ABSTRACT
A new rust genus, Quasipucciniastrum, typified by Q. agrimoniae sp. nov., is proposed based on distinct morphological characters and phylogenetic placement. This genus is characterised by its uredinial ostiolar peridial cells with rough surface and sessile, multicellular teliospores with apparently thickened apical wall. Molecular phylogenetic analyses using internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS) and the large subunit (LSU) rDNA revealed that this genus is sister to the genus Cronartium (Cronartiaceae), but morphologically it is distinct from Cronartium in the sessile teliospores that are divided by vertical septa. Morphologically, Quasipucciniastrum should be compared to Pucciniastrum (Pucciniastraceae) in its multicellular teliospores, but they were phylogenetically distant.
Introduction
Agrimonia Ldb. species, known as “common agrimony”, are perennial herbaceous flowering plants widely distributed in the temperate regions of the Northern Hemisphere, and they have been commonly planted for ornamentation and medicinal use (Lu Citation2001). Common agrimony is economically and horticulturally important, but their growths have been frequently threatened by rust diseases. Hitherto, seven rust species have been recorded on Agrimonia species (Farr and Rossman Citation2018), among which, Pucciniastrum agrimoniae (Dietel) Tranzschel (Pucciniastraceae) and its synonymies, P. agrimoniae-eupatoriae (DC.) Lagerh. (Pucciniastraceae), Thekopsora agrimoniae Dietel (Pucciniastraceae), Uredo agrimoniae J. Schröt. (Anamorphic genera) are frequently assigned names (Tai Citation1979; Zhuang Citation1983, Citation2001, Citation2005; Guo Citation1989; Zhang et al. Citation1997; Cao and Li Citation1999; Zhuang and Wei Citation1999; Cao et al. Citation2000), or in fewer cases as Melampsora agrimoniae Dietel (Melampsoraceae), Puccinia agrimoniae (Arthur) Arthur (Pucciniaceae) and Uropyxis agrimoniae Arthur (Uropyxidaceae) (Arthur Citation1910; Maneval Citation1937).
During our study on rust fungi in China, a morphologically distinct species was found on Agrimonia pilosa. It produces Milesia-type uredinia and ostiolar peridial cells with rough surface, and have subglobose teliospores divided by vertical septa. These characters are to some extent, similar to Pucciniastrum spp. (Pucciniastraceae), but the rDNA ITS and LSU sequences showed its close relationship to genus Cronartium Fr. (Cronartiaceae) rather than Pucciniastrum. Our critical morphological and molecular comparisons of this fungus with Cronartium, Pucciniastrum and other related genera suggested that this rust fungus represents a new genus herein described as Quasipucciniastrum agrimoniae gen. et sp. nov.
Materials and methods
Fungal specimens
A total of 204 dried herbarium specimens on Agrimonia pilosa were loaned from the Mycological Herbarium of Institute of Microbiology, CAS, China (HMAS). Several fresh specimens on Agrimonia species were collected from different provinces in China during last three years. Herbarium specimens of Cronartium and Peridermium (Link) J.C. Schmidt & Kunze involved in this study were loaned from University of Florida Herbarium, USA (FLAS), HMAS, University of Michigan Herbarium, USA (MICH) and New York Botanical Garden, USA (NY). In this study, the Roman numerals of II and III referred to uredinial and telial stages in the rust fungi.
Morphological examinations
Morphological characteristics of all specimens were observed using a dissecting microscope (DM), the light microscope (LM) and the scanning electron microscope (SEM). The methods for morphological analyses as outlined by Zhao et al. (Citation2013), Zhao et al. (Citation2014), Citation2017) were followed. Fifty measurements of sori and spores from each specimen were recorded, and morphological characteristics in uredinial and telial stages, i.e. the position of uredinia and telia, the ornamentation and dimension of urediniospores, the dimension and shape of ostiolar cells, the position and shape of peridial cells, the dimension of teliospores, the position and shape of teliospores were examined.
DNA extraction, amplification and sequencing
For the fungal specimens, several sori from each specimen were excised and DNA were extracted from all studied herbarium specimens using Gentra Puregene Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. From the crude extracts, 1–3 μl DNA templates were directly used for the polymerase chain reaction (PCR) amplification of the internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS) and the large subunit (LSU) rDNA. rDNA ITS regions were amplified using the primer pairs ITS1F (Gardes and Bruns Citation1993)/ITS4 (White et al. Citation1990), Rust2inv (Aime Citation2006)/ITS4BR (Feau et al. Citation2009), and fragment of LSU was amplified using the primer pairs ITS4BRf/LR6 (Vilgalys and Hester Citation1990), LR1R/LR3 and LR3R/LR6 (Vilgalys and Hester Citation1990). PCR was performed under the following conditions: denaturation at 95°C for 5 min; followed by 35 cycles of denaturation at 95°C for 45 s, annealing at 55°C for 1 min and elongation at 72°C for 1 min, finally with an extension step at 72°C for 10 min. Purification and sequencing of PCR amplicons were carried out at the Tianyi Huiyuan Company, Beijing.
Molecular phylogeny
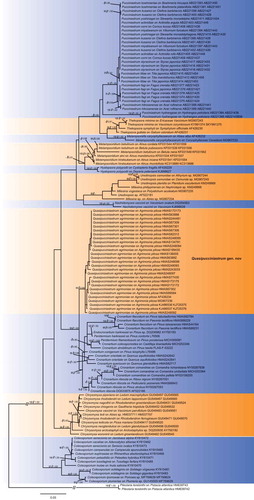
Full-length ITS and partial LSU regions were amplified from 23 specimens, and we included 16 specimens from genus Cronartium and Peridermium for comparable studies because they have high similarity to rust fungus on Agrimonia in rDNA sequences. Their herbarium numbers, host plants, geographical origins and GenBank accession numbers are listed in . Besides, a total of 148 sequences from closely related species were retrieved from GenBank for phylogenetic comparison (). Pileolaria terebinthi (DC.) Castagne was used as outgroup. In the final dataset, ITS and LSU were concatenated and the final alignment includes 124 specimens with a length of 1836 characters (743 for ITS, 1093 for 28S).
Table 1. Herbarium specimens used for molecular phylogenetic analyses.
Table 2. Sequence data retrieved from GenBank and used for phylogenetic analyses.
Sequences were manually aligned by using BioEdit 7.0.9 (Hall Citation1999), and multiple alignments were performed using MAFFT 7 (Katoh and Standley Citation2013). Gaps were treated as missing data for all analyses. The Akaike Information Criteria (AIC) in Modeltest 3.7 (Posada and Crandall Citation1998) was used to estimate the best-fit substitution models, and GTR+I + G was selected as the best evolutionary model. Maximum Likelihood (ML) analyses were performed using RAxML 8.0.0 (Stamatakis and Alachiotis Citation2010), and Bayesian Markov chain Monte Carlo (MCMC) analyses were performed by MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001). Supported values of ML and Bayesian posterior probability (Bpp) were indicated in the phylogenetic tree.
Results
Molecular phylogeny
The combined ITS and LSU dataset included 106 sequences of ITS and 123 sequence of LSU from 125 rust samples. The dataset comprised aligned length of 1836 characters, of which, 1162 characters are constant, and 640 are variable with 533 parsimony informative sites. Both ML and Bayesian inference resulted in a highly concordant topology (). Our specimens on Agrimonia pilosa constituted a strongly supported clade (ML/Bpp = 93/0.98), close to Cronartium, Endocronartium Y. Hirats. of the family Cronartiaceae. Genera Hyalopsora Magnus, Melampsorella J. Schröt., Melampsoridium Kleb., Milesia F.B. White, Naohidemyces S. Sato, Katsuya & Y. Hirats., Pucciniastrum, Thekopsora Magnus and Uredinopsis Magnus from the family Pucciniastraceae, which produce Milesia-type uredinia and multicellular teliospores, clustered together but are phylogenetically distant from our rust specimens on A. pilosa.
Figure 1. Bayesian 50% majority-rule consensus tree based on concatenated data of rDNA ITS and LSU sequences. Pileolaria terebinthi was used as outgroup. Values on the branches indicate maximum likelihood bootstrap values and Bayesian posterior probabilities. Hyphen indicates that bootstrap values were less than 75% and Bayesian posterior probabilities less than 0.80.
TaxonomyQuasipucciniastrum X.H. Qi, P. Zhao & L. Cai, gen. nov.MycoBank no.: MB828669
Etymology
Quasipucciniastrum (Lat.) referring to the morphological characters (uredinial and telial stages) similar to genus Puccinistrum.
Type species
Quasipucciniastrum agrimoniae X.H. Qi, P. Zhao & L. Cai
Generic diagnosis
Spermogonia and aecia unknown. Uredinia hypophyllous, subepidermal, round, scattered or crowded in groups, Milesia-type, erumpent with peridium opening by a pore delimited by well-developed, globose and verrucose ostiolar cells, peridial cells small, irregular. Urediniospores borne singly, no pedicels, ovoid, globoid or ellipsoid, spore wall colourless, echinulate. Telia hypophyllous, subepidermal, one spore deep, light yellow. Teliospores subglobose, sessile, multicellular, separated by vertical or oblique septa, wall apparently thickened at apex.
Notes
The new genus Quasipucciniastrum is characterised by its Milesia-type uredinia with well-developed ostiolar cells, well-developed peridial cells, hypophyllous telia producing subglobose teliospores which is divided by vertical and oblique septa under host epidermis. This genus resembles Pucciniastrum (Pucciniastraceae, Pucciniales) but differs in producing hypophyllous telia, subglobose teliospores with apparently thickened apical wall. Within family Pucciniastraceae, other genera clearly differed from Quasipucciniastrum in uredinial ostiole and teliospores. Genera Calyptospora, Hyalopsora, Milesina and Uredinopsis differ from this new genus in the type of ostiole, position of telia and type of teliospores, while Melampsorella and Melampsoridium differ from Quasipucciniastrum mainly in their unicellular teliospores without septa (Cummins and Hiratsuka Citation2003). In addition, Melampsorella has Milesia-type uredinia with discrete ostiole, which also clearly differs from Quasipucciniastrum. Phylogenetic results supported the separation of Quasipucciniastrum from Pucciniastrum, Melampsorella and other genera in the family Pucciniastraceae ().
Quasipucciniastrum agrimoniae X.H. Qi, P. Zhao & L. Cai, sp. nov. ()MycoBank no.: MB828670
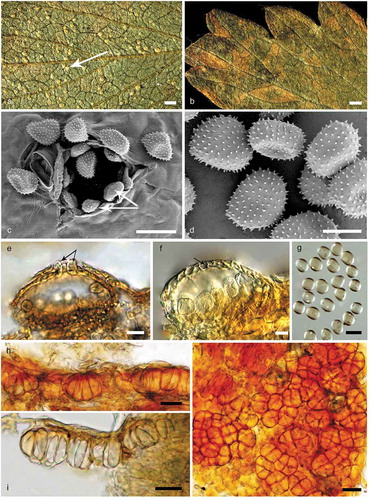
Figure 2. Morphological characters of Quasipucciniastrum agrimoniae. A: Uredinia (white arrow) on the hypophyllous leaf surface. B: No uredinia and telia on eiphyllous leaf surface. C: Uredinium with echinulate ostiolar cells (white arrow) observed by SEM. D: Urediniospores with echinulate spines observed by SEM. E: Uredinium with well-developed ostiolar cells (black arrow) observed by LM. F: Uredinium with peridium cell (black arrow). G: Urediniospores observed by LM. H: Vertical section of hypophyllous telia. I: Subepidermal teliospores with apparently thickened apical wall. J: Subglobose teliospores with vertical or oblique septa. Bars: A, B = 0.6 mm; C, E, G, H, I, J = 20µm; D, F = 10µm.
Typification
CHINA. HEILONGJIANG: Greater Higgnan Mountains, Tahe, II, III on A. pilosa, 6 September 2015, P. Zhao (Holotype designated here, HMAS248095), GenBank: ITS = MK208281; LSU = MK193852.
Etymology
Named after the host plant of the type specimen.
Description from holotype
Spermogonia and aecia unknown. Uredinia hypophyllous, subepidermal, round, pale-yellow, scattered or rarely grouped, 0.1–0.4 mm in diameter, erumpent with peridium with well-developed ostiolar cells, usually six to nine, arranged at the apex of uredinia. Peridial cells small, irregular, walls smooth. Ostiolar cells well-developed, oblong or ellipsoid, with rough surface. Urediniospores borne singly, no pedicels, scattered, globoid, ovoid or ellipsoid, 16.5–22 × 12.5–17.3 µm, wall 0.7–2.1 µm thick, evenly echinulate without smooth area on surface, spinules 0.5–1.2 µm in length, and the distance between spinules 1.1 and 2.2 µm, germ bizonated. Telia hypophyllous, subepidermal, one spore deep, light yellow. Teliospores consisting of several cells adhering laterally under the epidermal cells, sessile, subglobose, 18.5–36.1 × 13.7–29.9 µm, 2–6 celled, mainly with vertical septa, walls apparently thickened at apex, up to 4.6 µm.
Additional specimens examined: CHINA. BEIJING: Dongling Mountain, II, III on Agrimonia pilosa, 16 September 1998, J.Y. Zhuang (HMAS82312); CHINA. GANSU: Gan Nan, Diebu, II, III on A. pilosa, 10 September 1992, J.Y. Zhuang (HMAS67301); CHINA. GANSU: Gan Nan, Diebu, II, III on A. pilosa, 12 September 1992, J.Y. Zhuang (HMAS67302); CHINA. GANSU: Gan Nan, Zhouqu, II, III on A. pilosa, 5 September 1992, J.Y. Zhuang (HMAS67306); CHINA. GANSU: Lan Zhou, Yongdeng, II, III on A. pilosa, 11 October 2003, J.Y. Zhuang (HMAS134791); CHINA. GANSU: Long Nan, Wenxian, II, III on A. pilosa, 21 September 1992, J.Y. Zhuang (HMAS67309); CHINA. GUIZHOU: Qian Nan, Maolan, II, III on A. pilosa, 21 June 2015, P. Zhao (HMAS248096); CHINA. HEILONGJIANG: Greater Higgnan Mountains, Jiagedaqi District, II, III on A. pilosa, 9 September 2015, P. Zhao (HMAS248094); CHINA. HEILONGJIANG: Khakan Nature Reserve, II, III on A. pilosa, 9 August 2003, J.Y. Zhuang (HMAS89584); CHINA. HEILONGJIANG: Mu Dan Jiang, II, III on A. pilosa, 9 August 2004, J.Y. Zhuang (HMAS136005); CHINA. INNER MONGOLIA: Tong Liao, Horqin, II, III on A. pilosa, 18 September 2000, J.Y. Zhuang (HMAS172175); CHINA. NINGXIA: Jin Yuan, II, III on A. pilosa, 31 August 2000, J.Y. Zhuang (HMAS172172); CHINA. NINGXIA: Jin Yuan, II, III on A. pilosa, 1 September 2000, J.Y. Zhuang (HMAS172173); CHINA. SICHUAN: Liangshan Yi Autonomous Prefecture, Meigu, II, III on A. pilosa, 8 October 1989, J.Y. Zhuang (HMAS63892); CHINA. SICHUAN: Liangshan Yi Autonomous Prefecture, Yanyuan, II, III on A. pilosa, 11 September 2010, J.Y. Zhuang (HMAS243033); CHINA. SICHUAN: Tibetan Qiang Autonomous Prefecture of Ngawa, Wolong, II, III on A. pilosa, 23 September 1989, J.Y. Zhuang & S.X. Wei (HMAS63888); CHINA. SICHUAN: Yi Bin, Xingwen, II, III on A. pilosa, 21 June 2016, P. Zhao (HMAS248093); CHINA. TIBET: Rikaze, Yadong, II, III on A. pilosa, 20 August 2011, J.Y. Zhuang & T.Z. Wei (HMAS244481); CHINA. YUNNAN: Hong He, Binbian, II, III on A. pilosa, 19 September 2007, J.Y. Zhuang (HMAS199430); CHINA. YUNNAN: Hong He, Mengzi, II, III on A. pilosa, 14 June 2016, P. Zhao (HMAS248092); CHINA. YUNNAN: Kun Ming, II, III on A. pilosa, 17 June 2016, P. Zhao (HMAS248097).
Hosts of uredinial and telial stages and geographical distribution: Agrimonia pilosa – China: Beijing, Gansu, Guangxi, Guizhou, Heilongjiang, Inner Mongolia, Ningxia, Sichuan, Tibet, Yunnan.
Discussion
In this study, we recognised a new genus Quasipucciniastrum on Agrimonia pilosa, and described a new species Q. agrimoniae based on morphological and molecular evidences. Hitherto, rust species in genera Puccinia Pers., Pucciniastrum, Thekopsora, Uredo Pers. and Uropyxis J. Schröt. have been recorded on Agrimonia species, but Quasipucciniastrum clearly differs from above-mentioned genera by its hypophyllous telia, subepidermal teliospores with subglobose shape, and multicellular teliospores with thickened apical wall. rDNA based phylogenies further supported the independence of Quasipucciniastrum from these genera, especially Pucciniastrum and other genera in Pucciniastraceae, which have similar uredinial and telial morphologies. Here we confirmed the close relationship of Quasipucciniastrum and Cronartium (). Quasipucciniastrum is currently best placed in Cronartiaceae, together with Cronartium, although this should be better confirmed after the examination of the morphological characters in spermogonial and aecial stages of Q. agrimoniae. Morphologically, genus Cronartium owned Milesia-type uredinia that is similar to Quasipucciniastrum, but its columnar telia and unicellular teliopsores are embedded in a common matrix (Cummins and Hiratsuka Citation2003). Hitherto, we are not successful to obtain the spermogonial and aecial stages of Q. agrimoniae. Further investigation on its life cycles and detailed morphological examination of spermogonia and aecia are necessary.
The rust fungus on Agrimonia pilosa was previously frequently recognised as Pucciniastrum agrimoniae due to its ostiolar cells and subepidermal teliospores divided by vertical septa (Tai Citation1979; Guo Citation1989; Zhang et al. Citation1997; Cao and Li Citation1999; Zhuang and Wei Citation1999; Cao et al. Citation2000; Zhuang Citation2001, Citation2005). P. agrimoniae was first described on A. pilosa from Western Siberia Borus Mountains, Altai in Russia by Tranzschel (Citation1895). Based on the original description of P. agrimoniae from its type specimens and other Russian materials (Tranzschel Citation1895; Sydow and Sydow Citation1915; Kuprevich and Tranzschel Citation1957), our novel species Quasipucciniastrum agrimoniae on A. pilosa resembles P. agrimoniae in several aspects but still clearly differs in its 2 to 6 celled and subglobose teliospores with apparently thickened apical wall. Similarly P. agrimoniae-like rust on A. eupatoria was once reported to bear phylogenetic affinities to Cronartium flaccidum and C. ribicola (Cronartiaceae) Maier et al. (Citation2003). Those specimens used by Maier et al. (Citation2003) have been shown to be conspecific to Q. agrimoniae in our study. As for P. agrimoniae, currently there is no type-derived sequence to confirm its phylogenetic affinities. Since the type specimen of P. agrimoniae was not obtained from all possibly deposited herbaria, it might have been lost. Neotypification is thus needed using a new and suitable specimen from the original host and locality.
Hitherto, several genera in Pucciniastraceae have been delimitated based on morphological characters in teliospores (Cummins and Hiratsuka Citation2003), even at generic level, the position of telia and morphology of teliospores have long been used as important taxonomic criteria (Hiratsuka Citation1958; Cummins and Hiratsuka Citation2003; Liang et al. Citation2005; Yang Citation2015). Although there have been much debates concerning the generic classification based on telial morphologies, recent phylogenetic studies and our study supported the monophyly of all sampled genera in Pucciniastraceae, thus, supported the effectiveness of telial morphologies as taxonomic criteria at generic level. In addition, our morphological and molecular studies further emphasised the importance of the uredinial morphology at generic level because Quasipucciniastrum and Cronartium with Milesia-type uredinia show a much closer relationship than those with Caeoma-type uredinia (e.g. Cronartium, Chrysomyxa). These overlooked morphological characters appeared to be very useful in delimiting taxa at generic and suprageneric level. Further comprehensive studies need to be conducted to evaluate the effectiveness of these morphological characters in rust taxonomy.
Acknowledgements
We express our gratitude to Dr. Yi-Jian Yao (State Key Laboratory of Mycology, Institute of Microbiology, CAS, Beijing, China), Dr. Matthew E. Smith (University of Florida Herbarium, USA), Dr. Timothy James (University of Michigan Herbarium, USA), Dr. Barbara M. Thiers (The New York Botanical Garden, NY, USA) for providing dried specimens for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aime MC. 2006. Toward resolving family-level relationships in rust fungi (Uredinales). Mycoscience. 47:112–122.
- Aime MC, Bell CD, Wilson AW. 2018. Deconstructing the evolutionary complexity between rust fungi (Pucciniales) and their plant hosts. Stud Mycol. 89:143–152.
- Alaei H, Mohammadi AH, Dehghani A. 2012. Molecular characterization of the rDNA-ITS sequence and a PCR diagnostic technique for Pileolaria terebinthi, the cause of pistachio rust. Phytopathol Mediterr. 1:488–495.
- Arthur JC. 1910. New species of Uredineae VII. Bull Torrey Bot Club. 37:569–580.
- Beenken L, Lutz M, Scholler M. 2017. DNA barcoding and phylogenetic analyses of the genus Coleosporium (Pucciniales) reveal that the North American goldenrod rust C. solidaginis is a neomycete on introduced and native Solidago species in Europe. Mycol Prog. 16:1073–1085.
- Blomquist CL, Scheck HJ, Haynes J, Woods PW, Bischoff J. 2014. First published report of rust on white alder caused by Melampsoridium hiratsukanum in the United States. Plant Dis. 98:155.
- Cao ZM, Li ZQ. 1999. 秦岭锈菌. [Rust fungi of Qinling Mountains]. Beijing (China): China Forestry Publishing House. Chinese.
- Cao ZM, Li ZQ, Zhuang JY. 2000. Uredinales from the Qinling Mountains. Mycosystema. 19:13–23.
- Cummins GB, Hiratsuka Y. 2003. Illustrated genera of rust fungi. 3rd ed. St. Paul: Minnesota: American Phytopathological Society.
- Farr DF, Rossman AY. 2018. Fungal databases, systematic mycology and microbiology laboratory, agricultural research service, United States department of agriculture. [accessed 2018 Aug. 10]. http://nt.ars-grin.gov/fungaldatabases/
- Feau N, Vialle A, Allaire M, Maier W, Hamelin RC. 2011. DNA barcoding in the rust genus Chrysomyxa and its implications for the phylogeny of the genus. Mycologia. 103:1250–1266.
- Feau N, Vialle A, Allaire M, Tanguay P, Joly DL, Frey P, Callan BE, Hamelin RC. 2009. Fungal pathogen (mis-) identifications: a case study with DNA barcodes on Melampsora rusts of aspen and white poplar. Mycol Res. 113:713–724.
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 2(2):113–118.
- Guo L. 1989. 神农架锈菌. [Uredinales of Shennongjia]. Beijing (China): World Book Inc. Chinese.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Hiratsuka N. 1958. Revision of taxonomy of the Pucciniastreae with special reference to species of the Japanese Archipelago. Tokyo (Japan): Kasai Publishing and Printing.
- Holcomb GE, Aime MC. 2010. First report of Plumeria spp. rust caused by Coleosporium plumeriae in Louisiana and Malaysia and Catheranthus roseus, a new host of this rust. Plant Dis. 94:272–273.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Kaitera J, Tillman-Sutela E, Kauppi A. 2010. Chrysomyxa ledi, A new rust fungus sporulating in cone scales of Picea abies in Finland. Scand J Forest Res. 5(3):202–207.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kuprevich VF, Tranzschel VG. 1957. Rust fungi. 1. Family Melampsoraceae. In: Savich VP, editor. Cryptogamic plants of the USSR (Vol. 4). Komarova: Botanicheskogo Instituta; p. 423–464.
- Liang YM. 2006. Taxonomic evaluation of morphologically similar species of Pucciniastrum in Japan based on comparative analyses of molecular phylogeny and morphology. Ibaraki (Japan): University of Tsukuba.
- Liang YM, Tian CM, Hiratsuka K, Kakishima M. 2005. A new species of Pucciniastrum on Enkianthus campanulatus from Japan. Mycotaxon. 92:371–376.
- Lu HS. 2001. Using value of Agrimonia pilosa and its cultivation. Chinese Wild Plant Resources. S6:45–52.
- Maier W, Begerow D, Weiss M, Oberwinkler F. 2003. Phylogeny of the rust fungi: an approach using nuclear large subunit ribosomal DNA sequences. Can J Bot. 81:12–23.
- Maneval WE. 1937. A list of the Missouri fungi. Univ of Missouri Studies. 12(3):1–150.
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, et al. 2007. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Biol Evol. 43:430–451.
- McKenzie EHC, Padamsee M, Dick M. 2013. First report of rust on Alnus in New Zealand is Melampsoridium betulinum, not M. hiratsukanum. Plant Pathol Quar. 3:59–65.
- McTaggart AR, Aime MC. 2018. The species of Coleosporium (Pucciniales) on Solidago in North America. Fungal Biol. 122:800–809.
- McTaggart AR, Geering ADW, Shivas RG. 2014. Uredinopsis pteridis and Desmella aneimiae, the first rust fungi (Pucciniales) reported on ferns (Pteridophyta) in Australia. Australia Plant Dis Notes. 9(1):149.
- Padamsee MMcKenzie EHC. 2014. A new species of rust fungus on the new zealand endemic plant, myosotidium, from the isolated chatham islands. Phytotaxa. 174(4):223–230.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Stamatakis A, Alachiotis N. 2010. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics. 26:132–139.
- Sydow P, Sydow H. 1915. Monographia Uredinearum. III: melampsoraceae, Zaghouaniaceae, Coleosporiaceae. Leipzig (Germany): Borntraeger.
- Tai FL. 1979. 中国真菌总汇. [Sylloge Fungorum Sinicorum]. Beijing (China): Science Press. Chinese.
- Tranzschel WA. 1895. Über die Teleutosporen von Urtlda arcticus, U. Agrimoniae u. Melampsora alni. Script Horti Bot Univ, Petropolis. 4:299–301.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of bacteriology. 172:4238–4246.
- Vogler DR, Bruns TD. 1998. Phylogenetic relationships among the pine stem rust fungi (Cronartium and Peridermium spp.). Mycologia. 90(2):244–257.
- White TJ, Bruns T, Lee SJWT, Taylor JL. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. 18(1): 315–322.
- Yang T. 2015. 广义膨痂锈菌属的系统分类学研究. [Phylogenetic and taxonomic studies of Pucciniastrum s.l.] [Dissertation]. Beijing (China): Beijing Forestry University. Chinese.
- Zhang N, Zhuang JY, Wei SX. 1997. Fungal flora of the Daba Mountains: uredinales. Mycotaxon. 61:49–79.
- Zhao P, Kakishima M, Wang Q, Cai L. 2017. Resolving the Melampsora epitea complex. Mycologia. 109(3):391–407.
- Zhao P, Tian CM, Yao YJ, Wang Q, Kakishima M, Yamaoka Y. 2014. Melampsora salicis-sinicae (Melampsoraceae, Pucciniales), a new rust fungus found on willows in China. Mycoscience. 55:390–399.
- Zhao P, Tian CM, Yao YJ, Wang Q, Yamaoka Y, Kakishima M. 2013. New records of Melampsora species on willows in China. Mycotaxon. 123:81–89.
- Zhuang JY. 1983. 福建锈菌初步名录. [A provisional list of Uredinales of Fujian Province, China]. Mycosystema. 2:146–158. Chinese.
- Zhuang JY, Wei SX. 1999. Fungal flora of tropical Guangxi, China: a preliminary checklist of rust fungi. Mycotaxon. 72:377–388.
- Zhuang WY. 2001. Higher Fungi of Tropical China. New York (NY): Mycotaxon, Ltd.
- Zhuang WY. 2005. Fungi of northwestern China. New York (NY): Mycotaxon Ltd.
