ABSTRACT
Medicinal plants are considered as sources of novel and unexplored groups of endophytic microorganisms. A study on endophytic fungal species from the medicinal plant Withania somnifera (L.) Dunal resulted in the isolation of a Penicillium isolate (WSR 62) with antibiotic activity. Phylogenetic analysis showed that the isolate belongs to section Lanata-divaricata, and it is most closely related to P. javanicum. Subsequent detailed phylogenetic analyses using partial β-tubulin (BenA), calmodulin (CaM) and DNA-dependent RNA polymerase II (RPB2) gene sequences of a larger number of related strains revealed the distinctiveness of the isolate in the P. javanicum-clade. The isolate grows fast on Czapek yeast autolysate agar (CYA) and malt extract agar (MEA) incubated at 25°C, 30°C and 37°C. The obverse colony colour is dominated by the conspicuous production of cleistothecia and is greyish yellow on CYA and yellowish brown on MEA. Production of cleistothecia containing prominent spinose ascospores was present on all tested agar media. Based on the phylogenetic analysis and the phenotypic characterisation, strain WSR 62 from Withania is described here as a novel species named Penicillium setosum.
1. Introduction
Endophytes are microorganisms living within a plant throughout its life cycle or complete at least a part of its life cycle without causing any apparent disease in the host plant (Petrini Citation1991). They are ubiquitous and can be involved in neutral, commensal and/or beneficial associations with the host plant and tend to become opportunistic saprobes or pathogens at a later phase of their life cycle (Compant et al. Citation2016). The endophytic mode of life helps them to enhance their dispersal and attain suitable substrates and environmental conditions for their survival (Thomas et al. Citation2017). Their associations with their host plant enable them to establish a microbial community in the plant endosphere (Hardoim et al. Citation2015). Such colonisation of endophytes offers a unique biological niche favouring their growth and grants them a distinctive ability to produce new bioactive metabolites. Hence, exhaustive searches are being made for discovering their diversity and usefulness in agriculture, environment and medicine (Jia et al. Citation2016).
Penicillium is one of the most common and diverse fungal genera, has a worldwide distribution and contains both beneficial and harmful members. This genus currently includes 429 accepted species (Visagie et al. Citation2014, www.aspergilluspenicillium.org). Species of this economically important genus are commercial sources of antibiotics, organic acids, alcohols, enzymes, pharmaceuticals, and other metabolites (Frisvad et al. Citation2004). Nonetheless, some of them cause food spoilage, produce mycotoxins, and cause human and animal diseases (Pitt Citation1994). The predominance of Penicillium in nature is due to their ability to thrive in multiple ecological niches such as soil, decomposing organic materials, food and feed, and marine environment. Some species of Penicillium follow an endophytic mode of existence and can reside on, e.g. rhizosphere, laminosphere, and phyllosphere. Penicillium features a prominent position among the culturable endophytic microbes (Potshangbam et al. Citation2017). For example, Penicillium cf. glaucoalbidum from section Thysanophora (previously the genus Thysanophora) is often isolated from Picea rubens (Sieber-Canavesi & Sieber Citation1993; Müller & Hallaksela Citation1998; Koukol et al. Citation2012). Some other reports for the endophytic occurrence of this genus are Penicillium coffeae from Coffea arabica (Peterson et al. Citation2005), P. citrinum from Ixeris repenes (Khan et al. Citation2008), P. goetzii from Pinus ponderosa and Pseudotsuga menziesii (Houbraken et al. Citation2012), P. glabrum from Punica granatum (Hammerschmidt et al. Citation2012), and P. fluviserpens from coffee plant (Peterson et al. Citation2015). Mechanisms behind invasion, establishment, endophytic association and also beneficial role or harmfulness of this genus are not yet clear (Compant et al. Citation2016).
Withania somnifera (common name: Ashwagandha, Family: Solanaceae) is a well-known medicinal plant found naturally in arid wastelands of the tropics. It is a perennial shrub growing up to 30 cm to 1.5 m in height. The whole plant including root, leaves, and berries has been used in the Ayurveda system of indigenous medicine for centuries as a Rasayana herb (Ven Murthy et al. Citation2010). Many bioactive substances are extracted from this plant and used for their immunomodulatory, anti-stress and anti-tumour activity (Bharti et al. Citation2016). The ayurvedic formulations of this plant exhibit multiple pharmacological actions, like general tonic, sedative, liver tonic and anti-inflammatory activity. Studies also showed that leaves and roots of Ashwagandha have antibacterial properties (Owais et al. Citation2005). Nevertheless, only scanty reports are available regarding the endophytes present in this plant.
In our search for endophytic fungi from W. somnifera, with the ability to produce antimicrobial compounds, we encountered a novel Penicillium species. The present study aims to describe this endophytic Penicillium isolate from the surface-sterilized roots of W. somnifera based on phenotypic and phylogenetic analysis.
2. Materials and methods
2.1. Collection of Withania somnifera
Withania somnifera (L.) Dunal () plantlets were collected from the nursery of Aromatic and Medicinal Plants Research Station (Kerala Agricultural University), Odakali, Ernakulam District, Kerala, India and a voucher specimen, RHK 6350, was deposited at the Regional Herbarium of Kerala, St. Berchman’s College, Kerala, India.
2.2. Isolation
Healthy, fully grown plants were used for the isolation of the endophytes. Plants were uprooted, brought to the laboratory in sealed sterile polythene bags and processed immediately. Surface sterilization of plant roots was performed according to a modified method of George et al. (Citation2015). Plant parts were washed in running tap water for 10 min followed by washing with Tween 40 (1%) solution. The detergent was completely removed by washing under running tap water. Thereafter, sequential rinsing of the plant material was done with 70% ethanol for 1 min, 4% sodium hypochlorite (NaOCl) for 2–3 min and again with 70% ethanol for 30 s. Final rinses were made with three changes of sterile distilled water, with total wash duration of 3 min. Successful surface sterilization was ensured by pressing randomly chosen plant parts onto culture plates for a few seconds and looking for any microbial growth for a period of 2 weeks. Surface-sterilised plant roots were cut into small pieces (≈1 cm) and each piece was placed on potato dextrose agar (PDA), malt extract agar (MEA) and yeast extract glucose agar (YEGA) supplemented with streptomycin (3 mg/100 mL) (HiMedia laboratories, Mumbai, India). Plates were incubated at 26 ± 2°C, till the appearance of fungal mycelia (2–3 weeks). A total of 73 fungal isolates were obtained. They were purified, inoculated onto PDA slants and kept as stock cultures at 4°C. Among these isolates, a Penicillium isolate (WSR 62) with antibacterial activity was studied in detail.
2.3. DNA extraction, sequencing and phylogenetic analysis
The total genomic DNA of WSR 62 grown in potato dextrose broth was isolated by using the fungal genomic DNA minispin kit RKN 19 (Chromous Biotech, India). The internal transcribed spacer (ITS) was amplified from the isolated fungal genomic DNA using the universal ITS primers (ITS1 and ITS4) (White et al. Citation1990). Additionally, partial β-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (regions 5–7) (RPB2) genes were amplified, using the PCR conditions and primers as described by Visagie et al. (Citation2014). PCR reaction volumes of 50 µL contained 2 µL (10 pmol/µL) of each primer, 5 µL of DNA template (30 ng) and 25 µL of 2X OrionX Taq PCR smart mix (Origin, India). Amplification was carried out in a Bio-Rad thermocycler (Bio-Rad, USA) programmed with an initial denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 45 s, with a final extension at 72°C for 10 min. The PCR product was electrophoresed on 1.2% agarose gel in 0.5X TBE buffer at 65 V for 1 h to check the amplicons. The Illustra GFX PCR DNA and gel band purification kit (GE Healthcare, USA) was used for the rapid purification and concentration of PCR products. Sequencing of the amplicons was done by Big dye terminator v3.2 cycle sequencing chemistry for ABI Bioprism (Applied Biosystems, USA). Forward and reverse strands of all four genomic regions were sequenced, reverse strands were reverse complemented, aligned by BioEdit (Hall Citation1999) and analysed manually for any errors. The sequences were queried using the BLAST (www.ncbi.nlm.nih.gov) search algorithm and deposited in the NCBI GenBank (ITS- KT852579, BenA- MF184995, CaM- MH105905 and RPB2- MH016196).
The phylogenetic position of isolate WSR 62 in section Lanata-divaricata was studied using a combined data set of ITS, BenA and CaM sequences. The generated sequences of isolate WSR 62 were compared with a reference data set containing sequences of the type strains belonging to section Lanata-divaricata (Houbraken et al. Citation2011; Visagie et al. Citation2014, Citation2015) (Supplementary Table 1). Sequence alignments were performed using the CLUSTAL W program in BioEdit and alignments were manually optimised. The Akaike information criterion (AIC) in the program jModelTest version 2.1.5 (Darriba et al. Citation2012) was employed to determine the most optimal substitution model. Penicillium glabrum CBS 125543T was used as outgroup. Data sets were analysed using MrBayes software v. 3.2.2 (Ronquist et al. Citation2012) by running 200,000 generations of two independent chains. At this point, the split frequencies fell below 0.05. The Bayesian posterior probability (pp) values were used to determine the node reliability. Trees were visualised in FigTree version 1.4.3 (Rambaut Citation2016). Maximum Likelihood (ML) analysis was performed using RaxML (Randomized Accelerated Maximum Likelihood) v 7.2.5 and the bootstrap support of the nodes was determined. The obtained bootstrap values are plotted on the Bayesian phylogram. In order to capture the intraspecies variation of P. javanicum in this study, the generated BenA, CaM, and RPB2 sequences of WSR 62 were compared with sequences derived from strains maintained as P. javanicum in the CBS culture collection or the internal working collection of the department Applied and Industrial Mycology (DTO) housed at the Westerdijk Fungal Biodiversity Institute. An overview of studied strains is given in . The single gene and combined data sets were aligned and Maximum Likelihood analysis was performed as described by Frisvad et al. (Citation2019). Sequences of P. oxalicum CBS 219.30T were used as outgroup. The alignments were submitted to TreeBase with submission ID 22415.
Table 1. Strains used in the phylogenetic analyses of the Penicillium javanicum clade.
2.4. Morphology
The colony descriptions of isolate WSR 62 were based on growth on MEA (media composition: malt extract – 50 gL−1 (HiMedia, India), trace elements stock solution – 1 mLL−1 (trace elements stock solution (100 mL): CuSO4.5H2O – 0.5 g, ZuSO4.7H2O – 0.1 g) (Samson et al. Citation2010), Czapek dox agar (CA), Czapek yeast autolysate agar (CYA), Czapek yeast autolysate agar with 5% NaCl (CYAS), and yeast extract sucrose agar (YES). The media composition, fungal inoculation procedure and incubation were performed as described in Visagie et al. (Citation2014). After incubation, colony characters such as colony diameter, pigment production, exudate production, obverse and reverse colony colour, hyphal and mycelial growth, surface texture and degree of sporulation were recorded thrice to confirm the stability of morphological characters. Identification methods proposed by Raper and Thom (Citation1949), Pitt (Citation1974), Pitt (Citation1980), Frisvad and Samson (Citation2004) and Visagie et al. (Citation2014) were employed for the morphological characterisation of the fungus. Ten-day old colonies from MEA were used for light microscopy and SEM imaging of cleistothecia, ascospores and conidial structures. The slides were examined using a Motic BA 310 (USA) light microscope. Conidiophore structures were measured using Motic Image Plus 2.0 ML software. Micrographs of individual cleistothecia were taken with a Labomed Lx400 microscope equipped with a Micaps digital camera run by Micaps software. To prepare the scanning electron microgram of the structures, fungal samples were fixed overnight in 2.5% glutaraldehyde at 4°C. Slides were washed in phosphate buffer (pH 7.4), followed by sequential dehydration by series of washes with acetone/alcohol (30%, 50%, 70%, 80%, 90% and 100%) (Hartmann et al. Citation2010). The dried material was mounted on stubs, sputtered with gold and examined with a scanning electron microscope (SEM; JSM-6490LA, JEOL, Tokyo, Japan). The ascospores and spines were measured on the SEM micrographs.
3. Results
3.1. Phylogenetic analyses
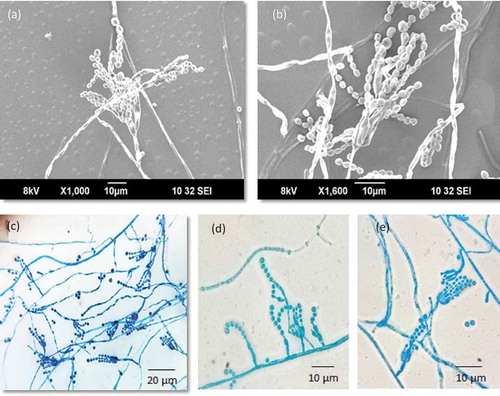
The phylogenetic relationship of isolate WSR 62 was studied using a combined multilocus sequence alignment of ITS, BenA and CaM sequences of members of Penicillium section Lanata-Divaricata (). The General Time Reversible (GTR) model using a discrete gamma distribution (+G) and invariant sites (+I) was optimal for the multilocus data set. The analysis showed that isolate WSR 62 is related to P. javanicum, P. elleniae, P. reticulisporum, P. malacosphaerulum, P. caperatum, and P. urugayense, a group of species previously referred to as the P. javanicum-clade (Visagie et al. Citation2015). Furthermore, this analysis showed that WSR 62 clustered with high statistical support (1 pp) to P. javanicum. The phylogenetic relationships and intraspecies variation within the P. javanicum-clade was studied in more detail using a larger selection of strains (). The length of the aligned BenA, CaM, and RPB2 data sets were 482, 465, and 755 bp, respectively (including gaps). The Kimura 2-parameter (K2) model using the gamma distribution (+G) was found to be optimal for the BenA data set, the GTR+G model was optimal for the CaM data set and the Tamura-Nei (TN) model using the gamma distribution (+G) was most optimal for the RPB2 data set. The topologies of the single locus phylogenies were largely congruent, though some inconsistencies were present among the strains identified as P. javanicum. The strains DTO 284-F3 and CBS 576.70 clustered in all analyses with high bootstrap support (>92 %) with isolate WSR 62 and this analysis showed that these can be identified as P. setosum as well. CBS 564.85 is also closely related to the P. javanicum and P. setosum. The phylogenetic position of this strain is unresolved and might represent a new species in this clade.
Figure 2. Phylogenetic tree based on a combination of partial ITS, BenA and CaM sequences of Penicillium sect. of Lanata-divaricata. The isolate Penicillium setosum is highlighted in red colour. Penicillium glabrum CBS 125543T was chosen as outgroup. Only BI posterior probability values higher than 0.95 and bootstrap value above 80% are shown at the branch nodes.

Figure 3. Maximum-likelihood trees based on BenA, CaM, RPB2 data sets and the combined data sets of afore mentioned genes showing the relationship of species belonging to the Penicillium javanicum-clade. Penicillium oxalicum was chosen as outgroup in the phylogenies. Numbers at branch nodes refer to bootstrap values (1,000 replicates), only values of >70% are shown. Names in red indicate strains belonging to the new species.
3.2. Morphology and physiology
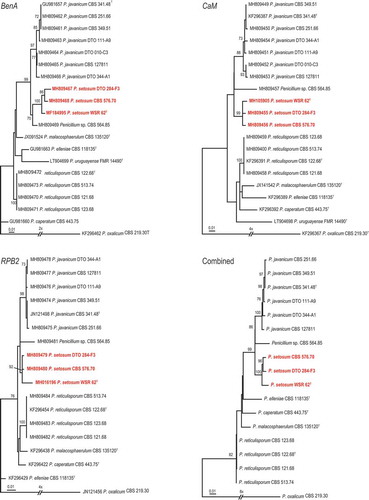
Among the isolated endophytes from Withania somnifera, the Penicillium isolate WSR 62 was studied in detail because of its antibacterial activity. The micro- and macro-morphological characters of this strain were compared to their closest relatives based on notes provided in previous studies (Pitt Citation1974; Stolk & Samson Citation1983; Visagie et al. Citation2014). WSR 62 grew well on MEA and CYA incubated at 25°C (CYA 50–52 mm, MEA 52–55 mm) and produced prominent spinose ascospores measuring 2.0–3.0 × 2.3–3.0 μm.
3.3. Taxonomy
Multigene phylogenetic analyses consistently placed isolate WSR 62 in a separate clade distinct from P. javanicum. This strain is phenotypically distinct from P. javanicum and other phylogenetically related species. Based on these data, we describe this isolate as a new species named Penicillium setosum.
Penicillium setosum Tijith, Houbraken, Linu & Jisha sp. nov., MycoBank MB818581. .
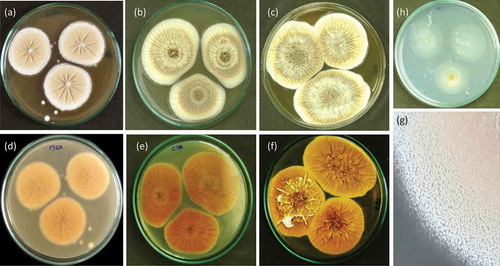
Figure 4. Penicillium setosum (WSR 62): a, b, c. Colonies on MEA, CYA and YES (all obverse); d, e, f. Colonies on MEA, CYA and YES (all reverse); g. Texture on MEA; h. Colony on CA.
In: Penicillium subgenus Aspergilloides, section Lanata-divaricata.
ITS barcode: KT852579, Alternative identification markers: BenA = MF184995, CaM = MH105905, RPB2 = MH016196.
Etymology
setosum (Latin), bristly, referring to the surface of ascospores having prominent spinulose structure.
Typification
India, Kerala, isolated from Withania somnifera (L.) Dunal, 21 November 2014, T. K. George (holotype WSR 62, culture ex-type CBS 144865 = MCC 1370 = NCFT NO 8222.16 = AMH-9974).
Colony diameter, 7 d (in mm) of incubation: CYA (at 25°C) 50–52; CYA (at 30°C) 50–53; CYA at (37°C) 40–42; CYA with 5% NaCl (at 25°C) 50–53, MEA (at 25°C) 52–55; CA (at 25°C) 36–40, YES (at 25°C) 48–53.
Macromorphology
CYA 25°C, 7 d: Colonies velvety, moderately deep, radially sulcate; margins low, wide, entire; mycelia white and inconspicuously yellow; cleistothecia abundantly produced mainly near the colony centre, maturing after 1–2 weeks, giving a greyish yellow colour to colony; sporulation absent or sparse; soluble pigments absent; exudates present as small hyaline to yellow droplets; reverse orange-brown. MEA 25°C, 7 d: Colonies granular, plane; margins low, wide, entire; mycelia yellowish white; cleistothecia abundantly produced in the centre, maturing within 1–2 weeks, giving an yellowish brown colour to colony; sporulation sparse; soluble pigments absent; exudates absent; reverse yellowish orange. YES 25°C, 7 d: Colonies deep, cerebriform at the centre, radially and concentrically sulcate, raised at centre; margins low, wide, entire; mycelia white turning slightly yellow in time; cleistothecia abundantly produced, maturing within 2 weeks, giving an yellow colour to colony; sporulation absent; soluble pigments absent; exudates present as small yellow droplets; reverse olive brown. CYAS 25°C, 7 d: Colonies moderately deep; margins low, wide, entire; mycelia white, slightly mauve in the centre; texture floccose to velutinous; cleistothecia produced at the centre, giving a yellowish brown colour to colony; sporulation absent; soluble pigments absent; exudates absent; reverse olive orange. CA 25°C, 7 d: Colonies plane, thin; margins low, narrow, entire; mycelia greyish white; sporulation absent; cleistothecia moderately produced, giving a yellowish colour to colony; soluble pigments absent; exudates absent; reverse whitish grey.
Micromorphology
Conidiophores divaricate, few solitary phialides observed. Stipes smooth walled, variable in length, 10–50 × 2.0–3.0 μm, vesiculate, 2.0–3.5 µm; phialides 1–5 per stipe/metula, ampulliform, 5–7.5 × 2–3 μm. Conidia in long, loose, irregular columns, finely roughened, broadly ellipsoidal, some globose 2–3 × 2–2.5 μm (). Cleistothecia globose to subglobose, abundantly produced on MEA, CYA and YES, 50–120 μm in diameter, solitary or clustered, maturing from centre to margins of the colony; surface covered with globose to hexagonal cells; outer 3–4 layers of cells are thin-walled, while the inner layer is brownish, with thick-walled cells. Asci 8-spored, pyriform when young, becoming subglobose to ellipsoidal at maturity, 8–12 × 5.0–8.5 μm. Ascospores globose to subglobose, 2.0–3.0 × 2.3–3.0 μm in diameter; hyaline to greenish yellow, spinose, spines measuring 0.2–0.4 μm ().
Notes
A combination of features is unique for Penicillium setosum. This species has a greyish yellow obverse due to the abundant production of cleistothecia, and an orange brown reverse colony on CYA. P. setosum produces solitary phialides and divaricate type conidiophores with smooth-walled stipes. Furthermore, it produces small-sized (2.0–3.0 × 2.3–3.0 μm) globose to subglobose ascospores, decorated with spines measuring 0.2–0.4 μm.
4. Discussion
Penicillium section Lanata–divaricata is taxonomically challenging due to the morphological similarities among members of this section (Visagie et al. Citation2015). This taxonomically difficult section currently comprises 56 species (Visagie et al. Citation2016; Guevara-Suarez et al. Citation2017). Species belonging to section Lanata–divaricata are mainly soil inhabitants. Some species were isolated from the rhizosphere and phyllosphere of native plant communities, especially grasses and cereals, and some are frequently found on rotting leaf litter (Houbraken and Samson Citation2011; Visagie et al. Citation2015). In the present study, a novel endophytic Penicillium species was isolated from the surface of sterilized roots of W. somnifera. The two additional P. setosum strains were isolated from soil in Thailand and Mexico, showing the worldwide distribution of this species. There are other reports on endophytic species belonging to section Lanata–divaricata. Penicillium janthinellum was prominently reported as an endophyte from various plants such as Melia azedarach (Marinho et al. Citation2005), Coffea arabica (Vega et al. Citation2006), Solanum lycopersicum (Khan et al. Citation2013) and Sonneratia caseolaris (Zhu et al. Citation2017). Other endophytic species in section Lanata-divaricata are P. oxalicum from Coffea (Vega et al. Citation2006), P. brasilianum from Melia azedarach (Fill et al. Citation2010), P. simplicissimum and P. javanicum from maize and rice (Potshangbam et al. Citation2017).
The molecular study showed that P. setosum is most closely related with P. javanicum. Other species related to P. setosum are P. elleniae, P. caperatum, P. malacosphaerulum, and P. reticulisporum. All these species shared the ability to produce a sexual state. Penicillium caperatum, P. malacosphaerulum, and P. reticulisporum have roughened ascospores with two longitudinal flanges, P. javanicum has roughened ascospores and P. elleniae has broadly ellipsoidal ascospores with spinose valves (Visagie et al. Citation2015, Citation2016). These ascosporic structures differed from P. setosum which has spinose, globose to subglobose ascospores. The ascospores of P. setosum (2.0–3.0 × 2.3–3.0 μm) are smaller than those of P. elleniae (3–3.5 × 2.5–3) and P. javanicum (3–3.5 × 2–3). The length of the spines on the ascospores of P. setosum is comparable with that of P. elleniae (0.2–0.4 µm). Both P. javanicum and P. setosum abundantly produce cleistothecia on agar media. Penicillium setosum has a greyish yellow obverse colour and orange–brown reverse colony on CYA at 25°C, while P. javanicum has a light-brown obverse and a pale orange reverse and P. elleniae has yellow-brown (obverse and reverse) coloured colonies. Penicillium setosum grows faster on CYA than its close relatives. After 7 days incubation at 25°C, P. setosum has a colony diameter between 50 and 52 mm, while the colony diameters of the other species are (slightly) smaller (in mm): P. javanicum CBS 341.48T (33–41), P. malacosphaerulum CBS 135120T (28–36), P. reticulisporum CBS 122.68T (45–46), P. brefeldianum CBS 235.81T (36–38), P. caperatum CBS 443.75T (38–40) and P. elleniae CBS 118135T (48–50) (Visagie et al. Citation2015, Citation2016). Sporulation was sparse to absent in P. setosum, while P. javanicum showed abundant sporulation. Solitary phialides and divaricate type conidiophores with smooth-walled stipes were observed in P. setosum, while P. javanicum is reported to have rough-walled stipes and monoverticillate, or occasionally biverticillate penicilli (Stolk and Samson Citation1983).
The taxonomic novelty of WSR 62, a Penicillium isolate from the root of Withania somnifera, was determined using a polyphasic approach combining molecular data and phenotypic characters (e.g. colony colour and diameter, ornamentation of conidiophores and ascospore size, shape and ornamentation). Based on this polyphasic approach, we concluded that isolate WSR 62 represents a novel species, here named Penicillium setosum Tijith, Houbraken, Linu & Jisha.
Acknowledgments
We thank NCFT at New Delhi, MCC and NFCCI at Pune for incorporation of WSR 62 in their culture collections; Amrita Center for Nanosciences is thanked for providing the SEM facility. We also acknowledge the reviewers, who showed immense patience for rectifying the flaws in our research article. The first author gratefully acknowledges the financial support received from the Kerala State Council for Science, Technology, and Environment (026/FSHP-LSS/2013/KSCSTE fellowship).
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Bharti VK, Malik JK, Gupta RC 2016. Ashwagandha. In: Nutraceuticals. [Internet]. [place unknown]: Elsevier. 717–733: http://linkinghub.elsevier.com/retrieve/pii/B9780128021477000528 [Accessed 2018 May 3].
- Compant S, Saikkonen K, Mitter B. 2016. Editorial special issue: soil, plants and endophytes. Plant Soil [Internet]. 405:1–11. doi:10.1007/s11104-016-2927-9
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods [Internet]. 9:772. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4594756/
- Fill TP, Da Silva BF, Rodrigues-Fo E. 2010. Biosynthesis of phenylpropanoid amides by an endophytic penicillium brasilianum found in root bark of melia azedarach. J. Microbiol. Biotechnol. 20:622–629.
- Frisvad J, Samson RA. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol [Internet]. 49: 1–174. http://www.mycobank.org/BioloMICS.aspx?TableKey=14682616000000061&Rec=19610&Fields=All [Accessed 2017 May 25].
- Frisvad JC, Hubka V, Ezekiel CN, Hong S-B, Nováková A, Chen AJ, Arzanlou M, Larsen TO, Sklenář F, Mahakarnchanakul W, et al. 2019. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol [Internet]. 93:1–63. https://www.sciencedirect.com/science/article/pii/S0166061618300289 [Accessed 2018 Sep 4].
- Frisvad JC, Smedsgaard J, Larsen TO, Samson RA. 2004. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004:201–241.
- George TK, Asok AK, Rebello S, Fathima PA. 2015. Diversity of Bruguiera cylindrica and Rhizophora candelaria from Ayiramthengu mangrove ecosystem. Ann Biol Res. 6:55–63.
- Guevara-Suarez M, García D, Cano-Lira JF, Guarro J, Gené J 2017. Penicillium uruguayense [Internet]. [place unknown]: Persoonia – lFungal Planet description sheets. http://www.fungalplanet.org/content/pdf-files/FungalPlanet645.pdf [Accessed 2018 Apr 30]
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser No41 [Internet].: 95–98. [accessed 2018 Mar 15]. http://brownlab.mbio.ncsu.edu/JWB/papers/1999Hall1.pdf.
- Hammerschmidt L, Wray V, Lin W, Kamilova E, Proksch P, Aly AH. 2012. New styrylpyrones from the fungal endophyte Penicillium glabrum isolated from Punica granatum. Phytochem Lett [Internet]. 5: 600–603. [accessed 2018 Jun 7]. http://linkinghub.elsevier.com/retrieve/pii/S1874390012001164.
- Hardoim PR, Van Overbeek LS, Berg G, Pirttilä M, Compant S, Campisano A, Döring M. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 79:293–320.
- Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 54:3132–3142.
- Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG, Samson RA. 2012. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia [Internet]. 29: 78–100. [accessed 2018 Apr 30]. http://www.ncbi.nlm.nih.gov/pubmed/23606767.
- Houbraken J, López-Quintero CA, Frisvad JC, Boekhout T, Theelen B, Franco-Molano AE, Samson RA. 2011. Penicillium araracuarense sp. nov., Penicillium elleniae sp. nov., Penicillium penarojense sp. nov., Penicillium vanderhammenii sp. nov. and Penicillium wotroi sp. nov., isolated from leaf litter. Int. J. Syst. Evol. Microbiol. 61:1462–1475.
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol [Internet]. 70: 1–51. [accessed 2017 May 25]. http://www.sciencedirect.com/science/article/pii/S0166061614600622?np=y&npKey=d78d97fde818b5905899c85b2da30a5b7681d863f5bfbf6a95b6800d322035b5.
- Jia M, Chen L, Xin H-L, Zheng C-J, Rahman K, Han T, Qin L-P. 2016. A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front Microbiol [Internet]. 7: 906. [accessed 2018 May 9]. http://www.ncbi.nlm.nih.gov/pubmed/27375610.
- Khan AL, Waqas M, Khan AR, Hussain J, Kang S-M, Gilani SA, Hamayun M, Shin J-H, Kamran M, Al-Harrasi A, et al. 2013. Fungal endophyte Penicillium janthinellum LK5 improves growth of ABA-deficient tomato under salinity. World J Microbiol Biotechnol [Internet]. 29:2133–2144. [accessed 2017 Jul 3]. http://www.ncbi.nlm.nih.gov/pubmed/23842755.
- Khan S, Hamayun M, Yoon H, Kim H-Y, Suh S-J, Hwang S-K, Kim J-M, Lee I-J, Choo Y-S, Yoon U-H, et al. 2008. Plant growth promotion and Penicillium citrinum. BMC Microbiol [Internet]. 8:231. [accessed 2018 Jun 7]. http://www.ncbi.nlm.nih.gov/pubmed/19099608.
- Koukol O, Kolařík M, Kolářová Z, Baldrian P. 2012. Diversity of foliar endophytes in wind-fallen Picea abies trees. Fungal Divers [Internet]. 54: 69–77. [accessed 2018 Jun 8]. http://link.springer.com/10.1007/s13225-011-0112-2.
- Marinho Am do R, Rodrigues-Filho E, Moitinho M da LR, Santos LS. 2005. Biologically active polyketides produced by Penicillium janthinellum isolated as an endophytic fungus from fruits of Melia azedarach. J Braz Chem Soc [Internet]. 16: 280–283. [accessed 2017 Jul 3]. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0103-50532005000200023&lng=en&nrm=iso&tlng=en.
- Müller MM, Hallaksela A-M. 1998. Diversity of Norway spruce needle endophytes in various mixed and pure Norway spruce stands. Mycol Res [Internet]. 102: 1183–1189. [accessed 2018 Jun 8]. https://www.sciencedirect.com/science/article/pii/S0953756208610072.
- Owais M, Sharad KS, Shehbaz A, Saleemuddin M. 2005. Antibacterial efficacy of Withania somnifera (ashwagandha) an indigenous medicinal plant against experimental murine salmonellosis. Phytomedicine [Internet]. 12: 229–235. [accessed 2018 May 1]. http://www.ncbi.nlm.nih.gov/pubmed/15830846.
- Peterson SW, Jurjević Ž, Frisvad JC. 2015. Expanding the species and chemical diversity of penicillium section cinnamopurpurea.Yu J-H, editor. PLoS One [Internet]. 10: e0121987. [accessed 2018 Apr 29]. 10.1371/journal.pone.0121987.
- Peterson SW, Vega FE, Posada F, Nagai C. 2005. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia [Internet]. 97: 659–666. [[accessed 2018 Apr 30]. http://www.ncbi.nlm.nih.gov/pubmed/16392254.
- Petrini O. 1991. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microb ecol leaves [Internet]. New York (NY): Springer New York; p. 179–197. doi:10.1007/978-1-4612-3168-4_9
- Pitt JI. 1974. A synoptic key to the genus Eupenicillium and to sclerotigenic Penicillium species. Can J Bot [Internet]. 52: 2231–2236. [accessed 2017 May 25]. http://www.nrcresearchpress.com/doi/abs/10.1139/b74-290.
- Pitt JI 1980. The genus Penicillium and its teleomorphic states Eupenicillium and talaromyces. [place unknown]. [accessed 2017 Jun 3]. http://doi.wiley.com/10.1002/jobm.19810210822.
- Pitt JI. 1994. The current role of Aspergillus and Penicillium in human and animal health. Med Mycol [Internet]. 32: 17–32. [accessed 2018 Feb 15]. 10.1080/02681219480000701.
- Potshangbam M, Devi SI, Sahoo D, Strobel GA. 2017. Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front Microbiol [Internet]. 8: 325. [accessed 2018 Jun 8]. http://www.ncbi.nlm.nih.gov/pubmed/28303127.
- Rambaut A. 2016. FigTree version 1.4.3 [Internet]. [accessed 2017 Nov 11]. http://tree.bio.ed.ac.uk/software/figtree/.
- Raper K, Thom C 1949. A manual of the penicillia, [Internet]. Baltimore: Williams & Wilkins. [accessed 2017 May 25]. http://www.worldcat.org/title/manual-of-the-penicillia/oclc/1522344.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol [Internet]. 61: 539–542. [accessed 2018 Mar 26]. http://www.ncbi.nlm.nih.gov/pubmed/22357727.
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B 2010. Food and indoor fungi [Internet]. [place unknown]: CBS-KNAW Fungal Biodiversity Centre. [accessed 2018 Jul 19]. https://www.nhbs.com/food-and-indoor-fungi-book.
- Sieber-Canavesi F, Sieber TN. 1993. Successional patterns of fungal communities in needles of European silver Fir (Abies alba Mill.). New Phytol [Internet]. 125: 149–161. [accessed 2018 Jun 8]. https://www.jstor.org/stable/2557915.
- Stolk A, Samson R. 1983. The Ascomycete genus Eupenicillium and related Penicillium anamorphs. Stud. Mycol. 23:1–149.
- Thomas D, Vandegrift R, Roy B. 2017. An agent-based model of the Foraging Ascomycete Hypothesis. BioRxiv. 1–21
- Vega FE, Posada F, Peterson SW, Gianfagna TJ, Chaves F. 2006. Penicillium species endophytic in coffee plants and ochratoxin A production. Mycologia [Internet]. 98: 31–42. [accessed 2018 Jun 5]. https://www.tandfonline.com/doi/full/10.1080/15572536.2006.11832710.
- Ven Murthy MR, Ranjekar PK, Ramassamy C, Deshpande M. 2010. Scientific basis for the use of Indian ayurvedic medicinal plants in the treatment of neurodegenerative disorders: ashwagandha. Cent Nerv Syst Agents Med Chem [Internet]. 10: 238–246. [accessed 2018 Mar 10]. http://www.ncbi.nlm.nih.gov/pubmed/20528765.
- Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol [Internet]. 78:343–371.
- Visagie CM, Houbraken J, Seifert KA, Samson RA, Jacobs K. 2015. Four new Penicillium species isolated from the fynbos biome in South Africa, including a multigene phylogeny of section Lanata-Divaricata(Mycol Progress. Mycol Prog. 14:1.
- Visagie CM, Renaud JB, Burgess KMN, Malloch DW, Clark D, Ketch L, Urb M, Assabgui R, Sumarah MW, Seifert KA. 2016. Fifteen new species of Penicillium. Persoonia 36: 247–280
- White TJ, Bruns T, Lee SJ, Taylor JL. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, JJS & TJW, editors. PCR protoc a guid to methods appl. New York: Academic Press; p. 315–322.
- Zhu M, Zhang X, Feng H, Dai J, Li J, Che Q, Gu Q, Zhu T, Li D. 2017. Penicisulfuranols A–F, Alkaloids from the mangrove endophytic fungus Penicillium janthinellum HDN13-309. J Nat Prod [Internet]. 80: 71–75. [accessed 2018 Jun 7]. http://www.ncbi.nlm.nih.gov/pubmed/27992183.