ABSTRACT
Several Usnea species in subgenus Eumitria (Parmeliaceae, lichenized Ascomycota) have been described from East Africa in the past decades. These have been based on morphology and chemistry data while molecular studies remain very limited. In this paper we are for the first time publishing phylogenetic analyses along with morphological and chemical data for Eumitria. A total of 62 new sequences of Eumitria (26 ITS, 20 nuLSU, 6 MCM7, 10 RPB1) were generated in this study. nuLSU, MCM7 and RPB1 sequences are here for the first time reported for U. baileyi. A phylogeny of subgenus Eumitria from Tanzania based on Bayesian and maximum likelihood analyses of a concatenated four-loci data set is presented, confirming the monophyly of Eumitria. Further, secondary chemistry and variation in characters, such as the pigmentation of the central axis and branch shape were investigated.
Introduction
Usnea is one of the largest genera in the family Parmeliaceae, comprising about 350 species (Thell et al. Citation2012). They are characterized by having radially symmetric branches with a central chord consisting of an elastic cartilaginous strand of longitudinally orientated hyphae (Wirtz et al. Citation2006) and the presence of usnic acid in the cortex. The morphological variability among the species is a major challenge in the delimitation of the species (Clerc Citation1998). Motyka (Citation1936) published a world monograph of Usnea in which the morphological features were the base for species recognition. However, morphological characters as used by modern taxonomists differ from those of Motyka (Clerc Citation1998). Revisions of the taxonomy of Usnea including descriptions of morphological characters and secondary chemistry as detected by thin layer chromatography have during the past 50 years been published from different parts of the world (Swinscow and Krog Citation1975; Citation1979, Citation1986, Citation1988; Clerc Citation1997; Citation1998, Citation2011; Ohmura Citation2001; Citation2012).
Molecular data have recently proved very useful in subgeneric and species recognition in Usnea, and the use of a combination of loci has resulted in good resolution (Truong et al. Citation2013).
Studies on Usnea species in Africa are few, but some species were described from Southern (Dodge Citation1956, Citation1957) and Eastern Africa (Swinscow and Krog Citation1975, Citation1979, Citation1986, Citation1988) based on morphology and chemistry data. Molecular work on African Usnea is limited, and only two sequences (ITS) have been published (Orock et al. Citation2012).
The genus Eumitria was described by Stirton (Citation1882). It was characterized by having a tubular central axis ± filled with loose hyphae throughout the branches of the thallus. It was later considered a subgenus of Usnea (Motyka Citation1936; Ohmura Citation2001, Citation2002; Truong and Clerc Citation2013). Articus (Citation2004), on the base of molecular data, resurrected it to generic level, which was also accepted by Divakar et al. (Citation2017). Here, we treat Eumitria as a subgenus of Usnea since the latter forms a strongly supported monophyletic group that is morphologically well characterized (Ohmura and Kanda Citation2004; Wirtz et al. Citation2006; Thell et al. Citation2018).
Eumitria has been studied in Australia (Rogers and Stevens Citation1988; Stevens Citation1999), East Asia (Ohmura Citation2001, Citation2012), and South America and the Galapagos (Truong and Clerc Citation2013). The knowledge of Eumitria in Africa, however, is rather limited and mainly restricted to Central and East Africa (Motyka Citation1936; Dodge Citation1956; Swinscow and Krog Citation1974, Citation1986; Krog Citation1994). In these papers morphology and secondary chemistry () were used in the descriptions.
Table 1. Table of Usnea subgenus Eumitria taxa from Africa (Dodge Citation1956; Swinscow and Krog Citation1974, Citation1986; Krog Citation1994) with their main chemistry indicated by X; protocetraric acid (PRO), fumarprotocetraic acid (FUM), diffractaic acid (DIF), salazinic acid (SAL), norstictic acid (NOR), constictic acid (CON), thamnolic acid (THA), psoromic acid (PSO), triterpenoids (TRI), virensic acid (VIR).
Very little molecular work has been done on Eumitria worldwide, and even less on African material. Up to date (retrieved on the 24th of January 2019) only one Eumitria ITS sequence (from Usnea baileyi), has been published from African material (Orock et al. Citation2012) and three ITS sequences are available from other parts of the world (Ohmura Citation2002). For Usnea pectinata Taylor so far only six verified sequences (three ITS, one mtSSU, one nuLSU and one MCM7) have been deposited in GenBank (Ohmura and Kanda Citation2004; Articus Citation2004; Truong et al. Citation2013), none of them from Africa. No other Eumitria species have so far been scrutinised by molecular work.
Mountain rainforests in Tanzania harbour a high diversity of Usnea species that remains understudied. This paper aims to present a phylogeny of Eumitria recently collected in Tanzania based on molecular information, supplemented by morphological and secondary chemistry data, providing up-to-date knowledge of this notoriously difficult and thus understudied group.
Material and methods
Taxon sampling
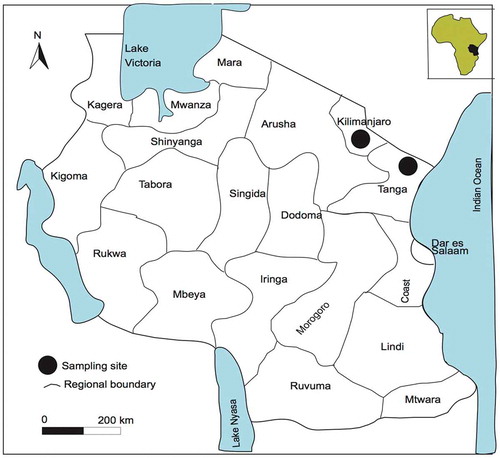
This study is mainly based on materials collected by the first author in Korogwe (Tanga) and Kilimanjaro, Tanzania () in 2016 and 2017. Voucher specimens collected during field trips have been deposited in UPS with some duplicates in UDSM and G.
For the first dataset, i.e. the large dataset (LD), the ITS, nuLSU, MCM7 and RPB1 loci were included. This dataset comprised 49 specimens representing 15 species of Usnea () selected for this study following the clades named by Truong et al. (Citation2013). In addition, two species belonging to Parmeliaceae that served as outgroup (Pleurosticta acetabulum (Neck.) Elix & Lumbsch and Lethariella cashmeriana Krog were included.
Table 2. Species and GenBank accession numbers of sequences used in the DNA analyses. Newly produced sequences in bold.
The second dataset, the small dataset (SD), comprised the subgenus Eumitria, where all the above mentioned loci were combined to form 23 sets of sequences representing Usnea baileyi and U. pectinata (). Based on the results of LD analyses Usnea longissima Ach. and U. trichodeoides Vain. from the subgenus Dolichousnea Y. Ohmura were chosen as outgroup for the analyses of the SD.
Morphological, anatomical and chemical studies
The morphology of specimens was investigated using a stereomicroscope. For each specimen, three measurements were made: cortex, medulla, and central axis. These were made on longitudinal sections of branches at ×50 magnification. The relative thickness of cortex/medulla/axis of the total branch diameter (CMA) and the ratio of axis/medulla (A/M) of all the studied specimens were calculated according to Clerc (Citation1984, Citation1987) and were ascribed to the categories defined by Clerc (Citation2011). Percentages of the tubular part of the axis (TBA) were calculated according to Truong and Clerc (Citation2013). In the species diagnostic features part, the CMA and TBA values are presented with their standard deviation. Observations of the anatomical structure of the cortex were made on thin hand-cut sections at ×400 as in Ohmura (Citation2001).
Chemical analyses of all the studied specimens were performed by thin layer chromatography (TLC) in solvent A, B and C following Culberson and Ammann (Citation1979), with solvent B modified according to Culberson and Johnson (Citation1982). The chemical substances were considered as a “main substance” if they were present in all specimens studied of a given species.
DNA extraction, PCR amplification and sequencing
Total DNA was extracted from freshly collected material less than three months old after having been kept at −20°C for a period of one month, using the DNeasy Plant Mini Kit (Quiagen, Hilden, Germany) following the manufacturer’s instructions. The material for extraction was selected carefully to avoid contamination. For each examined specimen, a branch piece about 1 cm long was used.
Total DNA was used for PCR amplifications. The primers used were ITS1F (Gardes and Bruns Citation1993), ITS4 (White et al. Citation1990); LROR and LR5 (Vilgalys and Hester Citation1990); MCM7-709 and MCM7-1349 (Schmitt et al. Citation2009); gRPB1-A and gRPB1-C (Matheny et al. Citation2002). The amplifications were carried out by using the AccuPower PCR PreMix (Bioneer, Daejeon, Korea), the reaction mixture consisting of 3 µl diluted DNA, 1.5 µl of each primer (10 mM), and water to a total volume of 20 µl. The thermal cycling parameters were: initial denaturation for 4 min at 95ºC, followed by 35 cycles of 1min at 94ºC, 1min at 54ºC, 45 s. at 72ºC, and a final elongation for 5 min at 72ºC. The PCR products were visualized by electrophoresis on 1.5% agarose gels. Products were purified using Illustra™ ExoStar buffer diluted 10×, following the manufacturer’s protocol. Sequencing was carried out by Macrogen (www.macrogen.com).
Alignments and phylogenetic reconstructions
DNA sequences of ITS, nuLSU, MCM7 and RPB1 representing species in each of the subgenera of Usnea according to Ohmura and Kanda (Citation2004) and Truong et al. (Citation2013) were downloaded from GenBank, after assessment of their quality. The selected DNA sequences downloaded from GenBank, along with the newly produced sequences (), were assembled and edited using AliView (Larsson Citation2014) and aligned with MAFFT v.7 (https//mafft.cbrc.jp/alignment/server/).
Phylogenetic relationships and their posterior probabilities, for both the LD and the SD, were inferred/calculated using a Bayesian approach, and additional support values were estimated using Maximum Likelihood Bootstrap Support (MLbs). For the Bayesian analyses, the most likely models of evolution were estimated for each region separately using the Akaike Information Criterion (AIC) as implemented in Modeltest 3.7 (Posada and Crandall Citation1998).
For the LD the GTR + G model of evolution was employed for nuLSU, MCM7 and RPB1 whereas GTR + I + G was used for ITS. For the SD the GTR + G model was implemented for ITS, nuLSU and MCM7 while the K80 was employed in RPB1.
The Bayesian analysis was executed using MrBayes 3.2.6 (Ronquist et al. Citation2012), where two analyses of two parallel runs were carried out for 10 M generations. Each run included four chains, and trees were sampled every 1000 generations and 25% were discarded as burn in. All runs converged on the same average likelihood score and topology. Single gene analyses were performed to test the congruence among the four datasets (ITS, nuLSU, MCM7 and RPB1). A test was considered incongruent if a well-supported monophyletic group with posterior probability (PP) ≥ 0.95 was found to have low support (non- monophyletic) when different loci were used. No significant incongruence among the single gene trees (Supplementary figures S2A, S2B, S2C and S2D) was detected, hence the four matrices were concatenated. Further analyses were performed after concatenation using Sequence Matrix (Vaidya et al. Citation2011). Maximum likelihood estimates were carried out by RAxML version 8.2.10 using the GTR + G + I model of site substitution (Stamatakis Citation2014). The branch support was acquired by maximum likelihood bootstrapping (MLbs) of 1000 replicates (Hillis and Bull Citation1993), and MLbs ≥ 70% were considered to be significant.
The trees were visualized in FigTree version 1.3.1 (Rambaut and Drummond Citation2010).
Results and discussion
Phylogenetic studies
Infrageneric clades in Usnea
This study generated 62 new sequences of Eumitria (26 ITS, 20 nuLSU, 6 MCM7, 10 RPB1).
Pleurosticta acetabulum and Lethariella cashmeriana were chosen as outgroups. A consensus tree based on the LD (ITS, nuLSU, MCM7 and RPB1), with an indication of infrageneric clades of Usnea, is shown in . Usnea contains well supported monophyletic clades agreeing with the circumscribed subgenera Dolichousnea, Eumitria and Neuropogon, in addition to Usnea 1 and Usnea 2, with Eumitria being the sister-clade to the other parts of Usnea. These results confirm those of Truong et al. (Citation2013).
Figure 2. Consensus tree based on a Bayesian and ML analyses of concatenated ITS, nuLSU, RPB1 and MCM7 showing infrageneric clades in Usnea. The tree was rooted using two species Pleurosticta acetabulum and Lethariella cashmeriana. The two support values associated with each internal branch correspond to posterior probability (PP) and bootstrap support (bs) respectively. Branches in bold indicate a support of PP ≥ 95% and a MLbs ≥ 70%. An asterisk on a bold branch indicates that this node has a support of 100 % for both support estimates. A dash instead of a MLbs value indicates that the node of the Bayesian tree was not recovered by ML bootstrapping. Species groups (within annotation marks) are in accordance with Truong et al (Citation2013). Eumitria is highlighted by a shaded box.
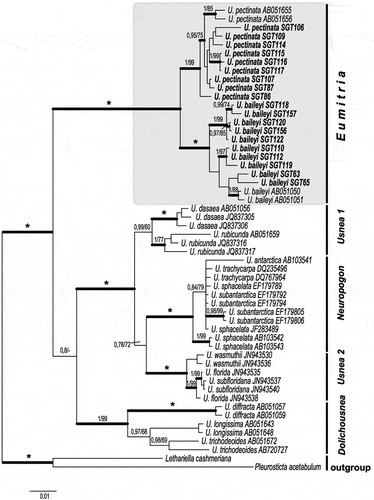
Eumitria in the SD is here represented by sequences of two species, Usnea baileyi and U. pectinata, with ten and nine samples respectively in the molecular study. nuLSU, MCM7 and RPB1 sequences are here for the first time reported for U. baileyi. Usnea baileyi and U. pectinata have previously been reported to belong in Eumitria based on molecular data (Ohmura and Kanda Citation2004; Articus Citation2004; Truong et al. Citation2013).
Phylogeny of Eumitria
A phylogeny of Eumitria in Tanzania is presented in where Usnea longissima and U. trichodeiodes, were chosen as outgroup. The subgenus Eumitria is strongly supported in the molecular phylogeny (). Eumitria was originally described as having a fistulose axis (Stirton Citation1882) and this feature was also emphasized by Motyka (Citation1936). Usnea baileyi has a typical fistulose axis ()). The close relationship between Eumitria baileyi and U. pectinata was, based on ITS, shown by Ohmura (Citation2002), Articus (Citation2004) and Wirtz et al. (Citation2006). Truong and Clerc (Citation2013), however stressed that the relationship of Eumitria needs more careful scrutiny. Usnea pectinata has a slightly fistulose axis on main branches ()) and an overall morphology rather different from that of U. baileyi, but Ohmura (Citation2002) considered it to belong to Eumitria as molecular data suggested a strong relationship with U. baileyi. The molecular data presented here as based on four loci strongly support the inclusion of Usnea pectinata in Eumitria ( and ).
Figure 3. Consensus tree based on Bayesian and ML analyses of Eumitria species in Tanzania (ITS, nuLSU, RPB1 & MCM7). The two support values associated with each internal branch correspond to posterior probability (PP) and bootstrap support (bs) respectively. Branches in bold indicate a support of PP ≥ 95% and a MLbs ≥ 70%. An asterisk on a bold branch indicates that this node has a support of 100% for both support estimates. A dash instead of a MLbs value indicates that the node of the Bayesian tree was not recovered by ML bootstrapping. A: America, I: Indonesia, J: Japan, T: Tanzania 1: main chemical substance, 0 accessory chemical substance, x: dark brown pigmentation, big black dots: terete branch shape, triangles: alate branch shape, pentagon: ridged branch shape.
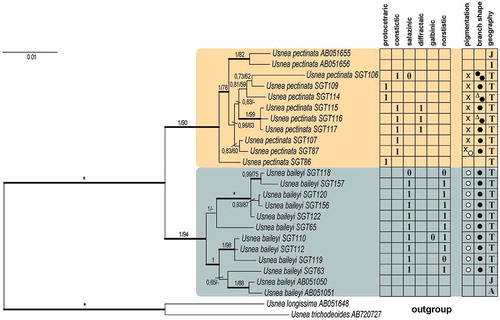
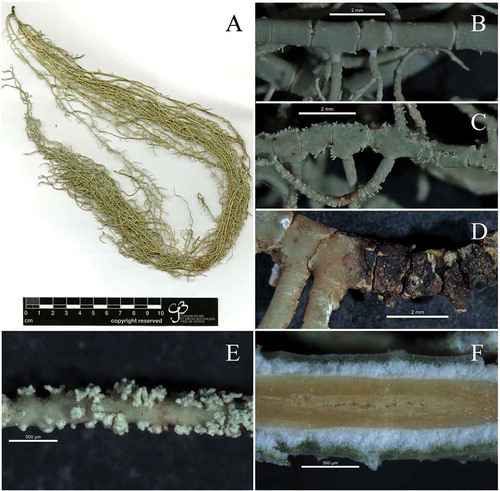
Figure 4. Usnea baileyi; (a): Usnea baileyi studied specimen (SGT 157), (b): blackish base, (c): soralia with short isidiomorphs (d): thin and shiny cortex, red subcortical pigment and tubular axis filled with loose hyphae.

Figure 5. Usnea pectinata; (a): Usnea pectinata studied specimen (SGT 114), (b): smain branch cylindrical with terete segments, (c): main branch irregular with alate segments, (d): blackish base, (e): soralia with short isidiomorphs, (e): dark brown pigmented axis of main branch with some fistulose areas in the central part of the axis.
RPB1 (Supplementary material (SM): Figure S2D) contributed to a better phylogenetic resolution than ITS (SM: Figure S2A) and MCM7 (SM: Figure S2C). The reliability of RPB1 as a phylogenetic marker has been emphasized in studies of Parmeliaceae such as in parmelioid and cetrarioid lichens (Crespo et al. Citation2010; Nelsen et al. Citation2011). The sequences of U. pectinata produced in this study form a strongly supported monophyletic clade (PP 1, MLbs 90) with the sequences U. pectinata available from GenBank (). The strong relationship between Usnea baileyi and U. pectinata is also demonstrated. This is a further step in treating Eumitria species from Tanzania and Africa, and for the first time comprehensive molecular data is supplied.
The species
Usnea baileyi (Stirt.) Zahlbr. ()
Diagnostic notes
Detailed descriptions of this species were given by Ohmura (Citation2001) and Truong and Clerc (Citation2013). Usnea baileyi has a sorediate, shrubby to sub-pendulous thallus with cylindrical branches often tapering towards the apices, and with few annulations ()). The base is concolorous or blackish with cracks below the first ramification ()). Numerous soralia with short isidiomorphs cover the branches ()). The cortex is shiny in section and thin to moderately thin (5.7–7.8%). A red subcortical pigment was observed ()). The medulla is thin ranging from 3.6 to 5.7%. The axis is thick (78–82%), most of it occupied by a tubular section (43–46%) filled with loose hyphae ()). These values differ from those reported by Ohmura (Citation2001) and Truong and Clerc (Citation2013) as shown in .
Table 3. Comparison of U. baileyi anatomical characters. Mean (italic), standard deviation and extreme values (in parenthesis) are shown.
Chemistry
In addition to usnic acid, all the specimens examined contained salazinic and norstistic acids as main substances. These substances have previously been reported to occur in Usnea baileyi from East Africa (Swinscow and Krog Citation1974). A trace of galbinic acid was detected in one specimen (SGT 110), this is for the first time reported for U. baileyi.
Distribution and ecology
Usnea baileyi has a subtropical-tropical distribution and a wide ecological range (Truong and Clerc Citation2013). The specimens studied were found on twigs of Isoberlinia scheffleri and Alablankia stomanii twigs in an altitude of 1227–1307 m in Korogwe (Tanga), and also on Makaranga kilimanjarika twigs at 1954 m in Marangu (Kilimanjaro).
Specimens examined: Tanzania, Tanga, about 16.4 km from Korogwe district in the Usambara Mountains, 5°04’15 “S 38°24’02“E, 1227 m (SGT 63, SGT 65), 5°04’19“S 38°24’16”E, 1307 m (SGT 110, SGT 112, SGT 118, SGT 119, SGT 120, SGT 122), 2017. Tanzania, Kilimanjaro, Marangu route in the upper montane Podocarpus forests, 5°02’31”S 37°30’56”E, 1954 m (SGT 156, SGT 157), 2017.
Usnea pectinata Taylor ()
Diagnostic notes
For a detailed description of this species see Ohmura (Citation2001).
Usnea pectinata has a pendulous sorediate thallus ()); branches have annulations and vary from smooth and cylindrical to irregular/ridged and more or less alate ( and c)). The base is blackish ()) or not. The branches are covered by numerous soralia with isidiomorphs ()). The cortex is shiny in section and moderately thin to moderately thick (6.6–8.8%). The medulla is thin ranging from 11.8–13.8%. The axis is moderately thin to moderately thick (44–46.3%), solid, varying in colour from white to pale to deep brown ()).
Chemistry
Usnic acid was found in all specimens, but three chemotypes were observed as detected by TLC in the eight studied specimens. Only main substances were considered. Two specimens contained constictic acid; three contained constictic and diffractaic acid while the other three specimens contained protocetraric acid only. Ohmura (Citation2001) reported norstictic, menegazziaic, stictic and constictic acids for U. pectinata specimens in Japan. Further investigations of the chemotypes in U. pectinata with additional material are in progress.
Distribution and ecology
Tropical (Ohmura Citation2001). In Tanzania found on twigs of Drypetes usambarica, Alablankia stomanii and Ficus soningiae at an altitude between 1227–1307 m.
Specimens examined: Tanzania, Tanga, about 16.4 km from Korogwe district in the Usambara Mountains, 5°04’15“S 38°24’02“E, 1227 m (SGT 86, SGT 87), 5°04’15”S 38°24’02”E, 1307 m (SGT 106, SGT 107, SGT 109, SGT 114, SGT 115, SGT 116, SGT117), 2017.
Conclusion
Phylogenies of Eumitria species from Tanzania based on concatenated data sets were presented using Bayesian and maximum likelihood analyses ( and ). A total of 62 new sequences of Eumitria (26 ITS, 20 LSU, 6 MCM7, 10 RPB1) were generated, with nuLSU, MCM7 and RPB1 reported for the first time for Usnea baileyi. Variation in secondary chemistry and anatomical characters for U. pectinata was documented. For the first time morphology, chemistry, and molecular data were used in the analyses of Eumitria.
Supplemental Material
Download Zip (661.8 KB)Acknowledgements
We are grateful to Mr. Frank Mbago (UDSM) for his help in identifying host plants, to Ms. Ana Palma de Figueiredo (CJBG) for her help with TLC and to local field guides and the local community for their cooperation throughout the study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Articus K. 2004. Neuropogon and the phylogeny of Usnea s.l. (Parmeliaceae, lichenized Ascomycetes). Taxon. 53(4):925–934.
- Clerc P. 1984. Contribution à la révision de la systématique des Usnées (Ascomycotina, Usnea) d’Europe. I. Usnea florida (L.) Wigg. emend. Clerc [Contribution to the revision of the systematics of Usneas (Ascomycotina, Usnea) of Europe. I. Usnea florida (L.) Wigg. emend. Clerc]. Cryptogam Bryol Lichénol. 5:333–360.
- Clerc P. 1987. Systematics of the Usnea fragilescens aggregate and its distribution in Scandinavia. Nord J Bot. 7(4):479–495.
- Clerc P. 1997. Notes on the genus Usnea Dill. ex Adanson. Lichenologist. 29:209–215.
- Clerc P. 1998. Species concepts in the genus Usnea (lichenized Ascomycetes). Lichenologist. 30(4–5):321–340.
- Clerc P. 2011. Usnea. – in: thell A, Moberg R. (eds). Nordic Lichen Flora. 4:107–127.
- Crespo A, Kauff F, Divakar PK, Del Prado R, Pérez-Ortega S, de Paz GA, Cubas P. 2010. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon. 59(6):1735–1753.
- Culberson CF, Ammann K. 1979. Standard- methode zur dünnschichtchromatographie von flechtensubstanzen [Standard method for thin-layer chromatography of lichen substances]. Herzogia. 5:1–24.
- Culberson CF, Johnson A. 1982. Substitution of methyl tert-butyl ether for diethyl ether in the standardized thin-layer chromatographic method for lichen products. J Chromatogr A. 238:483–487.
- Divakar PK, Cresp A, Kraichak E, Leavitt SD, Singh G, Schmitt I, Lumbsch HT. 2017. Using a temporal phylogenetic method to harmonize family-and genus-level classification in the largest clade of lichen-forming fungi. Fungal Divers. 84(1):101–117.
- Dodge CW. 1956. Some lichens of tropical Africa. II. Usnea. Ann Missouri Bot Gard. 43:381–396.
- Dodge CW. 1957. Some lichens of tropical Africa. II. Usnea (continued). Ann Missouri Bot Gard. 44:1–76.
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes‐ application to the identification of mycorrhizae and rusts. Mol Ecol. 2(2):113–118.
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 42(2):182–192.
- Krog H. 1994. New observations on Usnea subgenus Eumitria in eastern and central Africa. In: Seyani JH, Chikuni AC, editors. Proceedings of XIIIth Plenary Meeting AETFAT, Malawi; p. 813–821.
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30(22):3276–3278.
- Matheny PB, Liu YJ, Ammirati JF, Hall BD. 2002. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot. 89(4):688–698.
- Motyka J. 1936. Lichenum generis Usnea studium monographicum [Lichen of genus Usnea study monography].
- Nelsen MP, Chavez N, Sackett-Hermann E, Thell A, Randlane T, Divakar PK, Lumbsch HT. 2011. The cetrarioid core group revisited (Lecanorales: Parmeliaceae). Lichenologist. 43(6):537–551.
- Ohmura Y. 2001. Taxonomic study of the genus Usnea (lichenized Ascomycetes) in Japan and Taiwan. J Hattori Bot Lab. 90:1–96.
- Ohmura Y. 2002. Phylogenetic evaluation of infrageneric groups of the genus Usnea based on ITS regions in rDNA. J Hattori Bot Lab. 92:23–243.
- Ohmura Y. 2012. A synopsis of the lichen genus Usnea (Parmeliaceae, Ascomycota) in Taiwan. Mem Natl Mus Nat Sci Tokyo. 48:91–137.
- Ohmura Y, Kanda H. 2004. Taxonomic status of section Neuropogon in the genus Usnea elucidated by morphological comparisons and ITS rDNA sequences. Lichenologist. 36(3–4):217–225.
- Orock E, Leavitt S, Fonge B, St Clair L, Lumbsch H. 2012. DNA-based identification of lichen-forming fungi: can publicly available sequence databases aid in lichen diversity inventories of Mount Cameroon (West Africa)? Lichenologist. 44(6):833–839.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Rambaut A, Drummond AJ. 2010. FigTree v1. 3.1 institute of evolutionary biology. Edinburgh: University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree
- Rogers RW, Stevens GN. 1988. The Usnea baileyi complex (Parmeliaceae, Lichenized Ascomycetes) in Australia. Aust Syst Bot. 1(4):355–361.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Schmitt I, Crespo A, Divakar PK, Fankhauser JD, Herman-Sackett E, Kalb K, Widhelm T. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia. 23:35–40.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Stevens GN. 1999. A revision of the lichen family Usneaceae in Australia. Bibl Lichenol. 72:1–128.
- Stirton J. 1882. Notes on the genus Usnea with description of new species. Scott Nat. 6:292–297.
- Swinscow TDV, Krog H. 1974. Usnea subgenus Eumitria in East Africa. Norw J Bot. 21:165–185.
- Swinscow TDV, Krog H. 1975. The Usnea undulata aggregate in East Africa. Lichenologist. 7:121–138.
- Swinscow TDV, Krog H. 1979. The Fruticose species of Usnea subgenus Usnea in East Africa. Lichenologist. 11:207–252.
- Swinscow TDV, Krog H. 1986. Usnea antiqua sp. nov. described from Tanzania. Lichenologist. 18(3):293–295.
- Swinscow TDV, Krog H. 1988. Macrolichens of East Africa. London: British Museum Natural History.
- Thell A, Crespo A, Divakar PK, Kärnefelt I, Leavitt SD, Lumbsch HT, Seaward MRD. 2012. A review of the lichen family Parmeliaceae – history, phylogeny and current taxonomy. Nord J Bot. 30:641–664.
- Thell A, Kärnefelt I, Seaward MRD. 2018. Splitting or synonymizing – genus concept and taxonomy exemplified by the Parmeliaceae in the Nordic region. Graphis Scripta. 30(6):130–137.
- Truong C, Clerc P. 2013. Eumitrioid Usnea species (Parmeliaceae, lichenized Ascomycota) in tropical South America and the Galapagos. Lichenologist. 45(3):383–395.
- Truong C, Divakar PK, Yahr R, Crespo A, Clerc P. 2013. Testing the use of ITS rDNA and protein-coding genes in the generic and species delimitation of the lichen genus Usnea (Parmeliaceae, Ascomycota). Mol Phylogenet Evol. 68(2):357–372.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172(8):4238–4246.
- White TJ, Burns T, Lee S, Taylor J. 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols A guide to methods and applications. San Diego (California): Academic Press; p. 315–322.
- Wirtz N, Printzen C, Sancho LG, Lumbsch TH. 2006. The phylogeny and classification of Neuropogon and Usnea (Parmeliaceae, Ascomycota) revisited. Taxon. 55(2):367–376.