes (Johnson and Sparrow Citation1961; Kohlmeyer and Kohlmeyer Citation1979). Marine fungi can be classified as true or obligate and facultative marine fungi. The former can complete their lifecycle in marine environments and the latter can grow in freshwater or terrestrial habitats as well as in marine environments (Kohlmeyer and Kohlmeyer Citation1979). However, Jones et al. (Citation2015) accept a wider interpretation of what can be considered marine.
Sponges are known inhabitants of coastal and deep-sea environments. They are filter feeders and known to harbour diverse groups of microbes (Hentschel et al. Citation2006; Taylor et al. Citation2007). Marine sponges are a rich source of compounds with bioactive potential. A review by Blunt et al. (Citation2012) reported that 296 new compounds from marine sponges were isolated only in 2011. But over the past decade, a consensus has developed among experts that these novel natural products from sponge extracts are synthesised, either in part or in entirely, by the symbiotic microbes that are intimately associated with these marine metazoans (Konig et al. Citation2006; Meenupriya and Thangaraj Citation2010). These microbes are thought to be involved in a variety of ecological functions including production of secondary metabolites that can contribute to their own ecological success and to that of their host (Höller et al. Citation2000). The previous work on marine microbial diversity in sponges focuses mostly on bacterial associates with proposed novel phylum candidate Poribacteria (Fieseler et al. Citation2004; Kamke et al. Citation2014). It is only in recent years that the fungal symbionts of marine sponges have received attention (Gao et al. Citation2008; Wang et al. Citation2008; Baker et al. Citation2009; Li and Wang Citation2009; Menezes et al. Citation2010; Debbab et al. Citation2011) mainly related to their capacity to produce novel bioactive metabolites (Proksch et al. Citation2003; Bugni and Ireland Citation2004; Amagata et al. Citation2006; Blunt et al. Citation2009; Aly et al. Citation2011; Jones Citation2011).
Fungi have been isolated from subtidal sponges in tropical, subtropical and temperate countries. (Höller et al. Citation2000; Wang et al. Citation2008; Caballero-George et al. Citation2010; Paz et al. Citation2010; Zhou et al. Citation2011; Thirunavukkarasu et al. Citation2012; Flemer Citation2013; Henriquez et al. Citation2013; Bolaños et al. Citation2015). In Asia, there have been published information on the diversity of sponge-associated fungi in countries like China (Zhang et al. Citation2009; Liu et al. Citation2010; Ding et al. Citation2011; Zhou et al. Citation2011; Yu et al. Citation2012; He et al. Citation2014; Jin et al. Citation2014), India (Meenupriya and Thangaraj Citation2010; Thirunavukkarasu et al. Citation2012) Indonesia (Namikoshi et al. Citation2002), Israel (Paz et al. Citation2010), Malaysia (Mahyudin Citation2008) and Russia (Pivkin et al. Citation2006). To date, there are no ecological studies of sponge-associated fungi or any information about the species richness and fungal biodiversity of sponges in the marine environment of the Philippines. Furthermore, there is no published information about the fungal diversity of mangrove-associated sponges. The present study aims to determine the diversity of fungi associated with different species of mangrove-associated sponges collected from the mangrove area of New Washington, Aklan, Philippines. The assessment of fungal diversity will be of great value to understand the fungal ecology in mangrove-associated sponges and to explore the biotechnological potentials of these fungi. The additional information obtained from this study will serve as baseline information on the fungal composition of the different sponge species in the Philippines.
Materials and methods
Study site
New Washington, Aklan is located on the north-eastern part of Panay Island. It is a coastal community composed of islets surrounded by brackish water rivers that forms part of the Batan Estuary and the northern coast of the island faces the Sibuyan Sea. Two sites, Kapispisan (11° 38ʹ 5.748” N, 122° 25ʹ2.388” E) and Boeo (11° 36ʹ 33.804 N, 122° 27ʹ 42.119 E), both in Pinamuk-an, New Washington, Aklan, were surveyed for mangrove sponges by mask and snorkel to a depth of 1–2 m in September 2015 (). The mangrove species that dominated Pinamuk-an River include Sonneratia alba, Avicennia umphiana and Avicennia marina (Ochavo et al. unpublished).
Collection and identification of mangrove-associated sponges
Sponges (Axinella sp., Halichondria cf. panicea, Haliclona sp., Tedania sp.) attached to the mangrove roots were collected and photographed for identification purposes. Latex glove was worn for the collection of sponges and samples were removed from the mangrove roots using scalpel or by directly cutting the roots with attached sponge. Sponge samples (~20–25 g) were transferred directly into zip-lock bags containing seawater from the collection sites to prevent direct contact with air. Sponges were stored and transported immediately in a cooler box back to the laboratory for processing within 24 h to avoid external microbial contamination and excessive proliferation of the samples. All the epiphytic faunas attached to the samples were removed.
Isolation of fungi
Sponge-associated fungi were isolated in the Microbiology Laboratory of University of the Philippines Visayas – National Institute of Molecular Biology and Biotechnology (UPV – NIMBB). Sponge tissue segments were rinsed three times with sterile artificial seawater to eliminate adherent surface debris and contaminants (Thirunavukkarasu et al. 2012). The inner tissue (middle internal mesohyl area) was excised with a sterile scalpel and was cut into small pieces. Ten grams of each sample was homogenised using a blender containing 20 ml sterile artificial seawater under aseptic conditions. One millilitre of the resulting homogenate was transferred to a sterile test tube containing 9 ml of 0.85% NaCl and mixed in the vortex (dilution 10−1). The serial dilution was repeated until 10−5 has been reached.
For fungal cultivation, 100 µl of dilutions 10−0 to 10−5 were plated in triplicate onto 15 ml of the selected isolation media supplemented with penicillin G and streptomycin (100 µg/ml each) using spread plate technique. All five media (Czapek Dox Agar, Cornmeal Agar, Mycobiotic Agar, Potato Dextrose Agar, Rose Bengal Agar) were added with 1.5% NaCl which is the salt concentration used in isolation of mangrove fungi in the Philippines. A control plate was prepared by exposing a blank plate in the middle of the working area for 15 min. The inoculated plates were sealed with ParafilmTM and incubated at room temperature (27°C) in inverted position and examined daily for the appearance of fungal colonies up to three weeks, depending on the growth of species. Edges from emerging fungal colonies growing out on culture medium were picked and transferred with a sterile inoculating loop onto culture tube containing fresh media supplemented with antibiotic solution. Mycelia or spores were transferred again in new culture media for purification. For yeasts, cells were streaked onto a fresh culture media. The resulting plates were incubated at room temperature (27°C) for pure culture and purification was done rigorously until a homogenous fungal isolate was obtained for identification.
Fungal density
The number of colony forming units (CFU) per gram of dry weight of sponge (CFU/g dw) was estimated for each identified fungal species.
Identification of fungal isolates
Filamentous fungi were identified based on their macroscopic (colonial) and microscopic characteristics. Colonial descriptions included colony characteristics such as colour (reverse and obverse), texture, margin, elevation and characteristics of aerial hyphae. Slide culture technique of Riddell (Citation1950) was employed for microscopic examination of fungal isolates that included spore morphology, colour, shape, wall ornamentation or texture, size, conidial formation and other relevant characteristics such as phialide and conidiation pattern. In the Mycology Laboratory of UPV- Freshwater Aquaculture Station, microscopic examination of colony colours and growth rates was assessed with a dissecting microscope. Microscopic characteristics of fungal isolates were determined by viewing slides with distilled water using a light microscope under Low Power Objective (LPO) and High Power Objective (HPO). Microphotographs of the reproductive structures were taken for identification purposes. Filamentous fungi were identified to at least genus level based on the identification scheme by Kohlmeyer (Citation1984), Kohlmeyer and Volkmann-Kohlmeyer (Citation1991), Barnett and Hunter Citation1998), Howard (Citation2003), Watanabe (Citation2010), Domsch et al. (Citation2007), Pitt and Hocking (Citation2009) and Campbell et al. (Citation2013) in addition to other available keys and monographs. Identification of yeast isolates was based on keys by Kurtzman and Fell (Citation1998) that included colonial and microscopic characteristics and by using API 20C Aux (bioMerieux, Rome, Italy) that was based on 19 carbohydrate assimilation tests with negative control. Mycelia sterilia or fungi that failed to grow and sporulate were given codes using cultural characteristics (e.g. colony surface, texture and hyphal pigmentation). The fungal descriptions in MycoBank (www.mycobank.com) were used as guide for further identification of the fungal isolates.
Preservation of fungal cultures
Continuous growth method was used for fungal culture preservation. After identification, all pure cultures of fungal isolates were grown on agar slant in a culture tube and stored at 5°C. Fungal cultures were maintained and preserved in National Institute of Molecular Biology and Biotechnology, University of the Philippines Visayas (UPV), Miagao, Iloilo and Mycology Laboratory of the Division of Biological Sciences, College of Arts and Sciences, UP Visayas.
Data analyses
Total frequency of occurrence (FOC, %) of fungi in sponge samples was computed using the formula:
Total FOC (%): number of presence/total sponges *100
Frequency of occurrence (FOC) of species A (%) per sponge species:
Based on the frequency of occurrence, the following groupings were made (Hyde Citation1989; Hyde and Sarma Citation2000; Sarma and Raghukumar Citation2013): very frequent (≥10%), frequent (5–10%), infrequent (1–5%), rare (≤1%).
(B) The diversity of fungi associated with the mangrove sponges was calculated following Ludwig and Reynolds (Citation1988).
(a) Shannon Index
where: pi is the proportion of individuals that species i contribute to the total number of individuals as shown in the formula below:
where: N = total number of individuals (records)
ni = number of individuals i1, i2, i3, i4, … ix
(b) Simpson Index of Dominance (D)
where: Pi = proportion of individuals in the ith species
As D increases, diversity decreases. Simpson’s index is usually expressed as:
(c) Shannon Evenness (J’)
where: H’max = maximum value of diversity for the number of species present
(d) Simpson’s Evenness (E1/D)
W3here: D = Simpson’ index of diversity
S = Species richness
Jaccard Index of Species Similarity was calculated pair-wise among the hosts based on the presence or absence of each fungal species using the formula
where: a is the number of fungal species occurring in both hosts
b is the number of fungal species unique to the first host and
c is the number of fungal species unique to the second host.
Results
Overall fungal populations and diversity
A total of 110 species of sponge-associated fungi belonging to 22 genera of ascomycetes with 18 genera of asexual morphs whose sexual stage is unknown, 2 genera of basidiomycetes, 21 morphospecies of Mycelia sterilia, 1 unidentified yeast species and 11 unidentified hyphomycetes were isolated from four species of mangrove-attached sponges collected from New Washington, Aklan, Philippines ().
Table 1. Overall frequency of occurrence (%) of sponge-associated fungi collected from a mangrove area in New Washington, Aklan, Philippines.
Based on the percentage of total isolates of all mangrove-associated sponges, Aspergillus dominated the fungal genera under asexual morphs (n = 23, 20.91%) followed by Mycelia sterilia (n = 21, 16.28%), Penicillium (n = 14, 12.73%), Paecilomyces (n = 10, 9.09%) and unidentified hyphomycete (n = 11, 8.53%) (). In addition, genera under Cladosporium and Acremonium have 5 (4.55%) and 4 (3.64%) species, respectively. Furthermore, two species (1.82%) were isolated under the genera Candida and Trichoderma while only 1 species each (0.91%) were isolated under the genera Acrodontium, Beauveria, Cryptococcus, Geotrichum, Gliomastix, Hortaea, Kloeckera, Mammaria, Pestalotiopsis, Pichia, Ramichloridium, Rhinocladiella, Scedosporium, Stachybotrys and Tritirachium. Sexual morphs were represented by four genera, Candida (2 species, 1.82%) Eupenicillium (1 species, 0.91%), Neosartorya (1 species) and Tritirchium (1 species).
Figure 2. Relative abundance of sponge-associated fungal genera isolated in mangrove-attached sponges collected from New Washington, Aklan, Philippines.
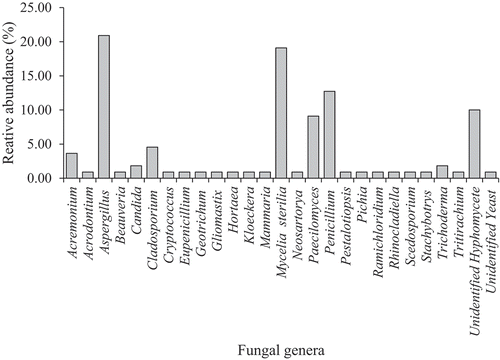
Based on the total frequency of occurrence (%), Acrodontium cf. crateriforme (11.39%) and Aspergillus niger (8.23%) were reported as very frequent and frequent species, respectively (). Seventeen species (Aspergillus sydowii, Aspergillus cf. fumisynnematus, Aspergillus sp. 11, Sect. Terreus, Candida guilliermondii, Hyphomycete 5, Hyphomycete 9, Mycelia sterilia 1, Mycelia sterilia 2, Mycelia sterilia 21, Paecilomyces cf. persicinus, Paecilomyces cf. roseolus, Paecilomyces victoriae, Penicillium cf. janthinellum, Penicillium chrysogenum, Penicillium sp. 3, Penicillium sp. 6, Tritirachium oryzae) were recorded as infrequent. Other 91 species were recorded as rare.
The mangrove sponge Halichondria cf. panicea harboured the most fungal isolates with 54 species, followed by Axinella sp. (45 isolates), Haliclona sp. (33 species) and Tedania sp. (27 species) (). Axinella sp. had the highest Shannon index of diversity value (H’: 3.57) while Haliclona sp. had the highest Shannon (J’) and Simpson (E1/D) index of evenness with a value of 0.95 and 1.22, respectively. Halichondria cf. panicea had the highest Simpson index of dominance (d’) with a value of 0.06.
The Jaccard’s coefficient of similarity (J’), based on the presence or absence of each fungus, was calculated among different sponge species to compare the composition of fungi on each sponge host (). Based on pair-wise comparison of similarities of 110 fungal isolates on four sponges, the similarity index was highest between Halichondria cf. panicea vs Axinella sp. (0.22) followed by both Halichondria cf. panicea vs Tedania sp. and Halichondria cf. panicea vs Haliclona sp. with the J’ value of 0.19. It was least between Axinella sp. and Haliclona sp. with J’ value of 0.13.
Table 2. Jaccard’s coefficient of similarity (J’) for fungal species among sponge species.
Association of fungi on the different mangrove-attached sponges
Three classifications were done based on the presence of fungal genera in certain number of sponge species, as adapted from Li and Wang (Citation2009). “Sponge-generalists” are genera that can be found in all sponge species and results showed that, the genera Acrodontium, Aspergillus, Candida, Paecilomyces and Penicillium could be classified under this group. The genera Acremonium, Cladosporium, Hortaea and Trichoderma are classified as sponge-associates since they were identified on more than one sponge. The “sponge-specialists” would include the genera Beauveria, Cryptococcus, EuPenicillium, Geotrichum, Gliomastix, Kloeckera, Mammaria, Neosartorya, Pestalotiopsis, Pichia, Ramichloridium, Rhinocladiella, Scedosporium, Stachybotrys and Tritirachium.
Fungal load of mangrove-associated sponges
The total fungal load of the four species of mangrove-associated sponges yielded varying counts on the various culture media used (). For Halichondria cf. panicea, the highest fungal density was recorded from cornmeal agar and lowest value from potato dextrose agar. The highest CFU in Axinella sp. was recorded from CMA and lowest fungal load from MBA. MBA had the highest fungal load in Tedania sp. and lowest value from CMA. For Haliclona sp., RBACl had the highest fungal load value and lowest CFU from MBA.
Table 3. Fungal density (CFU g-1) of different mangrove-associated sponges in five culture media.
Halichondria cf. panicea had the highest fungal load value (7.13 × 102) while the lowest CFU/g was recorded from Axinella sp. (6.00 × 101)
Discussion
Fungal diversity in mangrove-associated sponges
The study on the ecological role, including its diversity and association, of fungi on marine sponges are still scarce and data were largely generated due to the diversity of novel bioactive metabolites produced with promising biotechnological applications (Höller et al. Citation2000; Bugni and Ireland Citation2004; Konig et al. Citation2006; Wang Citation2006; Raghukumar Citation2008; Aly et al. Citation2011; Debbab et al. Citation2011; Jones Citation2011). Fungi were mostly isolated from subtidal sponges but there is no published information on the fungal associates of mangrove-associated sponges. To date, only bacterial communities from mangroves sponges collected from the Caribbean Sea were reported (Yang et al. Citation2011). Information on the role of fungi in mangrove sponges has not yet been studied but some mycologists suggest that the role of fungi in sponges include nutrient transfer and chemical defence (Bugni and Ireland Citation2004; Taylor et al. Citation2007; Ding et al. Citation2011). On the other hand, sponge-associated bacteria enhance the endemism of this invertebrate by degrading mangrove-derived DOM and other organic compounds which is important in organic matter assimilation leading to the survival of mangrove species and the exclusion of typical reef species (Hunting et al. Citation2010, Citation2013).
The present study gives an insight on the diversity of fungi associated with mangrove sponges. The fungal communities of mangrove sponges were composed mainly of asexual morphs with hyphomycetes represented by 87 species. Previous studies also observed asexual morphs dominating the fungal assemblages of subtidal sponges (Wang et al. Citation2008; Ding et al. Citation2011; Thirunavukkarasu 2012). The genera Aspergillus (21.52%), Penicillium (14.24%) and Acrodontium (11.39%) were recorded as very frequent while Paecilomyces (9.81%) was a frequently occurring genus (). Infrequently occurring genera include Cladosporium (3.16%), Acremonium (2.53%), Trichoderma (2.53%) and Candida (1.90%). Other genera were recorded as rare. It includes Beauveria and Tritirachium where previous works (Höller et al. Citation2000; Paz et al. Citation2010; Wiese et al. Citation2011) considered it also to be isolated rarely from marine sponges.
These mangrove sponges were collected in the same location but the composition of fungal genera differs from one another except Acrodontium, Aspergillus, Candida, Paecilomyces and Penicillium that were isolated in all sponge species. The genera Beauveria, Cryptococcus, Eupenicillium, Kloeckera, Pichia, Stachybotrys were only isolated in Tedania sp.; Geotrichum and Gliomastix in Axinella sp.; Mammaria, Neosartorya, Tritirachium in Haliclona sp.; Pestalotiopsis, Ramichloridium, Rhinocladiella, Scedosporium in Halichondria cf. panicea. The differences in the fungal composition on the different sponge species suggest host-preference of the different fungal taxa. Furthermore, the results in this study and previous works in subtidal sponges (Höller et al. Citation2000; Wang Citation2006; Wang et al. Citation2008; Liu et al. Citation2010; Yu et al. Citation2012) showed that the differences in the fungal composition and its diversity may be attributed to the species of sponges with various morphological structures. Ding et al. (Citation2011), on his work on South China Sea sponges (Clathrina luteoculcitella and Holoxea sp.) sampled in the same location, also observed this wherein orders Agaricales, Boliniales, Microascales, Mucorales, Pleosporales, Saccharomycetales and Xylariales were only isolated from sponge Clathrina luteoculcitella but not in Holoxea sp. Thus, even the sponges were collected on the same location, they harbour different isolates which suggested that these isolates were not spores from seawater column and trapped during the filter feeding process of sponges. Previous studies by Gao et al. (Citation2008), Li and Wang (Citation2009) and Jin et al. (Citation2014) demonstrated that fungal communities isolated from sponges differ from the surrounding water. For example, Penicillium janthinellum, Fusarium solani and P. chrysogenum which were isolated from seawater samples but not present within sponges (Li and Wang Citation2009). However, it is insufficient to disprove sponge-specific nature of a microbe by merely proving the presence of microbe outside a sponge. A predator or storm for example may damage sponge and microbes associated with it may disintegrate and spread into the seawater column (Taylor et al. Citation2007).
Diversity of fungi associated with marine sponges remains an understudied area and more evidence is required to elucidate their possible ecological role (Gao et al. Citation2008; Wang et al. Citation2008). The present investigation does not show any direct evidence that the isolated fungi have been actively growing on the sponge tissues. As a result, the difficulty also arises on how to determine whether they are sponge-symbiotic fungi or not. So far, there is little evidence regarding the symbiotic relationship between sponges and fungi. For example, Maldonado et al. (Citation2005) showed direct evidence of sponge-endosymbiotic yeasts in a marine sponge Chondrilla sp. that is transmitted maternally through fertilised eggs based using immunocytochemical technique to label the β-1,4-N-acetyl-D-glucosamine residues of chitin walls. There is also indirect evidence of the putative fungal original intron in a sponge Tetilla sp., as observed by Rot et al. (Citation2006), perhaps because of horizontal gene transfer. In addition, marine ascomycetes of the genus Koralionastes have been reported to be in some way associated with crustaceous sponges wherein it forms fruiting bodies only in close association with the sponges associated with corals (Kohlmeyer and Volkmann-Kohlmeyer Citation1990). Furthermore, Perovic-Ottstadt et al. (Citation2004) demonstrated the presence of receptor proteins for fungal cell wall components (e.g. (1→3)-β-d-glucan-binding proteins), in the marine sponge Suberetis domuncula, which indicates that sponges are biochemically equipped for dealing with fungi. Several bacteria and archaea, along with the ubiquitous fungus Penicillium, as reported by Simister et al. (Citation2012), are symbionts of sponges. Using immunocytochemistry, transmission electron microscopy (TEM) technique and non-cultivation-dependent analysis, the real association between fungi and marine sponges will be confirmed including vital role or functions of fungi in the sponge (Maldonado et al. Citation2005; Passarini et al. Citation2013). Höller et al. (Citation2000) proposed that investigation of a larger number of samples and surrounding water should be done to determine if sponge-associated fungi are not terrestrial fungi filtered from the surrounding waters but adapted to the marine habitat and on its host.
Even employing diverse culture media, 21 species remained sterile. Höller et al. (Citation2000) also isolated 37 strains of Mycelia sterilia in 14 sponges even using diverse culture media and culture conditions to induce sporulation of fungi. Molecular analysis can be of great help to identify fungi with no reproductive structures (e.g. conidia and ascomata) Furthermore, 11 species of hyphomycetes remained unidentified and requires further investigation including molecular analysis for identification.
Comparison of culturable fungal diversity of sponges
In this study, the genera Acrodontium, Aspergillus, Candida, Paecilomyces and Penicillium were considered sponge-generalists while Acremonium, Cladosporium, Hortaea and Trichoderma are classified as sponge-associates. The sponge-specialists include the genera Beauveria, Cryptococcus, Eupenicillium, Geotrichum, Gliomastix, Kloeckera, Mammaria, Neosartorya, Pestalotiopsis, Pichia, Ramichloridium, Rhinocladiella, Scedosporium, Stachybotrys and Tritirachium. Aspergillus, Penicillium and Eupenicillium were considered by Li and Wang (Citation2009) as “sponge-generalists” but Eupenicillium was “sponge-associate” in this study. Candida, on the other hand, was “sponge-generalist” but it was “sponge-specialist” on the study of Li and Wang (Citation2009). Trichoderma was considered “sponge-associate” in this study and this is in line with result of Höller et al. (Citation2000) but Menezes et al. (Citation2010) classified this as “sponge-generalist”. shows the summary of fungal genera associated with marine sponges recovered from various locations. The genera Cladosporium (39 sponge species), Aspergillus (47 sponge species), Penicillium (53 sponge species), Acremonium (41 sponge species) and Trichoderma (29 sponge species) were common marine fungi recovered from the marine sponges and can be considered sponge-generalists (Höller et al. Citation2000; Morrison-Gardiner Citation2002; Gesner et al. Citation2005; Proksch et al. Citation2008; Wang et al. Citation2008; Ein-Gil et al. Citation2009; Li and Wang Citation2009; Liu et al. Citation2010; Paz et al. Citation2010; Ding et al. Citation2011; Thirunavukkarasu et al. Citation2012).
Table 4. Fungal genera associated with sponges recovered in different locations (genus level).
It is difficult to suggest that the present study isolated sponge-specialist based on the differences of fungi recovered from four mangrove sponges because there is no direct evidence and there are limited studies on fungal associates of mangrove sponges. Extensive survey of fungi in more species of sponges including comparison on the same species in this study but different geographical locations and using biochemical and molecular methods (e.g. 454 pyrosequencing) could reveal the sponge-fungal association.
In addition, the isolated fungal genera are common to terrestrial habitats, suggesting that these isolates may also be of terrestrial origin and can be considered, based on the definition of Kohlmeyer and Kohlmeyer (Citation1979), facultative marine fungi but on the latest definition of Pang et al. (Citation2016), these isolates were considered marine fungi. Marine fungi, as defined by Pang et al. (Citation2016), are fungi that are recovered repeatedly from marine habitats because: (1) it can grow and/or sporulate (on substrata) in marine environments; (2) it forms symbiotic relationships with other marine organisms or (3) it is shown to adapt and evolve at the genetic level or be metabolically active in marine environments. If we based on the list of marine fungi by Jones et al. (Citation2015), 16 species were considered marine fungi that include 9 species of Aspergillus (A. candidus, A. niger, A. ochraceus, A. cf. penicilloides, A. restrictus, A. sclerotiorum, A. sydowii, A. tamarii, A. terreus), 3 species of Penicillium (P. cf. citreonigrum, P. cf. citrinum, P. spinulosum), 1 Trichoderma species (T. aureoviride), 1 species of Candida (C. guilliermondii) and 2 species of Cladosporium (C. cladosporioides, C. sphaerospermum).
Environment-dependent fungal diversity
The results of the present work and earlier studies show that the diversity sponge-associated fungi are more dependent on the surrounding environment where the sponge species thrives. For instance, no ascomycetes were isolated in mangrove sponge Halichondria cf. panicea collected from Aklan, Philippines while previous works of Höller et al. (Citation2000) in Helgoland, Germany and Pivkin et al. (Citation2006) in Sakhalin Island, Russia recovered ascomycetes in the subtidal sponge Halichondria panicea. Both present work and other published studies on Halichondria harbours Acremonium, Aspergillus, Penicillium, Mycelia sterilia and Trichoderma. Only Höller et al. (Citation2000) isolated Mucor, a zygomycete. Furthermore, Flemer (Citation2013) and Bolaños et al. (Citation2015) recovered ascomycetes in subtidal sponges under the genus Axinella while no ascomycetes were isolated from mangrove sponge Axinella sp. collected in Aklan, Philippines. Only Cladosporium was isolated in the present work and two former studies in Axinella dissimilis (Flemer Citation2013) and Axinella sp. 1 (Bolaños et al. Citation2015). There is no similar species from the present work were recovered in Axinella sp. 2 and Axinella sp. 3 (Bolaños et al. Citation2015). Furthermore, between the sponge Haliclona simulans, collected from the coastal waters of Ireland (Baker et al. Citation2009), and Hainan Province of China (Liu et al. Citation2010). The fungal diversity between two different regions is quite different. For instance, the orders Capnodiales, Dothideales, Agaricostilbales, Wallemiales, which were present in the “Hainan” sample are not found in the “Irish” sample. In the “Irish” sample, fungi under the orders Chaetosphaeriales, Chaetothyrailes, Helotiales, Mucorales and Agaricomycotina, were isolated but were absent in the “Hainan” sample. Furthermore, no shared identical fungal species were observed in the two collections. The results of the comparison of the abovementioned studies supports the notion that being filter feeders, sponges enrich various fungal species from the surrounding seawater that are merely washed into the sea from their terrestrial habitats and just happen to survive in their “host organisms” (Höller et al. Citation2000; Taylor et al. Citation2007; Proksch et al. Citation2008; Wang et al. Citation2008; Liu et al. Citation2010; Wiese et al. Citation2011; Zhou et al. Citation2011). These remain dormant until plated onto a suitable culture medium (Wang et al. Citation2008). If such has been the case, the metabolic activities of fungi from sponges should be the same as that of those in other terrestrial environments. Surprisingly, these facultative marine fungi produce novel compounds that are different and not produced from their terrestrial conspecifics (Proksch et al. Citation2003; Konig et al. Citation2006; Wang Citation2006). Secondary metabolites in producing fungi play an important role in ecological interactions with other organisms allowing it to survive in its ecological niche while on its host, it enhance the defence mechanisms against pathogens and predators (Fox and Howlett Citation2008; Thomas et al. Citation2010).
Conclusion
The fungal composition differs in each species of mangrove sponges even though they were collected in the same location suggesting that the isolates recovered were not merely seawater contamination and suggest sponge-preference by various fungal taxa that can be classified as true marine fungi. The development of marine fungi on these hosts appeared to be strongly influenced by the characteristics or nature of immediate environment.
Acknowledgment
This work was supported by the Korea Institute of Ocean Science and Technology (KIOST), Department of Science and Technology – Science Education Institute (DOST-SEI) and University of the Philippines Visayas – Office of the Vice Chancellor for Research and Extension (UPV-OVCRE). The authors are grateful to the Department of Science and Technology (DOST) - Accelerated Science and Technology Human Resource Development Program (ASTHRDP) for providing the scholarship to Mr. Calabon. The support of the researchers and staff of the different laboratories (UPV-NIMBB, Mycology Laboratory of the Division of Biological Sciences (DBS) and OceanBio and MarineBio Laboratories - DBS) is also highly appreciated.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aly AH, Debbad A, Proksch P. 2011. Fifty years of drug discovery from fungi. Fungal Divers. 50:3–19.
- Amagata T, Minoura K, Numata A. 2006. Gymnastatins F–H, cytostatic metabolites from the sponge-derived fungus Gymnascella dankaliensis. J Nat Pord. 69:1384–1388.
- Baker PW, Kennedy J, Dobson ADW, Marchesi JR. 2009. Phylogenetic diversity and antimicrobial activities of fungi associated with Haliclona simulans isolated from Irish coastal waters. Mar Biotechnol. 11:540–547.
- Barnett HLHunter BB. 1998. Illustrated Genera of Imperfect Fungi. 4th ed. Minneapolis, MN: Burgess.
- Blunt JW, Copp BR, Hu WP, Munro MH, Northcote PT, Prinsep MR. 2009. Marine natural products. Nat Prod Rep. 26:170–244.
- Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. 2012. Marine natural products. Nat Prod Rep. 29(2):144–222.
- Bolaños J, De León LF, Ochoa E, Darias J, Raja HA, Shearer CA, Miller AN, Vanderheyden P, Porras-Alfaro A, Caballero-George C. 2015. Phylogenetic diversity of sponge-associated fungi from the Caribbean and the Pacific of panama and their in vitro effect on angiotensin and endothelin receptors. Mar Biotechnol. 17(5):533-564.
- Bugni TS, Ireland CM. 2004. Marine derived fungi: a chemically and biologically diverse group of microorganisms. Nat Prod Rep. 21:143–163.
- Caballero-George C, Bolaños J, Ochoa E, Carballo JL, Cruz JA, Arnold AE. 2010. Protocol to isolate sponge-associated fungi from tropical waters and an examination of their cardioprotective potential. Curr Trends Biotechnol Pharm. 4(4):881–899.
- Campbell CK, Johnson EM, Warnock DW. 2013. Identification of pathogenic fungi. West Sussex (UK): John Wiley.
- Debbab A, Aly AH, Proksch P. 2011. Bioactive secondary metabolites from endophytes and associated marine derived fungi. Fungal Divers. 49:1–12.
- Ding B, Yin Y, Zhang F, Li Z. 2011. Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Holoxea sp. in the South China Sea. Mar Biotechnol. 13:713–721.
- Domsch KH, Gams W, Anderson TH. 2007. Compendium of soil fungi. 2nd ed. Eching (Germany): IHW-Verlag.
- Eamvijarn A, Gomes NM, Dethoup T, Buaruang J, Manoch L, Silva A, Pedro M, Marini I, Roussis V, Kijjoa A. 2013. Bioactive meroditerpenes and indole alkaloids from the soil fungus Neosartorya fischeri (KUFC 6344), and the marine-derived fungi Neosartorya laciniosa (KUFC 7896) and Neosartorya tsunodae (KUFC 9213). Tetrahedron. 69(40):8583–8591.
- Ein-Gil N, Ilan M, Carmeli S, Smith GW, Pawlik JR, Yarden O. 2009. Presence of Aspergillus sydowii, a pathogen of gorgonian sea-fans in the marine sponge Spongia obscura. ISME J. 3:752–755.
- Fieseler L, Horn M, Wagner M, Hentschel1 U. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl Environ Microbiol. 70(6):3724–3732.
- Flemer B 2013. Antimicrobial activities and diversity of sponge derived microbes [dissertation]. Ireland (IE): University of College Cork.
- Fox FM, Howlett BJ. 2008. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 11(6):481–487.
- Gao Z, Li B, Zheng C, Wang G. 2008. Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol. 74:6091–6101.
- Gesner S, Cohen N, Ilan M, Yarden O, Carmeli S. 2005. Pandangolide 1a, a metabolite of the sponge-associated fungus Cladosporium sp., and the absolute stereochemistry of pandangolide 1 and iso-cladospolide B. J Nat Prod. 68:1350–1353.
- Gomes NM, Bessa LJ, Buttachon S, Costa PM, Buaruang J, Dethoup T, Silva AM, Kijjoa A. 2014. Antibacterial and antibiofilm activities of tryptoquivalines and meroditerpenes isolated from the marine derived fungi Neosartorya paulistensis, N. laciniosa, N. tsunodae, and the soil fungi N. fischeri and N. siamensis. Mar Drugs. 12(2):822–839.
- He L, Liu F, Karuppiah V, Ren Y, Li Z. 2014. Comparisons of the fungal and protistan communities among different marine sponge holobionts by pyrosequencing. Microb Ecol. 67(4):951–961.
- Henriquez M, Vergara K, Norambuena J, Beiza A, Maza F, Ubilla P, Araya I, Chavez R, San-Martin A, Darias J, et al. 2013. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J Microbiol Biotechnol. 30(1):65–76.
- Hentschel U, Usher KM, Taylor MW. 2006. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 55:167–177.
- Höller H, Wright AD, Matthhee GF, Konig G, Draeger MS, Aust HJ, Schulz B. 2000. Fungi from marine sponges: diversity, biological activity and secondary metabolites. Mycol Res. 104:1354–1365.
- Howard DH. 2003. Pathogenic fungi in humans and animals. 2nd ed. New York (NY): Marcel Dekker.
- Hunting ER, de Goeij JM, Asselman M, van Soest RWM, van der Geest HG. 2010. Degradation of mangrove-derived organic matter in mangrove associated sponges. Bull Mar Sci. 86(4):871–877.
- Hunting ER, Franken O, Knopperts F, Kraak MHS, Vargas R, Roling WFM, van Ser Geest HG. 2013. Substrate as a driver of sponge distributions in mangrove ecosystems. Mar Ecol Prog Ser. 486:133–141.
- Hyde KD. 1989. Ecology of tropical marine fungi. Hydrobiologia. 178:199–208.
- Hyde KD, Sarma VV. 2000. Pictorial key to higher marine fungi. In: Hyde KD, Pointing SD, eds. Marine mycology: a practical approach. Hong Kong (HK): Fungal Diversity Press. p. 205–270.
- Jin L, Liu F, Sun W, Zhang F, Karuppiah V, Li Z. 2014. Pezizomycotina dominates the fungal communities of South China Sea Sponges Theonella swinhoei and Xestospongia testudinaria. FEMS Microbiol Ecol. 90:935–945.
- Johnson TWJ, Sparrow FKJ. 1961. Fungi in oceans and estuaries. Weinheim: J. Cramer.
- Jones EBG. 2011. Are there more marine fungi to be described? Bot Mar. 54:343–354.
- Jones EBG, Pang KL. 2012. Marine fungi and fungal-like organisms. Berlin: Walter de Gruyter.
- Jones EBG, Suetrong S, Sakayaroj J, Bahkali AH, Abdel-Wahab MA, Boekhout T, Pang KL. 2015. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 73(1):1–72.
- Kamke J, Rinke C, Schwientek P, Mavromatis K, Ivanova N, Sczyrba A, Woyke T, Hentschel U. 2014. The candidate phylum Poribacteria by single-cell genomics: new insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS ONE. 9(1):e87353.
- Kohlmeyer J. 1984. Tropical marine fungi. Mar Ecol. 5:329–378.
- Kohlmeyer J, Kohlmeyer E. 1979. Marine mycology: the higher fungi. New York: Academic Press.
- Kohlmeyer J, Volkmann-Kohlmeyer B. 1990. New Species of Koralionastes (Ascomycotina) from the Caribbean and Australia. Can J Bot. 68:1554–1559.
- Kohlmeyer J, Volkmann-Kohlmeyer B. 1991. Illustrated key to the filamentous higher marine fungi. Bot Mar. 34:1–35.
- Konig GM, Kehraus S, Seiber SF, Abdel-Lateff A, Muller D. 2006. Natural products from marine organisms and their associated microbes. Chem Bio Chem. 7:229–238.
- Kurtzman CP, Fell JW. 1998. The yeast, a taxonomic study. 4th ed. Amsterdam (NL): Elsevier.
- Li Q, Wang G. 2009. Diversity of fungal isolates from three Hawaiian marine sponges. Microbiol Res. 164:233–241.
- Liu WC, Li CQ, Zhu P, Yang JL, Chen KD. 2010. Phylogenetic diversity of culturable fungi associated with two marine sponges: haliclona simulans and Gelliodes carnosa, collected from the Hainan Island coastal waters of the South China Sea. Fungal Divers. 42:1–15.
- Ludwig JA, Reynolds JF. 1988. Statistical ecology: a primer on methods and computing. Totonto (CA): Wiley.
- Mahyudin NA 2008. Actinomycetes and fungi associated with marine invertebrates: a potential source of bioactive compounds [dissertation]. New Zealand: University of Canterbury.
- Maldonado M, Cortadellas N, Trillas MI, Rutzler K. 2005. Endosymbiotic yeast maternally transmitted in a marine sponge. Biolog Pharma Bull. 209:94–106.
- Meenupriya J, Thangaraj M. 2010. Isolation and molecular characterization of bioactive secondary metabolites from Callyspongia spp. associated fungi. Asian Pac J Trop Med. 3(9):738–740.
- Menezes CBA, Bonugli-Santos RC, Miqueletto PB, Passarini MRZ, Silva CHD, Justo MR, Leal RR, Fantinatti-Garboggini F, Oliveira VM, Berlinck RGS, et al. 2010. Microbial diversity associated with algae, ascidians and sponges from the north coast of Sao Paulo state, Brazil. Microbiol Res. 165:466–482.
- Morrison-Gardiner S. 2002. Dominant fungi from Australian coral reefs. Fungal Divers. 9:105–121.
- Namikoshi M, Akano K, Kobayashi H, Koike Y. 2002. Distribution of marine filamentous fungi associated with marine sponges in coral reefs of Palau and Bunaken Island, Indonesia. J Tokyo Univ Fish. 88:15–20.
- Pang KL, Overy D, Jones EBG, Calado MDL, Burgaud G, WAlker AK, Johnson JA, Kerr RG, Cha HJ, Bills JF. 2016. ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: toward a new consensual definition. Fungal Biol Rev. 30(4):163-175.
- Passarini MR, Santos C, Lima N, Berlinck RG, Sette LD. 2013. Filamentous fungi from the Atlantic marine sponge Dragmacidon reticulatum. Arch Microbiol. 195(2):99–111.
- Paz Z, Komon-Zelazowska M, Druzhinina IS, Aveskamp MM, Shnaiderman A, Aluma Y, Carmeli S, Ilan M, Yarden O. 2010. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 42:17–26.
- Perovic-Ottstadt S, Adell T, Proksch P, Wiens M, Korzhev M, Gamulin V, Mueller IM, Mueller WEG. 2004. A (1β3)-βD-glucan recognition protein from the sponge Suberites domuncula: mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Europe J Biochem. 271(10):1924–1937.
- Pitt JI, Hocking A. 2009. Fungi and food spoilage. 3rd ed. New York (NY): Springer.
- Pivkin MV, Aleshko SA, Krasokhin VB, Khudyakov YV. 2006. Fungal assemblages associated with sponges of the Southern Coast of Sakhalin Island. Russian J Mar Biol. 32(4):207–213.
- Proksch P, Ebel R, Edrada R, Riebe R, Liu H, Diesel A, Bayer M, Li X, Lin WH, Grebenyuk V, et al. 2008. Sponge-associated fungi and their bioactive compounds: the Suberites case. Bot Mar. 51:209–218.
- Proksch P, Ebel R, Edrada RA, Schupp P, Lin WH, Sudarsono WV, Steube K. 2003. Detection of pharmacologically active natural products using ecology. Selected examples from Indo Pacific marine invertebrates and sponge-derived fungi. Pure Appl Chem. 75:343–352.
- Raghukumar C. 2008. Marine fungal biotechnology: an ecological perspective. Fungal Divers. 31:19–35.
- Riddell RW. 1950. Permanent stained mycological preparation obtained by slide culture. Mycologia. 42:265–270.
- Rot C, Goldfarb I, Ilan M, Huchon D. 2006. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol Biol. 6:71.
- Sarma VV, Raghukumar S. 2013. Manglicolous fungi from Chorao mangroves, Goa, West coast of India: diversity and frequency of occurrence. Nova Herwidgia. 97(3–4):533–542.
- Simister RL, Deines P, Botté ES, Nicole S, Webster NS, Taylor MW. 2012. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol. 14:517–524.
- Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Molec Biol Rev. 71(2):295–347.
- Thirunavukkarasu N, Suryanarayanan TS, Girivasan KP, Venkatachalam A, Geetha V, Ravishankar JP, Mukesh D. 2012. Fungal symbionts of marine sponges from Rameswaram, southern India: species composition and bioactive metabolites. Fungal Divers. 55:37–46.
- Thomas TRA, Kavlekar DP, LokaBharathi PA. 2010. Marine drugs from sponge-microbe association- a review. Mar Drugs. 8(4):1417–1468.
- Vasanthabharathi VJayalakshmi S. 2012. Bioactive potential of symbiotic bacteria and fungi from marine sponges. African J Biotechnol. 11(29):7500–7511.
- Wang G. 2006. Diversity and biotechnological potential of the sponge-associated microbial consortia. J Ind Microbiol Biot. 33:545–551.
- Wang G, Li Q, Zhu P. 2008. Phylogenetic diversity of culturable fungi associated with the Hawaiian Sponges Suberites zeteki and Gelliodes fibrosa. Anton Leeuw. 93:163–174.
- Watanabe T. 2010. Pictorial atlas of soil and seed fungi: morphologies of cultured fungi and key to species. 3rd ed. Boca Raton (FL): Taylor and Francis.
- Wiese J, Ohlendorf B, Blümel M, Schmaljohann R, Imhoff JF. 2011. Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Mar Drugs. 9:561–585.
- Yang J, Sun J, Lee OO, Wong YM, Qian PY. 2011. Phylogenetic diversity and community structure of sponge-associated bacteria from mangroves of the Caribbean Sea. Aquat Microb Ecol. 62:231–240.
- Yu Z, Zhang B, Sun W, Zhang F, Li Z. 2012. Phylogenetically diverse endozoic fungi in the South China Sea sponges and their potential in synthesizing bioactive natural products suggested by PKS gene and cytotoxic activity analysis. Fungal Divers. 58(1):127-141.
- Zhang Y, Mu J, Feng Y, Kang Y, Zhang J, Gu P, Wang Y, Ma L, Zhu Y. 2009. Broad-spectrum antimicrobial epiphytic and endophytic fungi from marine organisms: isolation, bioassay and taxonomy. Mar Drugs. 7:97–112.
- Zhou K, Zhang X, Zhang F, Li Z. 2011. Phylogenetically diverse cultivable fungal community and polyketide synthase (PKS), non-ribosomal peptide synthase (NRPS) genes associated with the South China Sea sponges. Microb Ecol. 62:644–654.