ABSTRACT
A new species of Pythiogeton isolated from wild rice exhibiting rot and dieback of roots and stalks in California is described. Pythiogeton manoomin sp. nov. is characterized by coenocytic hyphae, club-like appressorium, and terminal or intercalary sporangia, which are often a short distance from the end of supporting hyphae. The protoplasm is discharged through a discharge tube into an elongate transient vesicle, which soon disappears, leaving the naked protoplasm to differentiate into reniform zoospores. Pythiogeton manoomin also produces thick-walled pigmented chlamydospores, not found in other Pythiogeton species. In greenhouse trials, Pg. manoomin did not infect economically important crops such as rice, bean, chard, corn, carrot, lettuce, oat, radish, sweet pepper, tomato, or wheat. Phylogenetic analysis based on ITS data supports the conclusion that this organism is a new species that is most closely related to Pg. ramosum. In this paper, we describe morphological characteristics, temperature–growth relationships, pathogenicity, and phylogenetic relationships that support the description of this taxon as a new species, Pythiogeton manoomin sp. nov.
urn:lsid:zoobank.org:pub:4C63AAA4-4D4A-4679-A344-79B75121A5C6
Introduction
The genus Pythiogeton (Pg.), together with Pythium and Phytophthora, belongs to the family Pythiaceae of the Oomycota. Minden first described members of the genus in 1916, and since then, Pythiogeton species have been reported in the USA (Drechsler Citation1932), Canada (Sparrow Citation1932), Germany and Denmark (Lund Citation1934), Japan (Ito and Tokunaga Citation1935), England (Sparrow Citation1936), China (Shen and Siang Citation1948), Africa (Gaertner Citation1954), India (Dayal Citation1968), Brazil (Beneke and Rogers Citation1970; Rocha et al. Citation2014), Taiwan (Watanabe Citation1974), Poland (Batko Citation1971), Pakistan (Lodhi et al. Citation2006), and Australia (Le et al. Citation2014). Pythiogeton species are characterized by production of zoospores outside of sporangia from a naked mass of protoplasm, not from a vesicle, which is characteristic of Pythium species. Members of the genus are facultative, anaerobic, ubiquitous inhabitants of soil and water. Pythiogeton can be readily isolated from plant materials submerged in stagnant water, and prefers anaerobic conditions (Huang et al. Citation2013).
Table 1. Averaged daily colony radial growth and standard deviation, in mm, on Rye B agar. Three plates were used for each temperature, and the experiment was repeated three times.
To date, the Pythiogeton genus encompasses 15 recognized species: Pythiogeton utriforme (Minden Citation1916), Pg. ramosum (Minden Citation1916), Pg. transversum (Minden Citation1916), Pg. autossytum (Drechsler Citation1932), Pg. uniforme (Lund Citation1934), Pg. dichotomum (Ito and Tokunaga Citation1935), Pg. nigrescens (Batko Citation1971), Pg. zeae (Jee et al. Citation2000), Pg. zizaniae (Ann et al. Citation2006), Pg. abundans (Huang et al. 2012), Pg. microzoosporum (Huang et al. 2012), Pg. oblongilobum (Huang et al. 2012), Pg. paucisporum (Huang et al. 2012), Pg. proliferatum (Huang et al. 2012), and Pg. puliensis (Huang et al. 2012). Most described Pythiogeton species are saprophytic, with only three species, Pg. zeae, Pg. zizaniae, and Pg. ramosum known to be pathogenic to corn, water bamboo, and ginger, respectively (Jee et al. Citation2000; Ann et al. Citation2006; Le et al. Citation2014).
Wild Rice (Zizania spp.) or manoomin, as it is called in the Ojibwe language, is a semi-aquatic grass species endemic to North America and China. Three species of wild rice, Zizania palustris, Z. aquatica, and Z. texana, are native to North America, and one species, Z. latifolia, is native to China. Wild rice is one of America’s oldest indigenous cultivated grains, with over 90 percent of cultivated wild rice grown in the United States produced in California and Minnesota (Hayes et al. Citation1989; Oelke Citation1993; Steeves Citation1952). Wild rice-growing regions in California include Shasta, Lake, Modoc, Lassen, Butte, Colusa, Yuba, Yolo and Sutter counties. In the summer of 2012, an outbreak of a newly discovered root and basal stalk rot of wild rice (Zizania palustris L.) cv. Franklin was observed in a 16-ha field in Big Valley, Lassen County, California. Infected plants exhibiting rot and dieback of roots and stalks were in various stages of decline, including death. A pythiaceous fungus was consistently isolated from the diseased plant and was identified as Pythiogeton sp. (Doan et al. Citation2014). The purpose of this study is to characterize the new Pythiogeton species isolated from wild rice and to confirm its pathogenicity.
Materials and methods
Isolation and cultivation
Wild rice (Zizania palustris L.) cv. Franklin exhibiting rot and dieback of roots and stalks were collected from Big Valley, Lassen County, California (GPS coordinates 41°08′41.93″ N 121°10′07.49″ W). Symptomatic stem and root tissues from affected plants were placed on 9 cm Petri dishes (four pieces per plate) of PARP agar (17 g Difco corn meal agar containing 5 ppm pimaricin, 250 ppm ampicillin, 10 ppm rifampicin, and 100 ppm pentachloronitrobenzene), which were then incubated at 25°C in the dark for 1 week. Agar blocks 8 mm in diameter containing hyphal tips were transferred onto 9 cm Petri dishes (4 pieces per plate) of potato dextrose agar (PDA) and modified Rye B agar (RBA) containing 60 g rye-grain extract, 20 g of Difco Bacto Agar, and 20 g of sucrose as described by Caten and Jinks (Citation1968).
Colonial morphology and mycelial growth
The effect of temperature on mycelial growth was evaluated on Rye B agar for three Pg. manoomin isolates (BV1, BV2, and BV3) from wild rice. A 3 day-old, 8 mm-diameter Rye B agar plug of each isolate was transferred onto Rye B agar plates for each of the nine temperatures evaluated. The plates were incubated at 4, 12, 15, 18, 21, 25, 28, 31, or 34°C in the dark, and the growth rate was determined daily by measuring the linear colony diameter in three places up to 5 d for each temperature. Three plates were used for each treatment, and the experiment was repeated three times with at least 24 h between each experiment. The average growth rate for each isolate from the three replications was calculated for each temperature. The average growth rate of three isolates was tested using analysis of variance (ANOVA) in R version 3.3.2 (The R Foundation).
Production of sporangia and release of zoospores
The morphology and development of sporangium were observed on Rye B agar using the method described by Chang (Citation1988) and Huang et al. (Citation2013). Hyphal tips were transferred onto modified Rye B agar. Agar blocks (5 x 5 x 3 mm) from 7 d-old Rye B agar cultures were placed in 9 cm Petri dishes (1 piece per plate) containing 20 mL of 10% clarified V-8 juice or deionized water and incubated in the dark at 24°C for 72 hours. The V-8 juice was then decanted and the mycelial mats were rinsed three times, at 20 min intervals, with sterile distilled water. For the development and release of zoospores, the mycelia mats with sporangia were incubated at 24°C for 48 h. Photographs were taken with a Leica DM5000 B microscope (Leica Microsystems, Wetzlar, Germany).
Pathogenicity tests
Pathogenicity of Pg. manoomin to Avena sativa L. cv. Cat Grass (Lake Valley Seed, Boulder, CO), Zea mays L. cv. Golden Bantam Improved (Seed Savers Exchange, Decorah, IA), Triticum aestivum cv. Liquid Sunshine (Botanical Interests, Broomfield, CO), Lactuca sativa L. cv. Great Lakes Iceberg (Lake Valley Seed), Beta vulgaris subsp. cicla L. cv. Fordhook Giant (Burpee Garden Product Co., Warminster, PA), Capsicum annum L. cv. California Wonder (Burpee Garden Product Co.), Daucus carota subsp. sativus L. cv. Short ‘n Sweet (Burpee Garden Product Co.), Solanum lycopersicum L. cv. Moneymaker (Everwilde Farms, Sand Creek, WI), Phaseolus vulgaris L. cv. Ejote silvestre Contender (Burpee Garden Product Co.), Raphanus sativus L. cv. Cherry Belle (Burpee Garden Product Co.), Oryza sativa L. cv. Calrose (USDA # C.I. 8988), and Zizania palustris L. cv. Franklin was determined in the greenhouse. Ten seeds of each were planted in sterilize sand in plastic pots measuring 10 cm in diameter (750 ml). After 5 days, six 8 mm-diameter agar discs from the margin of a 7day-old culture growing on modified Rye B agar were placed in each pot. Pots inoculated with six 8 mm-diameter agar discs from a modified Rye B agar plate were used as controls. After 20 d post inoculation, root and crown tissues were examined for symptoms and disease was recorded. The experimental design was a randomized block with ten replications (blocks) in the greenhouse on a 13-h photoperiod provided by high pressure sodium bulbs with daytime temperatures ranging from 80–85°C and nighttime temperatures ranging from 65–70°C. The pathogenicity test was repeated three times. Re-isolation of the pathogen from crown and root tissues was attempted for all inoculated plants, as described above. Damping-off assay was performed as above with the exception that 10 seeds were planted into sterilized sand that was pre-inoculated with 12 8 mm-diameter agar discs from the margin of a 7day-old culture grown on modified Rye B agar. After 10 d post inoculation, germinated seeds were counted, and re-isolation of the pathogen from seeds was attempted, as described above.
DNA extraction, PCR amplification, sequencing ITS rDNA
Total genomic DNA from three Pythiogeton manoomin isolates (BV1, BV2, and BV3) was extracted from mycelia using Qiagen® DNeasy Plant Mini Kit ™ (Valencia, CA) according to the manufacturer’s protocol. The internal transcribed spacer (ITS) region was amplified by PCR and sequenced using universal ITS5 and ITS4 primers (White et al. Citation1990). Three μL of genomic DNA were directly added to a 50 μL reaction that consisted of: 10 μL 5× Colorless GoTaq Reaction buffer (Promega Corp., Madison, Wisconsin), 5 μL 25 mM MgCl2, 4 μL containing 2.5 mM each dNTP, 0.2 μL GoTaq Taq polymerase (Promega Corp.), 0.1 μL each of a 50 μM concentration of primers, and 27.6 μL sterile deionized water. The thermal cycling parameters were initial denaturation at 94°C for 5 min followed by 34 cycles consisting of denaturation at 94°C for 1 min, annealing at 55°C for 2 min, and extension at 72°C for 2.5 min. A final extension at 72°C for 10 min was done at the end of the amplification followed by 4°C. The PCR products were purified with GeneJet PCR Purification Kit (Cat. No.: K0702, Thermo Fisher Scientific, Waltham, MA). Sequences of the forward and reverse strand were conducted by the College of Biological Sciences UCDNA Sequencing Facility (University of California, Davis, CA).
Sequence alignment and phylogenetic analyses
DNA sequences were edited and assembled using the BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The ITS sequence for all three Pythiogeton manoomin isolates was identical. Therefore, the sequences of Pythiogeton manoomin isolate BV1 was submitted to GenBank (KF719169). The ITS sequence (JQ610201) of Pythium aphanidermatum was selected as an outgroup. Sequences were aligned using ClustalW, and a phylogenetic tree was constructed with MEGA 5.03 (Tamura et al. Citation2011), using the maximum parsimony method with 1000 bootstrap replication and evolutionary distance analyzed according to Tamura-Nei model (Tamura et al. 2011). The maximum parsimony tree was obtained using the Tree-Bisection-Regrafting algorithm with search level 5 in which the initial trees were obtained by the random addition of sequences (Nei and Kumar Citation2000). All positions containing gaps and missing data were eliminated. The aligned sequence data set was deposited in TreeBASE (No. 24,156).
Taxonomy
Pythiogeton manoomin H. K. Doan and R. M. Davis, sp. nov.
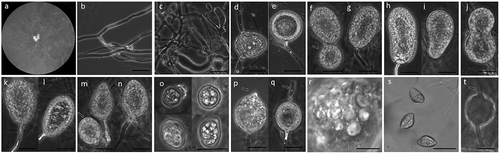
Figure 1. Pythiogeton manoomin. (a), Colony on PDA; (b–c), hyphae and appressoria (arrows) on RSA; (d–e), sporangia, terminal or intercalary and short distance from the end of supporting hyphae; (f–g), globose (arrow) and ellipsoid sporangia, (h–i), reniform sporangia, (j), bilobate sporangia, (k–l), pyriform sporangia, (m–n), globose (arrow) and ovoid sporangia; (o), chlamydospores, (p–q), sporangia initiating formation of the discharge tube (arrowheads), (r), formation of zoospores from a protoplasm mass, and, (s), discharged zoospores; (t), empty sporangia, showing empty discharge tubes (arrows). Scale bars = 20 μM.
MycoBank MB 830264
Typification
United States, California: Lassen County. Isolate BV1, designated as the holotype, was recovered from root tissue of wild rice (Zizania palustris L.) cv. Franklin from a commercial agricultural field on 20 June 2012 and deposited in the University and Jepson Herbaria (UC 2060102), Berkeley, California. The living culture of the isolate was deposited and maintained at the Centraalbureau voor Schimmelcultures (CBS 145436), Fungal Biodiversity Centre, Utrecht, The Netherlands. Description of the species was submitted to MycoBank (MB 830264) and the ITS sequence was deposited in GenBank (GenBank Accession No. KF719169).
Etymology
manoomin, referring to wild rice in the Ojibwe language. Wild rice or manoomin, is an important native grain with spiritual and subsistence significance to many indigenous people of North America.
Description
Colonies on Rye B agar at 25°C with daily growth rate of 23.0 mm per day, with no distinct pattern on PDA and Rye B agar. Hyphae coenocytic, 5–7 mm in diameter. Appressoria knob-like, 3.3–5.9 × 7.7–10.2 μM, or club-like, 7.9–15.7 × 42.1–72.0 μM. Sporangia terminal or intercalary on supporting hyphae, abundant, various in shape, globose, 21–33 μM in diam., ellipsoid, 20–35 × 32–80 μM, reniform, 25–35 × 35–80 μM, bilobate, 30–50 × 50–80 μM, pyriform, 25–35 × 40–60 μM, or ovoid, 25–45 × 35–50 μM. Chlamydospores, various in size and shape, 10–15 μM in diameter, thick-walled, intercalary, pigmented (golden-brown). Discharge tube 5–100 μM long and 5 μM wide, protruding from different positions around sporangia. Encysted zoospores 15–25 μM in diameter. Sexual structures absent. Cardinal temperatures () for growth: minimum, 12°C; optimum, 21°C; maximum, 31°C.
Results
Pathogenicity tests
– Pathogenicity of Pg. manoomin to wild rice was confirmed by inoculation followed by re-isolation. After 14 days, 84% of inoculated wild rice plants in all tests developed root and basal stalk rot, consistent with the symptoms observed in diseased wild rice in the field. No symptoms were observed on other inoculated plants. In addition, Pg. manoomin caused damping-off of 71% of wild rice seedlings tested. Pythiogeton manoomin was consistently re-isolated on PARP from symptomatic wild rice plants and seeds, but not from other inoculated plants or seeds and control plants, thus fulfilling Koch’s postulate. Pythiogeton manoomin did not infect the seedlings of Asian rice, bean, chard, corn, carrot, lettuce, oat, radish, sweet pepper, tomato, or wheat. All control plants grown in noninoculated soil remained healthy at the end of the experiment.
Sequence analysis
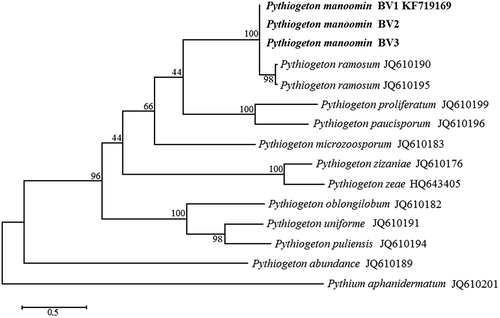
– The internal transcribed spacers (ITS) 1 and 2 flanking the 5.8S rRNA regions were amplified by PCR and sequenced using universal ITS5 and ITS4 primers. A BLAST search of the 855 bp sequences revealed 98% similarity with a sequence of Pg. ramosum isolate Pg-164 (GenBank Accession No. JQ610190.1). The 21 nucleotide differences suggest that the isolate from wild rice may be an unreported species. Based on previous published phylogeny, all Pythiogeton species belong to the same clade, separated from Pythium, Phytophthora, and downy mildew clades. Within the Pythiogeton clade, 10 subgroups (A1-A10) have been reported (Huang et al. Citation2013). The 21 nucleotide differences in the 5.8S rRNA region place Pythiogeton manoomin isolate BV1 into a new subgroup with high bootstrap value ().
Discussion
The genus Pythiogeton was erected in Germany by Minden in 1916. Since then, many species have been reported around the world (Jee et al. Citation2000). Of the described species, only three, Pg. zeae, Pg. zizaniae, and Pg. ramosum, are known to be plant pathogens (Jee et al. Citation2000; Ann et al. Citation2006; Le et al. Citation2014). Here, we characterized a new species of Pythiogeton capable of causing root and basal stalk rot of wild rice (Zizania palustris L.). Pythiogeton manoomin can be readily isolated from plant materials (in this case, wild rice) submerged in stagnant water during crop cultivation. Host range tests indicate that the pathogenicity of Pg. manoomin may be limited to wild rice and does not infect seedlings of economically important crops such rice, bean, carrot, chard, corn, lettuce, oat, radish, sweet pepper, tomato, or wheat. However, Pg. manoomin can possibly infect some wild grasses or weeds not tested here.
Morphologically, Pythiogeton manoomin is characterized by ovoid to ellipsoid sporangia in terminal or intercalary positions. Like all members of the genus Pythiogeton, Pg. manoomin sporangia release undifferentiating protoplasm through a discharge tube into an elongate transient vesicle, which soon disappears, leaving the naked protoplasm to differentiate into reniform zoospores. Appressoria form sparsely in contact with hard surfaces, (i.e. bottom surface of Petri dish). Pythiogeton manoomin is morphologically and genetically related to Pg. ramosum. Initially, descriptions of most Pythiogeton species were based exclusively on uncultured material (Minden Citation1916; Ito and Tokunaga Citation1935; Batko Citation1971; Jee et al. Citation2000). Isolation of some Pythiogeton species is difficult because other soilborne oomycetes outcompete it in culture. Recently, some species have been successfully grown in laboratory cultures (Jee et al. Citation2000; Ann et al. Citation2006; Huang et al. Citation2013). Pythiogeton manoomin and Pg. ramosum can be distinguished from other members of the genus by their readily culturable characteristics, as they can be cultured on a number of media, and have nonspecific requirements for growth media (Jee et al. Citation2000; Ann et al. Citation2006; Huang et al. Citation2013). In addition, both species are viable after subcultering and under storage conditions for up to two years (Jee et al. Citation2000; Le et al. Citation2015). The morphology of both species is variable and dependent on the type of water and medium used, so identification can be problematic if based on morphology alone (Le et al. Citation2015). However, Pythiogeton manoomin can be distinguished from Pg. ramosum based on pathogenicity and optimal growth temperature. Pg. ramosum, like other described Pythiogeton species, grows optimally at temperatures above 30°C, and does not grow at lower temperatures of 10–12°C (Huang et al. Citation2013; Le et al. Citation2015). In contrast, Pg. manoomin can grow at 10–12°C, with optimal growth at 25°C. Unlike other described Pythiogeton species, Pg. manoomin produces chlamydospores, which may allow for long-term survival (Jee et al. Citation2000). Chlamydospores are important survival structures of many plants of the family Pythiaceae, such as members of the Pythium and Phytophthora species (Hendrix and Campbell Citation1973; Mitchell Citation1978). Pythiogeton manoomin can also be distinguished from other Pythiogeton species based on pathogenicity tests. Pg. ramosum is pathogenic on ginger, bean, cauliflower, pepper, and lettuce (Le et al. Citation2015), Pg. zeae is pathogenic on corn and carrot (Jee et al. Citation2000), and Pg. zizaniae is pathogenic specifically to water bamboo, and does not infect seedlings of corn, rice, wheat, sorghum, cucumber, tomato, soybean or water spinach. (Ann et al. Citation2006). Pythiogeton manoomin causes disease only on wild rice among the crops tested here, but may be able to infect some wild grasses or weeds not tested here. Although Py. manoomin is phylogenetically similar to Pg. ramosum, a complete sequence comparison of the ITS revealed 98% similarity with a sequence of Pg. ramosum isolate Pg-164 (GenBank Accession No. JQ610190.1). However, the 21 nucleotide differences suggest that the isolate from wild rice is an unreported species.
Wild rice, the only cultivated cereal native to North America, is a staple food of indigenous peoples (Hayes et al. Citation1989; Oelke Citation1993). Today, wild rice is cultivated and commercialized for its unique flavor, texture, and nutritional value (Oelke et al. Citation1997). Several fungal diseases are considered important factors that limit wild rice production. For instance, fungal brown spots caused by Bipolaris oryzae (Bean and Schwartz Citation1961; Malvick and Percich Citation1993), sheath and stem rots caused by Sclerotium hydrophilum and S. oryzae (Punter et al., Citation1984), Fusarium head blight caused by Fusarium graminearum (Nyvall et al. Citation1999), crown and root rot caused by Phytophthora erythroseptica (Gunnell and Webster Citation1988), and damping-off caused by Pythium torulosum (Marcum and Davis Citation2006) have been associated with 100% losses in fields where disease was especially severe. This is the first report of the pathogenicity of a Pythiogeton species on wild rice. Infected plants exhibiting rot and dieback of roots and stalks were in various stages of decline, including death. Depending on disease severity, Pg. manoomin can cause losses in individual fields, and can vary from slight to near total stand failure. Since Pg. manoomin produces chlamydospores, management may be difficult. In conclusion, based on pathogenicity tests and evaluation of its morphological and molecular taxonomy, we confirm that Pg. manoomin (H. K. Doan and R. M. Davis), isolated from wild rice plants exhibiting rot and dieback of roots and stalks in California, is a distinct new Pythiogeton species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ann PJ, Huang JH, Wang IT, Ko WH. 2006. Pythiogeton zizaniae, a new species causing basal stalk rot of water bamboo in Taiwan. Mycologia. 98:116–120.
- Batko A. 1971. A new species of Pythiogeton showing Achlya- like diplanetism. Acta Mycol. 7:241–249.
- Bean GA, Schwartz R. 1961. A severe epidemic of Helminthosporium brown spot disease on cultivated wild rice in Minnesota. Plant Dis Rep. 45:901.
- Beneke ES, Rogers AL. 1970. Aquatic fungi of parque nacional de itatiaia in the state of Rio de Janeiro. Rickia. 5:51–64.
- Caten CE, Jinks JL. 1968. Spontaneous variability of single isolates of Phytophthora infestansI. Cultural variation. . Can J Bot. 46:329–348.
- Chang HS. 1988. Phytophthora species associated with strawberry fruit rot in Taiwan. Bot Bull Acad Sinica. 29:61–67.
- Dayal R. 1968. Studies in aquatic fungus of Varanasi. Part 4. New records and notes of taxonomy. Nat Acad Sci India Annu. 37:108.
- Doan HK, Davis RM, Sartori FF, Marcum DB. 2014. First report of a Pythiogeton sp. causing root and basal stalk rot of wild rice in California. Plant Dis. 98:851.
- Drechsler C. 1932. A species of Pythiogeton isolated from decaying leaf-sheaths of the common cat-tail. J Wash Acad Sci. 22:421–449.
- Gaertner AV. 1954. Uber das Vorkommen niederer Erd- phycomyceten in Afrika, Schweden und an einigen mit- teleuropaischen Standortem. Arch Microbiol. 21:4–56.
- Gunnell PS, Webster RK. 1988. Crown and root rot of cultivated wild rice in California caused by Phytophthora erythroseptica sensu lato. Plant Dis. 72:909–910.
- Hayes PM, Stucker RE, Wandrey GG. 1989. The domestication of American wild rice. Econ Bot. 43:203–214.
- Hendrix FF, Campbell WA. 1973. Pythiums as plant pathogens. Annu Rev Phytopathol. 11:77–98.
- Huang JH, Chen CY, Lin YS, Ann PJ, Huang HC, Chung WH. 2013. Six new species of Pythiogeton in Taiwan, with an account of the molecular phylogeny of this genus. Mycoscience. 54:130–147.
- Ito S, Tokunaga Y. 1935. Notae mycologicae Asiae orientalis I. Trans Sapporo Nat Hist Soc. 14:1–33.
- Jee HJ, Ho HH, Cho WD. 2000. Pythiogeton zeae sp nov. causing root and basal stalk rot of corn in Korea. Mycologia. 92:522–527.
- Le DP, Smith MK, Aitken EA. 2014. First report of Pythiogeton ramosum (Pythiales) in Australia. Australas Plant Dis Notes. 9:130.
- Le DP, Smith MK, Aitken EA. 2015. Pythiogeton ramosum, a new pathogen of soft rot disease of ginger (Zingiber officinale) at high temperatures in Australia. Crop Prot. 77:9–17.
- Lodhi AM, Shahzad S, Ghaffar A, Levesque CA. 2006. Oomycete species of Pakistan – a morphological and molecular study. Inoculum. 57:27.
- Lund A. 1934. Studies on Danish freshwater Phycomycetes and notes on their occurrence particularly relative to the hydrogen ion concentration of the water. Memoirs R Aca Sci Lett Denmark, Ser. 9:1–97.
- Malvick DK, Percich JA. 1993. Hydroponic culture of wild rice (Zizania palustris L.) and its application to studies of silicon nutrition and fungal brown spot disease. Can J Plant Sci. 73:969–975.
- Marcum DB, Davis RM. 2006. First report of damping-off of wild rice in California caused by Pythium torulosum. Plant Dis. 90:523.
- Minden MV. 1916. Beitrage zur biologie und systematik einheimsche submerser Phycomyceten. Falk Mykolog Untersuch Berichte. 2:146–255.
- Mitchell DJ. 1978. Relationships of inoculum levels of several soilborne species of Phytophthora and Pythium to infection of several hosts. Phytopathology. 68:1754–1759.
- Nei MKumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press; p. 115–140.
- Nyvall RF, Percich JA, Mirocha CJ. 1999. Fusarium head blight of cultivated and natural wild rice (Zizania palustris) in Minnesota caused by Fusarium graminearum and associated Fusarium spp. Plant Dis. 83:159–164.
- Oelke EA. 1993. Wild rice: domestication of a native North American genus. New York: New crops. Wiley; p. 235–243.
- Oelke EA, Porter RA, Grombacher AW, Addis PB. 1997. Wild rice-new interest in an old crop. Cereal Food World. 42:234–247.
- Punter D, Reid J, Hopkin AA. 1984. Notes on sclerotium-forming fungi from Zizania aquatica (Wildrice) and other hosts. Mycologia. 76:722–732.
- Rocha JR, Sousa ND, Santos LA, Pereira AA, Negreiros NC, Sales PC, Trindade Júnior OC. 2014. The genus Pythiogeton (Pythiogetonaceae) in Brazil. Mycosphere. 5:623–634.
- Shen SC, Siang WN. 1948. Studies in the aquatic Phycomycetes of China. Sci Rep Nat Tsing Hua Univ, Ser B. 3:179–206.
- Sparrow FK. 1932. Observations on the aquatic fungi of Cold Spring Harbor. Mycologia. 24:268–303.
- Sparrow FK Jr. 1936. A contribution to our knowledge of the aquatic Phycomycetes of Great Britain. Bot J Linn Soc. 50:417–478.
- Steeves TA. 1952. Wild rice—indian food and a modern delicacy. Econ Bot. 6:107–142.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei MKumar S. 2011. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Watanabe T. 1974. Fungi isolated from the underground parts of sugar cane in relationship to the poor ratooning in Taiwan. (2) Pythium and Pythiogeton. Trans Mycol Soc Japan. 15:343–357.
- White TJ, Bruns T, Lee S, Taylo JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a guide to methods and applications. New York: Academic Press, Inc; p. 315–322.