?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The island of Borneo is a global biodiversity hotspot. However, its limestone caves are one of its least-studied ecosystems. We report for the first time the fungal species richness, diversity and abundance from Madai cave, situated in north-eastern Borneo. Environmental samples from inside the cave environment were collected (guano, speleothem, and cavern water) via opportunistic sampling. The dilution method was performed for isolation of fungi. Morphological characterisation and molecular analysis of the ITS region were utilised for the identification of isolates. Fifty-five pure cultures of fungi were attained, comprising 32 species from 15 genera, eight orders, and two divisions, Ascomycota and Basidiomycota. Ascomycetes dominated the fungal composition, accounting for 53 (96%) out of 55 total isolates. Penicillium spp. accounted for more than 47.1% of fungal abundance in all sample types. However, Aspergillus spp. had the highest occurrence rate, being isolated from all environmental samples except one. Purpureocillium lilacinum was isolated most frequently, appearing in five separate samples across all three substrates. Annulohypoxylon nitens, Ganoderma australe, Pyrrhoderma noxium, and Xylaria feejeensis were discovered and reported for the first time from the cave environment. This study provides additional data for further research on the mycoflora of Sabah’s various ecosystems, especially limestone caves.
Introduction
Caves are unique ecosystems that are relatively underexplored in Borneo. This is particularly true for its microorganisms, especially the diversity of fungi in cave environments. In contrast to the external environment, caves are dark, relatively cool, humid, and nutrient-limited (Gabriel and Northup Citation2013; Zhang et al. Citation2017, Citation2018). The lack of photosynthetic organisms influences the oligotrophic nature of the cave, which in turn influences its mycofloral diversity (Gunde-Cimerman et al. Citation1998; Hose et al. Citation2000; Barton and Jurado Citation2007; Kuzmina et al. Citation2012; Gabriel and Northup Citation2013; Ogórek et al. Citation2013). Fungi are some of the most dominant organisms in caves due to the high rate of spore dissemination, colonisation capability in various substrates, and tolerance to a wide range of pH values (Nováková Citation2009; Bastian et al. Citation2010; Wang et al. Citation2010; Ogórek et al. Citation2013). Over 1150 species of fungi have been recorded from caves throughout the world, with the most species-rich division being Ascomycota, followed by Basidiomycota and Zygomycota with fewer species (Vanderwolf et al. Citation2013). Many of the fungi found in cave systems are not native to caves but are likely introduced and dispersed by humans, fauna, water sources, and air currents (Jablonsky et al. Citation1993; Ikner et al. Citation2007; Shapiro and Pringle Citation2010; Vanderwolf et al. Citation2016; Nováková et al. Citation2018). Some suspected obligate troglobitic fungi have been reported, such as Acaulium caviariforme (Vanderwolf et al. Citation2013), A. baecitus (Nováková et al. Citation2012), and A. thesauricus (Nováková et al. Citation2012). However, the existence of true obligate troglobitic fungi remains contentious. It is estimated that only 3–8% of all fungi on earth have been identified and described, and an overwhelming majority of extant fungi remain to be discovered (Hawksworth and Lücking Citation2017). Furthermore, the cave-dwelling fungi of the tropics, and of Malaysian Borneo in particular, remains very poorly documented.
Fungi play vital roles in the ecosystems they inhabit, whether as saprophytes, symbionts, parasites, or food sources (Bastian et al. Citation2010; Araújo and Hughes Citation2016). Mycoses are rapidly becoming one of the leading threats to wildlife with numerous epidemics around the world, especially in tropical regions due to their warm, humid climates (Jurado et al. Citation2010; Fisher et al. Citation2012; Hsu et al. Citation2012). White-nose Syndrome (WNS) (Puechmaille et al. Citation2011), chytridiomycosis (Fisher et al. Citation2009) and snake fungal disease (Lorch et al. Citation2016) are all severe and often fatal fungal disease towards their host animal populations. However, of these diseases, only WNS is found exclusively in caves.
Cave fungi have been isolated from various substrates, including sediment, wall, speleothem, guano, water, air, and various fauna (Jurado et al. Citation2008; Vanderwolf et al. Citation2013). While cave fungal studies specific to speleothem fungi remain scant, studies on the relationship of fungi with cave walls and overall cave morphology have been conducted. Endolithic growth of lithobiontic fungi can biologically weather rock surfaces, but in the long term they can help stabilise and preserve rock surface morphology (Hoppert et al. Citation2004). Lithogenic fungi can be detrimental to anthropological sites, especially due to cave wall staining of ancient paintings (Bastian et al. Citation2010). Cavern water indirectly affects cave mycoflora in various ways (Cunningham et al. Citation1995; Vanderwolf et al. Citation2013). Non-native fungal species and organic material may be introduced into the cave via running water through the cave floor or vertical filtration of rainwater from the soil above (Dupont et al. Citation2007; Ikner et al. Citation2007). The moist microclimate, stable temperatures, and abundance of nutrients of guano make it one of the most dominant fungal substrates in caves, effectively serving as a separate micro-ecosystem (Paulson Citation1972; Nieves-Rivera et al. Citation2009). In Domica cave, Slovakia, guano had the greatest fungal diversity when compared to eight other substrate types (Nováková Citation2009).
Ecological factors that affect levels of fungal diversity in caves include seasonal changes, temperature, humidity, rainfall, human activity, sunlight, and distance from cave entrance (Wang et al. Citation2010; Taylor et al. Citation2014). Furthermore, the frequency of human visitation and other anthropogenic influences may affect the cave mycoflora. Increased human traffic into cave systems can cause contamination of indigenous fungal communities by repeated introductions of non-indigenous microorganisms and nutrients (Ikner et al. Citation2007; Chelius et al. Citation2009; Porca et al. Citation2011; Griffin et al. Citation2014). Shapiro and Pringle (Citation2010) reported that increased human visitation is correlated with lower levels of fungal diversity in caves. Human disturbance in caves is usually localised to the specific areas of interaction. However, increased human visitation is correlated with increased fungal abundance (Wang et al. Citation2010; Porca et al. Citation2011). Regular visits by humans allow for certain fungi to survive in contaminated areas because visitors tend to leave behind food waste and other organic materials (Ogórek et al. Citation2013; Griffin et al. Citation2014). Even fungi native to the cave could be dispersed to other areas of the cave they did not occupy before human disturbance.
Subterranean ecosystems are inhabited by organisms that have adapted to tolerate relatively unfavourable and niche conditions, including fungi (Ogórek et al. Citation2017. Madai cave is known to have accommodated early humans based on excavation of faunal remains (Harrison Citation1998). A variety of bats roost in the cave, with some colonies having populations over 100,000 individuals (Kobayasi et al. Citation1980). These bats bring in nutrient sources from outside the cave daily, and their guano and cadavers are known to harbour fungi (Nieves-Rivera et al. Citation2009; Nováková et al. Citation2018). Madai cave is also subject to major anthropogenic influence due to land use for palm-oil plantations surrounding the forest reserve, seasonal swiftlet farming harvesting by the local community for generations, and visits by tourists from all over the world as a major eco-tourism attraction. The aim of this study was to establish baseline data that will determine the fungal diversity existing in Sabah’s caves and its ecological relationships. This study is the first of its kind in Madai cave, as most studies have been archaeological or faunal. Better understanding of the ecological roles and interactions of fungi and reporting on the potential existence of pathogenic fungi will allow improved management practices to cave caretakers and stakeholders, especially with the influx of tourists and professionals that often visit cave areas in Sabah.
Materials and methods
Site description
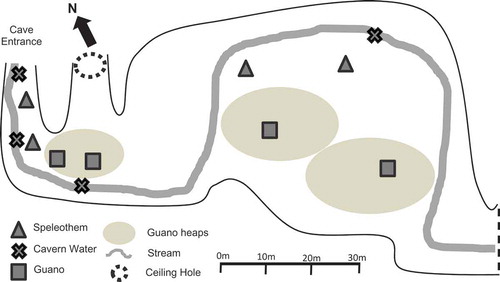
Madai Cave, Baturong Madai Forest Reserve, Class VI (Virgin Forest), Kunak, Sabah (4°41ʹ10.01”N 118°15ʹ4.12”E) were visited on 28–29 November 2017. A small village is located immediately outside the cave entrance, and there are two main chambers of the cave. The first one has an entrance at the ground level, and it is in this lower chamber that the sampling for this study took place (). A second chamber is located about a 100 m hike up the limestone formation, past some ancient burial sites. The air temperature in the cave on the day of sampling fluctuated from 27°C and rose to around 29°C near the cave entrance. The air humidity ranged between 92% and 100%. The main chamber of the cave is more than 400 m in length, not including the multiple branches of the main chamber (Wilford Citation1961). In this study, only the first 100 m of the main chamber was sampled and explored.
Figure 1. Madai Cave. A. Cave entrance. B. Researcher collecting guano sample. C. Speleothem. D. Guano pile. E. Cave stream deep in the cave. F. Village children playing near the cave entrance. G. Visible graffiti on cave wall. H. Land use for palm oil surrounding Baturong Madai Forest Reserve

The cave is open to the public during the off seasons of swiftlet nest harvesting and is often visited by large groups of both foreigners and locals. Immediately outside of the forest reserve exist monocrop farms for palm oil production (), which is what most of the unprotected forest areas have been converted into in this region.
For the purposes of this study, the cave was categorised into three separate zones based on light intensity. From the entrance, the first 20 m of the cave was designated as the lighted zone due to its exposure to direct sunlight. The twilight zone exists around 20–40 m within the cave, and it is defined as dimly lit areas that are not exposed to direct sunlight. Past the twilight zone area, the remainder of the cave is pitch black since there are no apertures to allow natural light into the cave. These unlighted areas are labelled as the dark zone of the cave, and it can only be properly traversed by humans if artificial light sources are available.
Sampling and identification
Opportunistic sampling of speleothem, cavern water, and guano was utilised, in which four samples of each substrate were acquired from the first 100 m from the cave entrance (). Speleothem was sampled using the swab method (25 cm2 area per swab) and stored in sterile sample tubes, capped, and sealed (Ikner et al. Citation2007; Vaughan et al. Citation2011). Guano samples (10 g) were collected using sterile scoops and stored in sterile sample tubes (Nieves-Rivera Citation2003). Samples of cavern water (10 ml) were also collected and sealed in sterile sample tubes. The distance from entrance was recorded for all collections immediately after sealing. Samples were labelled with a letter designating substrate type and a number in order of increasing distance from entrance (e.g. speleothem sample 1 = S1; speleothem samples 2 = S2). All samples were collected in triplicate and chilled in ice (<4°C) until transported to the mycology laboratory in the Institute for Tropical Biodiversity and Conservation, Universiti Malaysia Sabah. In the laboratory, samples were immediately stored in a chiller at <4°C until isolation.
Samples were serially diluted 10-fold up to 10−4, and 1 ml aliquots of the 1 10−2 1
10−3, and 1
10−4 dilutions were selected for plating. The samples were placed in Petri dishes via serial dilution onto Potato Dextrose Agar (PDA) and Malt Extract Agar (MEA) infused with 40 mg/L streptomycin sulphate, which was incubated from 7 to 28 days at 25 ± 1°C in the dark. Dilutions and plating were performed in triplicate. The colonies that appeared on the media were categorised by Morphological Taxonomic Units (MTU) and was counted. Pure isolates were obtained using the three–point method on PDA and MEA before morphological and genetic analysis. All pure isolates underwent morphological identification based on universal identification keys described by Raper and Fennel (Citation1965), Domsch et al. (Citation1980), and Klich (Citation2002).
DNA from pure cultures (7–14 days old) was extracted using the E.Z.N.A. DNA Fungal Kit (Omega Bio-Tek, USA) following the manufacturer’s instructions after seven days of incubation. The internal transcribed spacer region of fungal rDNA was amplified using the primers ITS1 (5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) (White et al. Citation1990). Polymerase chain reaction (PCR) amplifications were performed in a Bio-Rad T100 Thermal Cycler according to Ogórek et al. (Citation2019). The amplification products were electrophoresed in a 1% agarose gel for 30 mins, which was stained with gel red for visualisation. The PCR products were purified using Column-Pure PCR Clean-Up Kit (Applied Biological Materials, Inc.) according to manufacturer protocols and sequenced by MyTACG Bioscience SDN. BHD. (Kuala Lumpur, Malaysia).
Data analysis
The ITS sequences were assembled by BioEdit Sequence Alignment Editor version 7.2.5. The top hit sequences were determined by comparing the obtained sequences with those deposited in the GenBank database using the BLASTn algorithm (http://www.ncbi.nlm.nih.gov). The fungal abundance data obtained from the serial dilution of fungal colonies cultured in Petri dishes were expressed as colony-forming units (CFU). This was calculated with the formula X = (a x p)/V, where “a” is the number of colonies, “V” is the inoculation aliquot volume, and “p” is the dilution factor. The final CFU count for each sample recorded is the average of nine dilution plates per sample. The fungal abundance data were then used for diversity and evenness analysis in PAST 3.10 software. The diversity analyses were run separately for each substrate type due to difference in units of abundance (i.e. CFU/cm2, CFU/ml, and CFU/g). Fungal species occurrence is defined as the number of times the same species was isolated as pure culture (maximum of once per sample).
Results
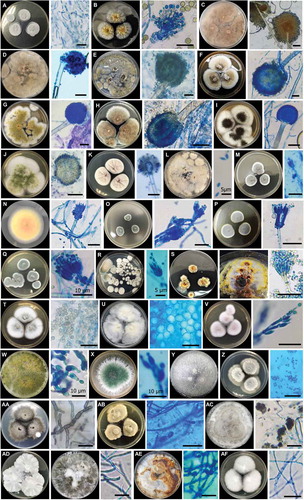
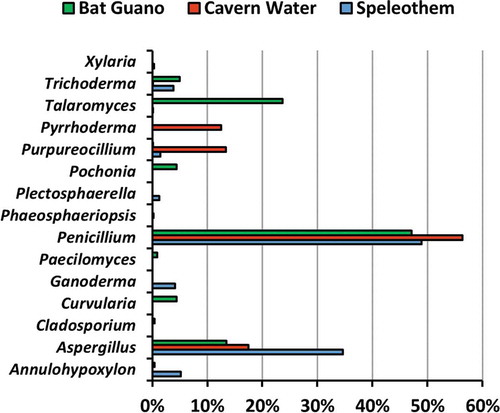
In total, 55 fungal isolates (pure cultures) were obtained. Twenty-three isolates were obtained from four speleothem samples, 15 isolates from four cavern water samples, and 17 isolates from four guano samples. The fungi were classified into 32 species 15 genera, eight orders, and two divisions, namely Ascomycota and Basidiomycota (), based on morphological characters () and molecular analysis of the ITS region for confirmation. Only two out of 32 species were identified as Basiodiomycota, Ganoderma australe and Pyrrhoderma noxium. Out of the 55 total isolates, 31 underwent DNA extraction (ITS) for subsequent molecular analysis to corroborate morphological identification and help identify cryptic taxa. All 31 extracted DNA samples were successfully sequenced, and BLASTn sequence analysis corroborated 24 out of 32 total species identified in this study (). All BLASTn results had identity matches over 96.7%, except for Talaromyces sp. (92.2%). The fungus that had the most frequent occurrence was Purpureocillium lilacinum, which was isolated from five different samples composed of all three substrate types. However, the most frequently isolated genus was Aspergillus, accounting for 36.4% of all isolates. Based on fungal abundance data, Penicillium spp. dominated fungal composition and accounted for 56.3%, 48.9%, and 47.1% of cavern water, speleothem and guano fungi, respectively ().
Table 1. Fungal taxa cultured from madai cave, malaysia
Table 2. Culturable cave fungi of madai cave, sabah BLASTn (genBank) analysis
Figure 4. Culturable fungi of Madai Cave, Sabah, 7–14 day-old cultures at 25°C, top view of colony and micromorphology under microscope on PDA media. A. C. cladosprioides. B. Aspergillus sp. 1. C. Aspergillus sp. 2. D. Aspergillus sp. 3. E. A. aculeatus. F. A. europaeus. G. A. flavus. H. A. japonicus. I. A. niger. J. A. nomius. K. A. sydowii. L. P. variotii. M. Penicillium sp. 1. N. Penicillium sp. 2. O. P. bilaiae. P. P. citrinum. Q. P. paxilli. R. P. simplicissimum. S. Talaromyces sp. T. T. minioluteus. U. P. chlamydosporia. V. P. lilacinum. W. T. asperellum. X. T. harzianum. Y. T. paraviridescens. Z. P. cucumerina. AA. C. lunata. AB. Phaeosphaeriopsis sp. AC. A. nitens. AD. X. feejeensis. AE. P. noxium. AF. G. australe. Scale bars = 20 µm (unless otherwise labelled)
In speleothem samples, the average fungal abundance was 229.3 CFU/cm2. The isolate with highest abundance count from speleothem was the Penicillium citrinum from sample S1 (272.4 CFU/cm2). The average fungal abundance of cavern water samples was 335.0 CFU/ml, and Penicillium sp. 1 from sample W1 had the highest abundance (716.7 CFU/ml). In guano samples, the average fungal abundance was 6,266.7 CFU/g, and the single isolate which had the highest abundance count was Penicillium paxilli from sample G1 (8,922.2 CFU/g) (). Both Shannon-Wiener and Simpson alpha diversity indices showed that speleothem samples had the most diverse fungal communities, followed by guano, and then cavern water (). Additionally, fungal abundance in speleothem seemed to decrease with increasing distance from the cave entrance, although there were only four samples with abundance of data for speleothem in this study.
Table 3. Average abundance of culturable fungi of madai cave, sabah (CFU per unit sample)
Table 4. Diversity index scores based on abundance counts of all taxa isolated
Discussion
This is the first study on fungi in Madai cave. Studies in limestone caves in Sabah have been limited to the exploration of their fauna, geology, or anthropogenic value, and fungi have often been overlooked. Thirty-four (61.8%) fungal isolates in this study were of the order Eurotiales, including Aspergillus spp. (36.4%), Penicillium spp. (18.2%), Talaromyces spp. (3.6%), and Paecilomyces variotii (3.6%). All isolates cultured were Ascomycota except for two Basidiomycota isolates. Fungi in the genera Penicillium and Aspergillus represented most abundant taxa for all substrates. Our results reflect those from previous fungal studies from cave ecosystems as Ascomycota fungi usually dominate the fungal composition of cave ecosystems, followed by Basidiomycota, Zygomycota, and then others (Vanderwolf et al. Citation2013). Four species of fungi were discovered from cave samples for the first time in this study, namely A. nitens, Ganoderma australe, Pyrrhoderma noxium, and Xylaria feejeensis (Vanderwolf et al. Citation2013; Nováková et al. Citation2018; Karunarathna et al. Citation2020; Zhang et al. Citation2020; Cunha et al. Citation2020).
Prior to the study, guano was hypothesised to have the largest diversity compared to speleothem surfaces and cavern water (Nováková Citation2009; Vanderwolf et al. Citation2013). Instead, the results showed that speleothem surfaces had the highest diversity indices (1-D = 0.84, H = 2.15), most pure isolates attained (23), and highest number of taxa identified (19). Speleothem surfaces yielded the highest proportion of isolates (41.8%), which included 19 species from nine genera and four orders. Only one isolate from speleothem was a basidiomycete (Ganoderma australe) and all others were ascomycetes. Fungi have been isolated from cave walls and sediment in previous studies, and fungi are postulated to participate in speleothem deposition (Engel et al. Citation2004; Bastian et al. Citation2010). Our results are congruent with a previous study on cave fungi, where Aspergillus and Penicillium fungi dominated the speleothem fungal community (Vaughan et al. Citation2011).
Fungi have been isolated from cave walls and sediment in previous studies, and these fungi are postulated to participate in speleothem deposition and biomineralization (Northup and Lavoie Citation2001; Engel et al. Citation2004; Barton and Northup Citation2007; Bastian et al. Citation2010). The speleothem in Madai cave, particularly those sampled, was wet due to vertical filtration of water from above. Wet, mouldy speleothem is an indication that they are biologically active (Dodge-Wan and Deng Citation2013). Fungal richness is often associated with substrate moisture (Schimel et al. Citation1999; Frey et al. Citation1999). Additionally, fungal spores have been shown to colonise and grow on virtually rock surface that has even minute traces of carbon (Wainwright et al. Citation1993; Barton and Jurado Citation2007). Vanderwolf et al. (Citation2013) showed in her world review that cave sediment (43%) and cave wall (18%) both individually composed of a higher percentage of fungal taxa than guano (16%). Although this is not speleothem, it does show that fungal diversity on cave rocks seems to be higher than that of guano.
Distance from cave entrance may affect fungal abundance on speleothem surfaces. This result concurs with previous studies that measured similar data, as fungal biodiversity, species occurrence, abundance, and biomass decreases with distance from cave entrance (Hsu and Agoramoorthy Citation2001; Kuzmina et al. Citation2012; Mulec et al. Citation2012; Taylor et al. Citation2014). There are a multitude of factors that may contribute to this relationship. Primarily, many of the fungi found in caves originate from the external environment and is introduced by a multitude of methods, such as air currents, humans, and fauna (Shapiro and Pringle Citation2010; Porca et al. Citation2011; Ogórek et al. Citation2013; Pusz et al. Citation2014, Citation2015; Vanderwolf et al. Citation2016). However, Zhang and Cai (Citation2019) reported that distance did not play a role in species richness. Instead, they showed that similarities of fungal communities inside and outside the cave decreased with increasing distance from the cave entrance. Since cosmopolitan soil fungi tend to be isolated at higher rates using culture-dependent methods, it may reflect on the results of this study and prior studies that rely on culture-based isolation. Nonetheless, if a cave has speleothem formations anywhere within the cave site, it should be assumed that fungi are active and present on these formations.
Guano yielded the second most isolates in this study (30.1%). A total of 16 species from 11 genera and 5 orders were identified from this substrate, and all of them were ascomycetes. Oligotrophy is a major limiting factor for fungi in caves, thus higher fungal diversity will likely be found on substrates with higher organic concentrations (Bastian et al. Citation2010; Jurado et al. Citation2010; Kuzmina et al. Citation2012). Guano is one of the major sources of organic matter in caves, and a broad spectrum of microfungal species are usually isolated from guano, including pathogenic yeasts, basidiomycetous yeasts, keratinophilic fungi, and ubiquitous ascomycetes (Larcher et al. Citation2003; Nováková Citation2009). Any cave that serves as bat roosts and is littered in one form or another with guano is likely to be reservoirs to a wide array for fungi.
Cavern water had the lowest proportion of fungal isolates compared to the other two substrates in this study (27.3%). A total of 12 species from six genera and four orders were identified from cavern water. Only one isolate was identified as a basidiomycete (Pyrrhoderma noxium), and the remaining are all ascomycetes. The water samples from this study came from a singular, minimally branched stream that ran through the cave towards the mouth of the cave. Man et al. (Citation2018) reported cavern water to contain the highest OTU count compared to other tested substrates, namely soil, rock, and air in caves. This is not the case in this study, but we sampled different substrates for comparison. Fungal dispersion and spore transmission via stream water are plausible explanations for our results. Fungi are expected to be isolated from cavern water because one of the main routes of organic matter transmission in caves is water flow (Ikner et al. Citation2007). Water flowing in and out of the cave serves as possible explanation for the transport and proliferation of fungi that can be isolated from water samples (Barton and Jurado Citation2007; Ortiz et al. Citation2014), especially since one of the main routes for organic matter to enter caves as dissolved organic carbon and colloidal organic matter through water flow (Ikner et al. Citation2007).
Some of the fungi identified in this study are known human pathogens. Aspergillus fungi are the most frequent cause of invasive mould infections in immunocompromised patients (Beck-Sagué and Jarvis Citation1993). While A. fumigatus, the most frequent cause of aspergillosis, was not reported in Madai cave, A. flavus, A. nomius, and A. niger can cause infection in humans and were isolated in this study (Marr et al. Citation2002; Caira et al. Citation2012). Paecilomyces variotii is another opportunistic human pathogen isolated from Madai cave, especially of immunocompromised patients (Sterflinger et al. Citation1999. Curvularia lunata is an opportunistic pathogen to humans and other animals, often infecting immunocompromised patients (Berman Citation2019). Any visitors or tourists entering in tropical caves should we aware of any potential risks posed by entering these ecosystems, especially those who may be immunocompromised.
There were several endophytic and phytopathogenic fungi isolated from Madai cave despite not having any autotrophs within the cave due to the lack of sunlight. Anthropogenic activity may explain how non-indigenous plant-associated fungi can be transported into novel ecosystems such as caves (Ikner et al. Citation2007; Shapiro and Pringle Citation2010). In Taiwan, distribution of Pyrrhoderma noxium is limited to areas with human activity, as no brown root rot has ever been found in undisturbed natural forests in Taiwan (Ann et al. Citation2002). Another possible method of introducing plant-associated fungi into the cave system is via arthropod vectors. The ability of some fungi to act as pathogens to both motile (fauna) and non-motile (flora) hosts could explain the role of arthropods as dispersers of endophytic and phytopathogenic fungi in caves. Annulohypoxylon nitens was isolated in this study. Annulohypoxylon fungi are associated with dead dicotyledonous wood as saprobes, but they are also found as endophytes and help promote growth in seed plants (Ju et al. Citation2017; Pereira et al. Citation2010; Ikeda et al. Citation2014). Penicillium bilaiae is another plant growth-promoting organism that was isolated in this study, as it has the ability to increase phosphorous nutrition in plants like wheat, medick, and lentil (Wakelin et al. Citation2007). On the other hand, phytopathogenic fungi identified in this study include Curvularia lunata (Liu et al. Citation2010), Phaeosphaeriopsis sp. (Golzar and Wang Citation2012; Thambugala et al. Citation2014), Talaromyces minioluteus (Stošić et al. Citation2019), P. noxium (Ann et al. Citation2002; Sahashi et al. Citation2014; Chung et al. Citation2015), and Plectosphaerella cucumerina (Carlucci et al. Citation2012; Li et al. Citation2017).
A number of fungi identified in this study are known as entomopathogens, many of which are being studied for their biological control potential. Madai cave is host to various invertebrates and acts as a roosting site for volant fauna such as bats and swiftlets. Animals are known to harbour fungi, and they are likely disseminators of fungal spores within caves, either as hosts, vectors, or cadavers (Vanderwolf et al. Citation2013; Nováková et al. Citation2018). Penicillium citrinum has been shown to cause mortality and reduced survival in Culex quinquefasciatus (mosquito) larvae after ingestion by the larvae (Maketon et al. Citation2013). Plectosphaerella cucumerina (Atkins et al. Citation2003), Pochonia chlamydosporia (Kerry Citation2000), and Purpureocillium lilacinum (Kepenekçi et al. Citation2018) are all being used and developed as biological control agents against plant pathogenic nematodes. Trichoderma asperellum and T. harzianum are both used as biological control agents against many plant disease-causing organisms, including Phytophthora megakarya (Deberdt et al. Citation2008), fungi (Watanabe et al. Citation2005; Alvindia and Hirooka Citation2011), and nematodes (Sharon et al. Citation2007). Insects feed on the fungi and bacteria that inhabit guano piles, which suggest their influence on the fungal community in caves both as consumers and dispersers (Šustr et al. Citation2005; Smrž et al. Citation2015). Arthropods are likely disseminators of cave fungi as many fungi isolated from cave environmental samples have been shown to include entomophilous, entomogenous, or entomopathogenic species (Ogórek et al. Citation2013; Vanderwolf et al. Citation2016).
This study on Madai cave’s fungal community serves to present baseline data with the purpose of serving a platform for future research of tropical mycota. Many fungi, especially microfungi, can only be identified with confidence to the genus level when using morphological analysis or when only utilising a single gene marker in phylogenetic analysis (Schoch et al. Citation2012). In this study, the utilisation of a combination of traditional morphological characterisation and molecular analysis allowed us to identify many of our specimens to the species level. However, our study only utilised culture-dependent methods of fungal isolation. Culture-dependent methods are known to only reveal as little as 0.6% to 8.0% of the total fungal species in a sample (Hibbett et al. Citation2009; Hawksworth and Lücking Citation2017). For a better understanding of cave mycobiota in Sabah, future studies should employ both morphological and molecular characterisation by implementing community-based culture-independent studies. Culture-independent methods, such as metagenomics and metabarcoding, can generate up to millions of raw sequences from a single sample and help eliminate bias towards fast-growing cosmopolitan fungi (Wiseschart et al. Citation2019; Zhang and Cai Citation2019). Hitherto, due to the lack of research on microfungi in Borneo, we are unaware of any deleterious fungal diseases in Sabah’s limestone caves that might infect fauna and humans.
Ongoing studies on fungi from various caves in Sabah are currently in progress. We urge more mycological studies and surveys to be conducted in caves in this region, not only to better understand fungal ecology, but to discover their enormous biological and industrial potential.
Wordcount
6,408 words.
Acknowledgements
This work was supported mainly by the Ministry of Education Malaysia (MOHE) under Grant FRGS/1/2017/WAB13/UMS/02/2; and Universiti Malaysia Sabah (UMSGreat) under grant GUG0143-1/2017. We are also grateful to Sabah Forestry Department and Sabah Wildlife Department for providing access to the sampling site. We also thank the Institute of Tropical Biology and Conservation, Universiti Malaysia Sabah for providing access to their laboratories and facilities. TMF was supported by a Czech Science Foundation Standard Grant (21-06446S). The authors thank the two anonymous reviewers for their comments and suggestions during the initial submission. The authors would also like to thank Sabah Forestry Department for granting the permit JPHTN/TKKH(PSH)100-14/18/2/JILID 36(47) to access the sampling site.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alvindia DG, Hirooka Y. 2011. Identification of clonostachys and trichoderma spp. from banana fruit surfaces by cultural, morphological and molecular methods. Mycol. 2(2):109–115. doi:https://doi.org/10.1080/21501203.2011.554904.
- .
- Ann PJ, Chang TT, Ko WH. 2002. Phellinus noxius brown root rot of fruit and ornamental trees in taiwan. Plant Dis. 86(8):820–826. doi:https://doi.org/10.1094/PDIS.2002.86.8.820.
- Araújo JPM, Hughes DP. 2016. Diversity of Entomopathogenic Fungi. Which Groups Conquered the Insect Body? Adv Genet. 94:1–39. doi:https://doi.org/10.1016/bs.adgen.2016.01.001
- Atkins SD, Clark IM, Sosnowska D, Hirsch PR, Kerry BR. 2003. Detection and quantification of plectosphaerella cucumerina, a potential biological control agent of potato cyst nematodes, by using conventional PCR, real-time PCR, selective media, and baiting. Appl Environ Microbiol. 69(8):4788–4793. doi:https://doi.org/10.1128/AEM.69.8.4788-4793.2003.
- Barton HA, Jurado V. 2007. What’s up down there? microbial diversity in caves microorganisms in caves survive under nutrient-poor conditions and are metabolically versatile and unexpectedly diverse. Microbe. 2(3):132–138.
- Barton HA, Northup DE. 2007. Geomicrobiology in cave environments: past, current and future perspectives. J Cave Karst Stud. 69(1):163–178.
- Bastian F, Jurado V, Nováková A, Alabouvette C, Saiz-Jimenez C. 2010. The microbiology of lascaux cave. Microbiology. 156(3):644–652. doi:https://doi.org/10.1099/mic.0.036160-0.
- Beck-Sagué CM, Jarvis WR, National Nosocomial Infections Surveillance System. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the united states, 1980-1990. The J Infect Dis. 167(5):1247–1251. doi:https://doi.org/10.1093/infdis/167.5.1247
- Berman JJ. 2019. Taxonomic guide to infectious diseases: understanding the biologic classes of pathogenic organisms. London (UK): Academic Press Ltd.
- Caira M, Posteraro B, Sanguinetti M, De Carolis E, Leone G, Pagano L. 2012. First case of breakthrough pneumonia due to aspergillus nomius in a patient with acute myeloid leukemia. Med Mycol. 50(7):746–750. doi:https://doi.org/10.3109/13693786.2012.660507.
- Carlucci A, Raimondo ML, Santos J, Phillips AJL. 2012. Plectosphaerella species associated with root and collar rots of horticultural crops in southern italy. Pers - Mol Phylogeny and Evol Fungi. 28(1):34. doi:https://doi.org/10.3767/003158512X638251.
- Chelius MK, Beresford G, Horton H, Quirk M, Selby G, Simpson RT, Horrocks R, Moore JC. 2009. Impacts of alterations of organic inputs on the bacterial community within the sediments of wind cave, south dakota, USA. Int J Speleol. 38(1):1. doi:https://doi.org/10.5038/1827-806X.38.1.1.
- Chung CL, Huang SY, Huang YC, Tzean SS, Ann PJ, Tsai JN, Yang CC, Lee HH, Huang TW, Huang HY, et al. 2015. The genetic structure of phellinus noxius and dissemination pattern of brown root rot disease in taiwan. PLoS ONE. 10(10):10. doi:https://doi.org/10.1371/journal.pone.0139445.
- Cunha AO, Bezerra JD, Oliveira TG, Barbier E, Bernard E, Machado AR, Souza-Motta CM. 2020. Living in the dark: bat caves as hotspots of fungal diversity. Plos One. 15(12):e0243494. doi:https://doi.org/10.1371/journal.pone.0243494.
- Cunningham KI, Northup DE, Pollastro RM, Wright WG, LaRock EJ. 1995. Bacteria, fungi and biokarst in lechuguilla cave, carlsbad caverns national park, new mexico. Environ Geol. 25(1):2–8. doi:https://doi.org/10.1007/BF01061824.
- Deberdt P, Mfegue CV, Tondje PR, Bon MC, Ducamp M, Hurard C, Begoude BAD, Ndoumbe-Nkeng M, Hebbar PK, Cilas C. 2008. Impact of environmental factors, chemical fungicide and biological control on cacao pod production dynamics and black pod disease (phytophthora megakarya) in cameroon. Biol Control. 44(2):149–159. doi:https://doi.org/10.1016/j.biocontrol.2007.10.026.
- Dodge-Wan D, Deng AHM. 2013. Biologically influenced stalagmites in niah and mulu caves (sarawak, malaysia). Acta Carsologica. 42(1):155–163. doi:https://doi.org/10.3986/ac.v42i1.634.
- Domsch KH, Gams W, Anderson TH. 1980. Compendium of soil fungi. volume 1. London (UK): Academic Press Ltd.
- Dupont J, Jacquet C, Dennetière B, Lacoste S, Bousta F, Orial G, Roquebert MF. 2007. Invasion of the french paleolithic painted cave of lascaux by members of the fusarium solani species complex. Mycol. 99(4):526–533. doi:https://doi.org/10.1080/15572536.2007.11832546.
- Engel AS, Stern LA, Bennett PC. 2004. Microbial contributions to cave formation: new insights into sulfuric acid speleogenesis. Geol. 32(5):369–372. doi:https://doi.org/10.1130/G20288.1.
- Fisher MC, Garner TWJ, Walker SF. 2009. Global emergence of batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 63(1):291–310. doi:https://doi.org/10.1146/annurev.micro.091208.073435.
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nat. 484(7393):186. doi:https://doi.org/10.1038/nature10947.
- Frey SD, Elliott ET, Paustian K. 1999. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol Biochem. 31(4):573–585. doi:https://doi.org/10.1016/S0038-0717(98)00161-8.
- Gabriel CR, Northup DE. 2013. Microbial ecology: caves as an extreme habitat. In: Cheeptham N, editor. Cave microbiomes: a novel resource for drug discovery. New York (NY): Springer; p. 85–108.
- Golzar H, Wang C. 2012. First report of phaeosphaeriopsis glaucopunctata as the cause of leaf spot and necrosis on ruscus aculeatus in australia. Australas Plant Dis Notes. 7(1):3–15. doi:https://doi.org/10.1007/s13314-011-0035-5.
- Griffin DW, Gray MA, Lyles MB, Northup DE. 2014. The transport of nonindigenous microorganisms into caves by human visitation: a case study at carlsbad caverns national park. Geomicrobiol J. 31(3):175–185. doi:https://doi.org/10.1080/01490451.2013.815294.
- Gunde-Cimerman N, Zalar P, Jeram S. 1998. Mycoflora of cave cricket troglophilus neglectus cadavers. Mycopathol. 141(2):111–114. doi:https://doi.org/10.1023/A:1006947524503.
- Harrison T. 1998. Vertebrate faunal remains from the madai caves (MAD 1/28), sabah, east malaysia. Bull Indo-Pac Prehis Assoc. 17:85–92.
- Hawksworth DL, Lücking R. 2017. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr. 5(4). FUNK–0052–2016. DOI: https://doi.org/10.1128/microbiolspec.FUNK-0052-2016.
- Hibbett DS, Ohman A, Kirk PM. 2009. Fungal ecology catches fire. New Phytol. 184(2):279–282. doi:https://doi.org/10.1111/j.1469-8137.2009.03042.x.
- Hoppert M, Flies C, Pohl W, Günzl B, Schneider J. 2004. Colonization strategies of lithobiontic microorganisms on carbonate rocks. Environ Geol. 46(3–4):421–428. doi:https://doi.org/10.1007/s00254-004-1043-y.
- Hose LD, Palmer AN, Palmer MV, Northup DE, Boston PJ, DuChene HR. 2000. Microbiology and geochemistry in a hydrogen-sulfide-rich karst environment. Chem Geol. 169(3–4):399–423. doi:https://doi.org/10.1016/S0009-2541(00)00217-5.
- Hsu LY, Wijaya L, Ng EST, Gotuzzo E. 2012. Tropical fungal infections. Infect Dis Clin North Am. 26(2):497–512. doi:https://doi.org/10.1016/j.idc.2012.02.004.
- Hsu MJ, Agoramoorthy G. 2001. Occurrence and diversity of thermophilous soil microfungi in forest and cave ecosystems of taiwan. Fungal Divers. 7:27–33.
- Ikeda A, Matsuoka S, Masuya H, Mori AS, Hirose D, Osono T. 2014. Comparison of the diversity, composition, and host recurrence of xylariaceous endophytes in subtropical, cool temperate, and subboreal regions in japan. Popul Ecol. 56:289–300.
- Ikner LA, Toomey RS, Nolan G, Neilson JW, Pryor BM, Maier RM. 2007. Culturable microbial diversity and the impact of tourism in kartchner caverns, arizona. Microb Ecol. 53(1):30–42. doi:https://doi.org/10.1007/s00248-006-9135-8.
- Jablonsky P, Kraemer S, Yett B 1993. Lint in caves. In: Proceedings 1993 National Cave Management Symposium, Carlsbad, New Mexico. Am Cave Conserv Assoc. 73–81.
- Ju Y, Rogers JD, Hsieh H, Ju Y, Rogers JD. 2017. New hypoxylon species and notes on some names associated with or related to hypoxylon. Mycol. 96(1):154–161. doi:https://doi.org/10.1080/15572536.2005.11833006.
- Jurado V, Laiz L, Rodriguez-Nava V, Boiron P, Hermosin B, Sanchez-Moral S, Saiz-Jimenez C. 2010. Pathogenic and opportunistic microorganisms in caves. Int J of Speleol. 39(1):15–24. doi:https://doi.org/10.5038/1827-806x.39.1.2.
- Jurado V, Sanchez-Moral S, Saiz-Jimenez C. 2008. Entomogenous fungi and the conservation of the cultural heritage: A review. Int Biodeterior Biodegrad. 62(4):325–330. doi:https://doi.org/10.1016/j.ibiod.2008.05.002.
- Karunarathna SC, Dong Y, Karasaki S, Tibpromma S, Hyde KD, Lumyong S, Xu J, Sheng J, Mortimer PE. 2020. Discovery of novel fungal species and pathogens on bat carcasses in a cave in yunnan province, china. Emerg Microbe Infect. 9(1):1554–1566. doi:https://doi.org/10.1080/22221751.2020.1785333.
- Kepenekçi İ, Toktay H, Oksal E, Bozbuğa R, İmren M. 2018. Effect of purpureocillium lilacinum on root lesion nematode, pratylenchus thornei. J Agric Sci. 24(3):323–328.
- Kerry BR. 2000. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu Rev Phytopathol. 38(1):423–441. doi:https://doi.org/10.1146/annurev.phyto.38.1.423.
- Klich MA. 2002. Biogeography of aspergillus species in soil and litter. Mycol. 94(1):21–27. doi:https://doi.org/10.1080/15572536.2003.11833245.
- Kobayasi T, Maeda K, Harada M. 1980. Studies on the small mammal fauna of sabah, east malaysia i. order chiroptera and genus tupaia (primates). Contributions from the Biological Laboratory, Kyoto University. 26(1):67–82.
- Kuzmina LY, Galimzianova NF, Abdullin SR, Ryabova AS. 2012. Microbiota of the kinderlinskaya cave (south urals, russia). Microbiology. 81(2):251–258. doi:https://doi.org/10.1134/S0026261712010109.
- Larcher G, Bouchara JP, Pailley P, Montfort D, Beguin H, De Bièvre C, Chabasse D. 2003. Fungal biota associated with bats in western france. J Mycol Méd. 13:29–34.
- Li PL, Chai AL, Shi YX, Xie XW, Li BJ. 2017. First report of root rot caused by plectosphaerella cucumerina on cabbage in china. Mycobiol. 45(2):110–113. doi:https://doi.org/10.5941/MYCO.2017.45.2.110.
- Liu T, Liu L, Jiang X, Hou J, Fu K, Zhou F, Chen J. 2010. Agrobacterium-mediated transformation as a useful tool for the molecular genetic study of the phytopathogen Curvularia lunata. Eur J Plant Pathol. 126(3):363–371. doi:https://doi.org/10.1007/s10658-009-9541-0.
- Lorch JM, Knowles S, Lankton JS, Michell K, Edwards JL, Kapfer JM, Staffen RA, Wild ER, Schmidt KZ, Ballmann AE, et al. 2016. Snake fungal disease: an emerging threat to wild snakes. Philos Trans R Soc B Biol Sci. 371(1709):1709. doi:https://doi.org/10.1098/rstb.2015.0457.
- Maketon M, Amnuaykanjanasin A, Kaysorngup A. 2013. A rapid knockdown effect of penicillium citrinum for control of the mosquito culex quinquefasciatus in thailand. World J Microbiol Biotechnol. 30(2):727–736. doi:https://doi.org/10.1007/s11274-013-1500-4.
- Man B, Wang H, Yun Y, Xiang X, Wang R, Duan Y, Cheng X. 2018. Diversity of fungal communities in heshang cave of central china revealed by mycobiome-sequencing. Front Microb. 9:1400. doi:https://doi.org/10.3389/fmicb.2018.01400.
- Marr KA, Carter RA, Crippa F, Wald A, Corey L. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin Infect Dis. 34(7):909–917. doi:https://doi.org/10.1086/339202.
- Mulec J, Vaupotič J, Walochnik J. 2012. Prokaryotic and eukaryotic airborne microorganisms as tracers of microclimatic changes in the underground (postojna cave, slovenia). Microb Ecol. 64(3):654–667. doi:https://doi.org/10.1007/s00248-012-0059-1.
- Nieves-Rivera ÁM. 2003. Mycological survey of río camuy caves park, puerto rico. J Cave and Karst Stud. 65(1):23–28.
- Nieves-Rivera ÁM., Santos-Flores CJ, Dugan FM, Miller TE. 2009. Guanophilic fungi in three caves of southwestern Puerto Rico. International Journal of Speleology. 38(1):61–70. https://doi.org/10.5038/1827-806X
- Northup DE, Lavoie KH. 2001. Geomicrobiology of caves: a review. Geomicrobiol J. 18(3):199–222. doi:https://doi.org/10.1080/01490450152467750.
- Nováková A. 2009. Microscopic fungi isolated from the domica cave system (slovak karst national park, slovakia). a review. Int J Speleol. 38(1):71–82. doi:https://doi.org/10.5038/1827-806X.38.1.8.
- Nováková A, Hubka V, Saiz-Jimenez C, Kolarik M. 2012. Aspergillus baeticus sp. nov. and aspergillus thesauricus sp. Nov., two species in section usti from spanish caves. Intern J Syst Evol Microb. 62(Pt_11):2778–2785. doi:https://doi.org/10.1099/ijs.0.041004-0.
- Nováková A, Kubátová A, Sklenář F, Hubka V. 2018. Microscopic fungi on cadavers and skeletons from cave and mine environments. Czech Mycol. 70(2):101–121. doi:https://doi.org/10.33585/cmy.70201.
- Ogórek R, Piecuch A, Višňovská Z, Cal M, Niedźwiecka K. 2019. First report on the occurence of Dermatophytes of Microsporum Cookei Clade and Close Affinities to Paraphyton Cookei in the Harmanecká Cave (Veľká Fatra Mts., Slovakia). Divers. 11(10): 191
- Ogórek R, Lejman A, Matkowski K. 2013. Fungi isolated from niedźwiedzia cave in kletno (lower silesia, poland). Int J Speleol. 42(2):161–166. doi:https://doi.org/10.5038/1827-806X.42.2.9.
- Ogórek R, Pusz W, Zagożdżon PP, Kozak B, Bujak H. 2017. Abundance and diversity of psychrotolerant cultivable mycobiota in winter of a former aluminous shale mine. Geomicrobiol J. 34(10):823–833. doi:https://doi.org/10.1080/01490451.2017.1280860.
- Ortiz M, Legatzki A, Neilson JW, Fryslie B, Nelson WM, Wing RA, Soderlund CA, Pryor BM, Maier RM. 2014. Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave. Isme J. 8(2):478–491. doi:https://doi.org/10.1038/ismej.2013.159.
- Paulson TL. 1972. Bat guano ecosystems. Bull of the Natl Speleol Soc. 34(2):55–59.
- Pereira J, Rogers JD, Bezerra JL. 2010. New annulohypoxylon species from brazil. Mycol. 102(1):248–252. doi:https://doi.org/10.3852/09-116.
- Porca E, Jurado V, Martin-Sanchez PM, Hermosin B, Bastian F, Alabouvette C, Saiz-Jimenez C. 2011. Aerobiology: an ecological indicator for early detection and control of fungal outbreaks in caves. Ecol Indic. 11(6):1594–1598. doi:https://doi.org/10.1016/j.ecolind.2011.04.003.
- Puechmaille SJ, Frick WF, Kunz TH, Racey PA, Voigt CC, Wibbelt G, Teeling EC. 2011. White-nose syndrome: Is this emerging disease a threat to european bats? trends in ecol & evol. 26(11):570–576. doi:https://doi.org/10.1016/j.tree.2011.06.013.
- Pusz W, Ogórek R, Knapik R, Kozak B, Bujak H. 2015. The occurrence of fungi in the recently discovered jarkowicka cave in the karkonosze mts. (poland). Geomicrobiol J. 32(1):59–67. doi:https://doi.org/10.1080/01490451.2014.925010.
- Pusz W, Ogórek R, Uklańska-Pusz CM, Zagożdżon P. 2014. Speleomycological research in underground osówka complex in sowie mountains (lower silesia, poland). Intern J Speleol. 43(1):3. doi:https://doi.org/10.5038/1827-806X.43.1.3.
- Raper KB, Fennel DI. 1965. . Aspergillus terreus group. the genus aspergillus. Baltimore (MD): The Williams & Wilkins Co.
- Sahashi N, Akiba M, Takemoto S, Yokoi T, Ota Y, Kanzaki N. 2014. Phellinus noxius causes brown root rot on four important conifer species in japan. Eur J Plant Pathol. 140(4):869–873. doi:https://doi.org/10.1007/s10658-014-0503-9.
- Schimel JP, Gulledge JM, Clein-Curley JS, Lindstrom JE, Braddock JF. 1999. Moisture effects on microbial activity and community structure in decomposing birch litter in the alaskan taiga. Soil Biol Biochem. 31(6):831–838. doi:https://doi.org/10.1016/S0038-0717(98)00182-5.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Consortium FB. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 109(16):6241–6246. doi:https://doi.org/10.1073/pnas.1117018109.
- Shapiro J, Pringle A. 2010. Anthropogenic influences on the diversity of fungi isolated from caves in kentucky and tennessee. Am Midl Nat. 163(1):76–87. doi:https://doi.org/10.1674/0003-0031-163.1.76.
- Sharon E, Chet I, Viterbo A, Bary-Eyal M, Nagan H, Samuels GJ, Spiegel Y. 2007. Parasitism of trichoderma on meloidogyne javanica and role of the gelatinous matrix. Eur J Plant Pathol. 118(3):247–258. doi:https://doi.org/10.1007/s10658-007-9140-x.
- Smrž J, Ľ K, Mikeš J, Šustr V. 2015. Food sources of selected terrestrial cave arthropods. 46:37–46. DOI:https://doi.org/10.3897/subtbiol.16.8609.
- Sterflinger K, DeHoog GS, Haase G. 1999. Phylogeny and ecology of meristematic ascomycetes. Stud Mycol. 43:5–22.
- Stošić S, Ristić D, Gašić K, Starović M, Ljaljević Grbić M, Vukojević J, Živković S. 2019. Talaromyces minioluteus–new postharvest fungal pathogen in Serbia. Plant Dis.
- Šustr V, Elhottová D, Krištůfek V, Lukešová A, Nováková A, Tajovský K, Tříska J. 2005. Ecophysiology of the cave isopod mesoniscus graniger (frivaldszky, 1865) (crustacea: isopoda). Eur J Soil Biol. 41(3–4):69–75. doi:https://doi.org/10.1016/j.ejsobi.2005.09.008.
- Taylor ELS, Ferreira RL, Cardoso PG, Stoianoff MAR. 2014. Cave entrance dependent spore dispersion of filamentous fungi isolated from various sediments of iron ore cave in brazil: a colloquy on human threats while caving. Ambient Sci. 1(1):16–28.
- Thambugala KM, Camporesi E, Ariyawansa HA, Phookamsak R, Liu Z, Hyde KD. 2014. Phylogeny and morphology of phaeosphaeriopsis triseptata sp. nov., and phaeosphaeriopsis glaucopunctata. 176(1):238–250. https://doi.org/10.1111/cei.12275.
- Vanderwolf KJ, Malloch D, McAlpine DF. 2016. Ectomycota associated with arthropods from bat hibernacula in eastern canada, with particular reference to pseudogymnoascus destructans. Insects. 7(2):16. doi:https://doi.org/10.3390/insects7020016.
- Vanderwolf KJ, Malloch D, Mcalpine DF, Forbes GJ. 2013. A world review of fungi, yeasts, and slime molds in caves. Int J Speleol. 42(1):77–96. doi:https://doi.org/10.5038/1827-806X.42.1.9.
- Vaughan MJ, Maier RM, Pryor BM. 2011. Fungal communities on speleothem surfaces in kartchner caverns, arizona, USA. Int J Speleol. 40(1):65–77. doi:https://doi.org/10.5038/1827-806X.40.1.8.
- Wainwright M, Ali TA, Barakah F. 1993. A review of the role of oligotrophic micro-organisms in biodeterioration. Intern Biodeterior & Biodegrad. 31(1):1–13. doi:https://doi.org/10.1016/0964-8305(93)90010-Y.
- Wakelin SA, Gupta VVSR, Harvey PR, Ryder MH. 2007. The effect of penicillium fungi on plant growth and phosphorus mobilization in neutral to alkaline soils from southern australia. Can J Microbiol. 53(1):106–115. doi:https://doi.org/10.1139/W06-109.
- Wang W, Ma X, Ma Y, Mao L, Wu F, Ma X, Feng H. 2010. Seasonal dynamics of airborne fungi in different caves of the mogao grottoes, dunhuang, china. Intern Biodeterior & Biodegrad. 64(6):461–466. doi:https://doi.org/10.1016/j.ibiod.2010.05.005.
- Watanabe S, Kumakura K, Kato H, Iyozumi H, Togawa M, Nagayama K. 2005. Identification of trichoderma SKT- 1, a biological control agent against seedborne pathogens of rice. J Gen Plant Pathol. 71(5):351–356. doi:https://doi.org/10.1007/s10327-005-0217-0.
- White TJ, Burns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 18(1):315–322.
- Wilford GE. 1961. The geology and mineral resources of brunei and adjacent parts of sarawak: with descriptions of seria and miri oilfields. geological survey department. Borneo Region, Malaysia, Memoir. 10:319.
- Wiseschart A, Mhuantong W, Tangphatsornruang S, Chantasingh D, Pootanakit K. 2019. Shotgun metagenomic sequencing from manao-pee cave, thailand, reveals insight into the microbial community structure and its metabolic potential. BMC Microbiol. 19(1):144. doi:https://doi.org/10.1186/s12866-019-1521-8.
- Zhang Z, Zhao P, Cai L. 2018. Origin of cave fungi. Front Microbiol. 9:1407. doi:https://doi.org/10.3389/fmicb.2018.01407
- Zhang ZF, Cai L. 2019. Substrate and spatial variables are major determinants of fungal community in karst caves in southwest china. J Biogeogr. 46(7):1504–1518.
- Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L. 2017. Culturable mycobiota from karst caves in china, with descriptions of 20 new species. Pers - Mol Phylogeny and Evol Fungi. 39(1):1–31. doi:https://doi.org/10.3767/persoonia.2017.39.01.
- Zhang ZF, Zhou SY, Eurwilaichitr L, Ingsriswang S, Raza M, Chen Q, Zhao P, Liu F, Cai L. 2020. Culturable mycobiota from karst caves in china II, with descriptions of 33 new species. Fungal divers. p. 1–108. doi: https://doi.org/10.1007/s13225-020-00453-7.