ABSTRACT
An entry postal parcel with mature nuts of Phytelephas macrocarpa from Togo was inspected at Dalian Customs (China) in December 2021, and four strains were isolated from symptomatic tissues of the nuts. Based on morphological observations and molecular phylogenetic analyses, above strains were identified as a new species which is mainly characterised by the verticillately branching conidiophores. Based on multi-locus phylogenetic analyses, this new species forms a monophyletic clade closely related to Corallomycetella, Paracremonium and Xenoacremonium but could not be accommodated in any known genera of Nectriaceae. Thus, a new genus Heteroverticillium is established to accommodate this new species (H. phytelephatis). To our knowledge, this is the first time that Chinese customs have intercepted a new fungal genus. In addition, we provided an updated backbone tree for the generic relationships in Nectriaceae, which may largely assist future identification of nectriaceous fungi to genus level in quarantine inspections. Based on our analysis, Varicosporellopsis is likely a late synonym of Paracremonium.
1. Introduction
The introduction of alien pathogens into a certain region may have profound negative effects on local native biota, ecological balance, human health and economic development (Capinha et al. Citation2015; Pyšek et al. Citation2020). With the growth of international trade, the risk of introducing invasive alien phytopathogens also increase (Zhao et al. Citation2021). Prevention the global spread of alien pathogens becomes an increasingly important concern for most countries. Alien pathogens often enter through cargo, passenger, conveyance, post, wood package and container, and the rate of pest interception is particularly high in parcels, as China is the world’s largest carrier of parcels. The quarantine pest list of China currently includes 130 fungal species, of which 10 belong to the Ascomycetes family Nectriaceae (http://dzs.customs.gov.cn/dzs/2746776/3699554/index.html; accessed 9 April 2022). Besides, 175 species in family Nectriaceae (e.g. species from Acremonium, Calonectria, Cylindrocladiella, Fusarium, Ilyonectria, Neocosmospora, Volutella) have been listed as quarantine organisms in 52 countries (Zhao et al. Citation2021). Nectriaceae species thus receive high attention in the quarantine departments of different countries.
The family Nectriaceae includes a variety of important plant and human pathogens (Rossman Citation1996; Luo and Zhuang Citation2008; Chaverri et al. Citation2011; Lechat et al. Citation2015; Lombard et al. Citation2015), such as pathogens of Fusarium head blight (Fusarium spp.), pathogens of vascular wilts of many economically important crops (members of the Fusarium oxysporum species complex), and pathogens of sudden death syndrome of soybeans and root rot of many diverse hosts (Neocosmospora spp.) (O’Donnell et al. Citation2000a; Aoki et al. Citation2003; Qiu et al. Citation2012; Lombard et al. Citation2019; Maryani et al. Citation2019; Sandoval-Denis et al. Citation2019; Medeiros Araújo et al. Citation2020; Xu et al. Citation2021). Recent advances in the taxonomy of fungi following the one fungus = one name initiative and the implementation of DNA phylogeny in taxonomic revisions, resulted in many name changes, particularly those applying to subspecies, varieties and formae speciales (Wingfield et al. Citation2012). For example, the pathogen of banana Fusarium wilt (previously collectively called F. oxysporum f. sp. cubense) has recently been splited into nine Fusarium species based on morphological characteristics and phylogenetic inferences (Maryani et al. Citation2019). Despite the recent good advances in the systematics and phylogeny of Nectriaceae (Lombard et al. Citation2015; Crous et al. Citation2021a), the identification of Nectriaceous species remains a challenge to both plant pathologists and phytoquarantine staffs.
Phytelephas macrocarpa is mainly distributed in South America, commonly named as vegetable ivory (Smith Citation2015). The very hard seeds of this plant are used as a substitute for ivory and are used to make buttons, chess pieces and a variety of ornamental antiques (Smith Citation2015). Mature nuts of Phytelephas macrocarpa are recently increasingly imported to China through international freight transport or entry postal parcel. In December 2021, an entry postal parcel with mature nuts of Phytelephas macrocarpa from Togo was intercepted at China Customs (Dalian), and four fungal strains were isolated from the symptomatic tissues of mature nuts. Employing multi-locus phylogeny and morphological features, above strains were identified as a novel species belonging to Nectriaceae but could not be accommodated in any known genera. Heteroverticillium phytelephatis gen. et sp. nov. is thus described in this paper and its phylogenetic position is determined through our construction of a new backbone tree of Nectriaceae.
2. Materials and methods
2.1. Strains isolation
A total of four strains were isolated from the symptomatic tissues of the mature nuts of Phytelephas macrocarpa using single spore isolation (Zhang et al. Citation2013). The type specimens of new species described in this study were deposited in the Fungarium of Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and the living ex-type cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC). All isolates examined in this study were also deposited in Lei Cai’s personal culture collection (LC), housed at Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. Taxonomic novelty descriptions and nomenclature were deposited in Fungal Names (Wang et al. Citation2023).
2.2. DNA extraction and amplification
Total genomic DNA was extracted from fresh fungal mycelia grown on PDA using the cetyltrimethyl ammonium bromide (CTAB) method (Porebski et al. Citation1997) and stored at −20 °C until use for polymerase chain reaction (PCR). PCR amplifications were performed in a reaction mixture consisting of 12.5 μL 2 × Taq PCR Master Mix (Vazyme Biotech Co., Ltd, Nanjing, China), 1 μL each of 10 μmol/L primers, 2 μL genomic DNA, adjusted to a final volume of 25 μL with distilled deionised water. Nine loci, including 28S large subunit (LSU) nrDNA, the internal transcribed spacer region and intervening 5.8S nrRNA gene (ITS), the large subunit of the ATP citrate lyase (acl1), the RNA polymerase II largest subunit (rpb1), RNA polymerase II second largest subunit (rpb2), β-tubulin (tub2), calmodulin (CaM), histone-3 (H3), and translation elongation factor 1-alpha (tef1) gene regions were amplified and sequenced, respectively. The PCR primer pairs and amplification conditions are listed in . The PCR products were visualised using 1% agarose electrophoresis gels. Sequencing was done by the Tianyi Huiyuan Company (Beijing, China) and SinoGenoMax Company (Beijing, China).
Table 1. Primers used in this study, with originating loci, sequences programme and references.
2.3. Phylogenetic analyses
Consensus sequences were obtained using MEGA v. 7 software (Kumar et al. Citation2016), and sequences for each locus were aligned using MAFFT v. 7.505 (Katoh and Standley Citation2013). Misalignments were corrected manually where necessary. Phylogenetic analyses were performed based on combined datasets, using Maximum-Likelihood (ML) and Bayesian Inference (BI) methods through the CIPRES Science Gateway portal (https://www.phylo.org/; Miller et al. Citation2012).
The ML analyses were carried out using RAxML-HPC v. 8.2.12 (Stamatakis Citation2014), with 1,000 replicates under the GTR+GAMMA model. The Bayesian analyses were carried out using MrBayes v. 3.2.7a (Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003), incorporating the best evolutionary models for each marker as determined by MrModelTest v. 2.4 (Nylander Citation2004). Bayesian analyses were computed with four simultaneous Markov Chain Monte Carlo chains for 20 M generations, and trees were sampled every 2,000 generations. The burn-in fraction was set to 0.25, after which the 50% majority rule consensus trees and posterior probability (PP) values were calculated.
The clade supported with RAxML Bootstrap ≥70%, and the Bayesian PP ≥ 0.9 were marked on the tree (). The tree was plotted using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree). All sequences and their alignments generated in this study were deposited in GenBank () and TreeBASE (submission ID 30339), respectively.
Table 2. Strains analysed in this study, with information about host/substrate, location and GenBank accessions of sequences.
2.4. Morphological observation
Obtained fungi were observed morphologically based on their macroscopic and microscopic features (Lombard et al. Citation2015). Plates were incubated for 7 days at 25 °C. Agar piece of approximately 5 × 5 mm was taken from the edge of colonies on synthetic nutrient-poor agar (SNA), and transferred onto different media for morphological observation. For macroscopic studies, potato dextrose agar (PDA), oatmeal agar (OA) and malt extract agar (MEA) media were used. The culture characteristics of the colony, including pigmentation and odour, were observed after 7 days of incubation in the dark on PDA, OA and MEA, respectively (Lombard et al. Citation2015). Colours were rated according to the colour charts (Kornerup and Wanscher Citation1978).
In the microscopic morphology study, SNA with sterile carnation leaves were used. Micromorphological characteristics, including conidiophores, phialides, and conidia, were observed after 7–14 days of incubation under a 12/12 h near-ultraviolet light/dark cycle at 25 °C (Lombard et al. Citation2015). Micromorphological characteristics were examined and photo-documented with water as mounting medium under a Nikon 80i microscope with differential interference contrast (DIC) optics, and a Nikon SMZ1500 dissecting microscope. For each species, 30 phialides and 50 conidia were randomly measured to calculate the mean value, standard deviation and minimum–maximum values. Descriptions and illustrations of taxonomic novelties were deposited in Fungal Names (Wang et al. Citation2023).
3. Results
3.1. Phylogenetic analysis
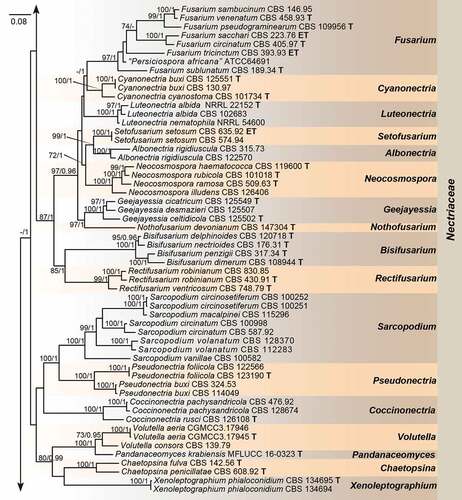
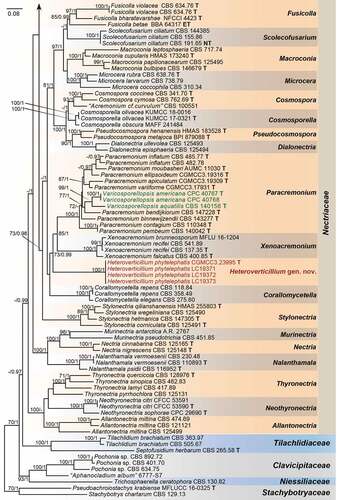
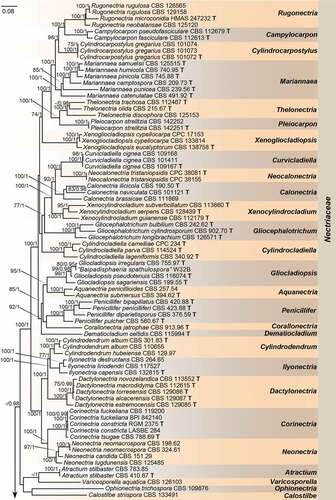
Employing 212 strains representing 169 species (five families), with Stachybotrys chartarum (CBS 129.13) and Pseudoachroiostachys krabiense (MFLUCC 16–0325) as outgroups, a high confidence phylogenetic tree was generated (). The nine-locus alignment was 7 497 bases in length including gaps. The best nucleotide substitution model for acl1, CaM and tub2 loci was HKY + I + G, while GTR + I + G was selected for H3, ITS, LSU, rpb1, rpb2, and tef1. The topology of multi-locus phylogenetic trees retrieved from ML and BI analyses were congruent. The results indicated that our new isolates formed a distinct clade in the Nectriaceae family but could not be included in any existing genera (). Moreover, we provided a hitherto largest backbone tree of generic relationships in Nectriaceae based on phylogenetic analyses using existing sequence data from nine-locus (acl1-CaM-H3-ITS-LSU-RPB1-RPB2-tef1-tub2), and phylogenetic relationship of 63 accepted genera in this family were revealed (). Most genera were monophyletic and supported by high bootstrap values. Our phylogenetic results showed that the ex-types of two Varicosporellopsis species, i.e. V. aquatilis and V. americana clustered in one clade together with 9 species of Paracremonium.
Figure 1. Phylogeny inferred based on the combined acl1-CaM-H3-ITS-LSU-RPB1-RPB2-tef1-tub2 gene regions of species from Nectriaceae. Stachybotrys chartarum (CBS 129.13) and Pseudoachroiostachys krabiense (MFLUCC 16–0325) were used as outgroups. Strains isolated in this study were indicated in red colour. Strains of Varicosporellopsis, which is likely a late synonym of Paracremonium were indicated in green colour. The RAxML Bootstrap support values (ML-BS ≥ 70%) and Bayesian posterior probabilities (BI-PP ≥ 0.9) were displayed at the nodes (ML-BS/BI-PP). Ex-type, ex-epitype and ex-neotype strains were indicated in bold with T, ET, and NT, respectively. Strains need to be further identified were indicated with double quotation marks (“”).
3.2. Taxonomy
New taxa
Heteroverticillium S.L. Han, L. Cai & P. Zhao, gen. nov. .
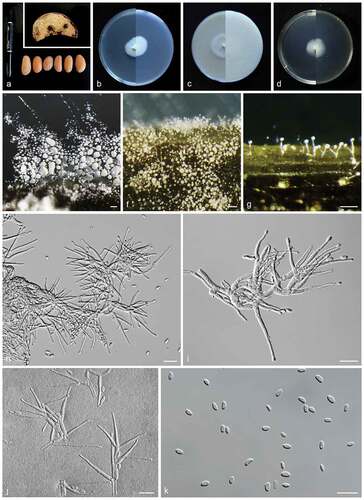
Figure 2. Heteroverticillium phytelephatis (CGMCC3.23995, ex-type culture). a: Disease symptoms on host plant. b: Front and reverse colony on PDA (7 d). c: Front and reverse colony on OA (7 d). d: Front and reverse colony on MEA (7 d). e–g: Aerial conidiophores. h–j: Conidiophores and phialides. k: Conidia. Scale bars: e–g = 50 μm; h–k = 10 μm.
Fungal name: FN 571491.
Etymology:
Heteros = έτερος in Greek, other; morphologically similar to but phylogenetically different from Verticillium.
Type species:
Heteroverticillium phytelephatis S.L. Han, L. Cai & P. Zhao.
Description:
Sexual morph: not observed. Asexual morph: Hyphae hyaline, smooth-walled, septate, branched, with inconspicuously swollen septa. Conidiophores arising laterally from somatic hyphae, verticillately branching at 2–3 levels, with a terminal whorl of 1–5 phialides, and 1–2 lower nodes of 1–3 phialides, rarely with single phialides. Phialides monophialide, subulate, smooth- and thin-walled, periclinal thickening inconspicuous or absent. Conidia hyaline, smooth- and thin-walled, aseptate, ellipsoidal to slightly reniform.
Heteroverticillium phytelephatis S.L. Han, L. Cai & P. Zhao, sp. nov.
Fungal name: FN 571494.
Etymology:
Named after the host genus from which the type strain was isolated, Phytelephas.
Description:
Sexual morph: not observed. Asexual morph: Hyphae 1.9–2.2 μm diam., hyaline, smooth-walled, septate, branched, with inconspicuously swollen septa. Conidiophores arises laterally from somatic hyphae, mostly 14.8–65.4 μm long, axis 1.7–2.9 μm wide, verticillately branching at 2–3 levels, with a terminal whorl of 1–5 phialides, and 1–2 lower nodes of 1–3 phialides, rarely with single phialides. Phialides monophialide, subulate, smooth- and thin-walled, periclinal thickening inconspicuous or absent, 11.7–39.1 × 1.2–2.3 μm (av. ± SD: 22.8 ± 1.9 × 2.1 ± 0.3 μm). Conidia hyaline, smooth- and thin-walled, aseptate, ellipsoidal to slightly reniform: 3.7–6.2 × 1.5–2.8 μm (av. ± SD: 4.4 ± 0.5 × 2.2 ± 0.3 μm). Chlamydospores not observed.
Culture characteristics:
Colonies on PDA slow growing, reaching 23–27 mm diam. in 7 d after incubation at 25 °C in the dark, flat, with almost invisible aerial mycelium, wrinkled, margin entire, surface ivory; reverse white; odour absent. On OA reaching 31–38 mm diam. in 7 d after incubation 25 °C in the dark; flat, with sparse aerial mycelium, margin entire; surface white; reverse cornsilk; odour absent. On MEA reaching 19–22 mm diam. in 7 d after incubation 25 °C in the dark; flat, with almost invisible aerial mycelium, margin entire; surface light beige; reverse beige; odour absent.
Material examined:
TOGO, intercepted at China Customs (Dalian), infected nuts of Phytelephas macrocarpa, Dec. 2021, X. Li, HMAS 352429; TOGO, intercepted at China Customs (Dalian), isolated from infected nuts of Phytelephas macrocarpa, Dec. 2021, X. Li (holotype HMAS 352423, dried culture; ex-holotype living culture CGMCC3.23995 = LC19374); ibid., LC19371; ibid., LC19372; ibid., LC19373.
Notes:
The genus Heteroverticillium is phylogenetically allied to genera Corallomycetella, Paracremonium and Xenoacremonium (). Morphologically, Heteroverticillium could be distinguished from these genera in the verticillately branching conidiophores, which is not observed in Corallomycetella, Paracremonium and Xenoacremonium (Lombard et al. Citation2015). Heteroverticillium also differs from Corallomycetella, Paracremonium and Xenoacremonium in lacking pigment production in culture, which is obvious in the later three.
4. Discussions
The growth of international trade, tourisms and post parcels in the past decades has facilitated the incidence of alien species invasion (Early et al. Citation2016). The number and frequency of intercepted quarantine species from post parcels are quickly accelerating, similar to those in cargo, passenger, conveyance, post, wood package and container (Lyu and Duan Citation2021). The Chinese customs has recently intercepted several alien insects, grass seeds and fungi from the post parcels (Yin and Ma Citation2004; Lin and Weng Citation2011; Ma et al. Citation2017; Wei Citation2019).
Based on the latest classification, four strains isolated from imported nuts of Phytelephas macrocarpas in post parcels were identified to be representing a new genus in Nectriaceae. This is, to our knowledge, the first time a new fungal genus has been intercepted by Chinese Customs. The new genus Heteroverticillium is mainly characterised by verticillately branching conidiophores, closely related to Corallomycetella, Paracremonium and Xenoacremonium (Nectriaceae). Notably, these three genera contain mostly human and plant pathogenic fungi (Gams Citation1971; Lombard et al. Citation2015). While the pathogenicity of Heteroverticillium phytelephatis need to be further confirmed in future studies when fresh specimens of Phytelephas macrocarpas were accessible.
Members of the family Nectriaceae are commonly found in various environments, and some are important plant or human pathogens (Lombard et al. Citation2015). Lombard et al. (Citation2015) conducted hitherto most comprehensive morphological and molecular phylogenetic assessments of available type and authentic strains representing known genera in Nectriaceae, and their data have resolved most taxonomic discordances within the family Nectriaceae. Thereafter, 15 new genera within this family have been published (Crous et al. Citation2015, Citation2016, Citation2020, Citation2021a; Lechat and Fournier Citation2015, Citation2016; Aiello et al. Citation2017; González and Chaverri Citation2017; Huang et al. Citation2018; Hyde Citation2020). In this study, we updated the phylogenetic tree based on Lombard et al. (Citation2015), with the addition of subsequently available data from 17 genera, which may greatly facilitate future identification of nectriaceous species in quarantine inspections. Several anamorph typified genera such as Cyanochyta, Cyanophomella, Dacryoma, and Pleurocolla were traditionally included in Nectriaceae but their phylogenetic relationships with other genera in Nectriaceae remain unclear due to the lack of molecular data. Compared with their original descriptions, our new genus could be well distinguishable from them by the verticillately branching conidiophores and the type of conidia (Höhnel von Citation1915, Citation1920; Petrak Citation1924; Samuels Citation1988).
Interestingly, the ex-types of two Varicosporellopsis species, i.e. V. aquatilis and V. americana proposed by Lechat and Fournier (Citation2016) and Crous et al. (Citation2021b) respectively, clustered within the Paracremonium clade (), as also indicated in Crous et al. (Citation2021b). Our phylogenentic analysis therefore indicated that the Varicosporellopsis is likely a late synonym of the anamorph typified genus Paracremonium. The genus Paracremonium, as circumscribed based on its type species P. inflatum (Lombard et al. Citation2015), could be distinguished from other acremonium-like genera by the formation of sterile coils (Lombard et al. Citation2015). However, none of the 9 subsequently described Paracremonium species produce sterile coils (Lynch et al. Citation2016; Crous et al. Citation2017, Citation2021b; Al-Bedak Citation2019; Zhang et al. Citation2020; DQ et al. Citation2021). The generic concept of Paracremonium might need to be updated with regard to the sterile coils, as well as the teleomorphic state observed in V. aquatilis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Aiello D, Polizzi G, Crous PW, Lombard L. 2017. Pleiocarpon gen. nov. and a new species of Ilyonectria causing basal rot of Strelitzia reginae in Italy. IMA Fungus. 8(1):65–76. doi:10.5598/imafungus.2017.08.01.05.
- Al-Bedak OA. 2019. Paracremonium moubasheri, a new species from an alkaline sediment of Lake Hamra in Wadi-El-Natron, Egypt with a key to the accepted species. Stud Fungi. 4:216−222.
- Aoki T, O’Donnell K, Homma Y, Lattanzi AR. 2003. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex—f. virguliforme in North America and F. tucumaniae in South America. Mycologia. 95(4):660–684. doi:10.1080/15572536.2004.11833070.
- Capinha C, Essl F, Seebens H, Moser D, Pereira HM. 2015. The dispersal of alien species redefines biogeography in the Anthropocene. Science. 348(6240):1248–1251. doi:10.1126/science.aaa8913.
- Chaverri P, Salgado C, Hirooka Y, Rossman AY, Samuels GJ. 2011. Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol. 68:57−78.
- Crous PW, Cowan DA, Maggs-Kolling G, Yilmaz N, Larsson E, Angelini C, Brandrud TE, Dearnaley JDW, Dima B, Dovana F, et al. 2020. Fungal planet description sheets: 1112–1181. Persoonia. 45(1):251–409. doi:10.3767/persoonia.2020.45.10
- Crous PW, Lombard L, Sandoval-Denis M, Seifert KA, Schroers HJ, Chaverri P, Gene J, Guarro J, Hirooka Y, Bensch K, et al. 2021a. Fusarium: more than a node or a foot-shaped basal cell. Stud Mycol. 98:1–184.
- Crous PW, Osieck ER, Ž J, Boers J, Van Iperen AL, Starink-Willemse M, Dima B, Balashov S, Bulgakov TS, Johnston PR, et al. 2021b. Fungal planet description sheets: 1284–1382. Persoonia. 47(1):178–374. doi:10.3767/persoonia.2021.47.06
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, et al. 2017. Fungal planet description sheets: 625–715. Persoonia. 39:270–467. doi:10.3767/persoonia.2017.39.11.
- Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Crane C, Barrett S, Cano-Lira JF, Le Roux JJ, Thangavel R, Guarro J, et al. 2016. Fungal Planet description sheets: 469-557. Persoonia - Molecular Phylogeny and Evolution of Fungi. 37(1):218–403. doi:10.3767/003158516X694499
- Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, et al. 2015. Fungal Planet description sheets: 371–399. Persoonia. 35(1):264–327. doi:10.3767/003158515X690269
- Mng DQ, Luo LY, He XX, Wang MS, Fang WX, Chen SF, Chen WH, Han YF, Liang ZQ. 2021. Paracremonium lepidopterorum, a new insect-associated fungus. Phytotaxa. 524:85–91. doi:10.11646/phytotaxa.524.2.2.
- Early R, Bradley BA, Dukes JS, Lawler JJ, Olden JD, Blumenthal DM, Gonzalez P, Grosholz ED, Ibanez I, Miller LP, et al. 2016. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat Commun. 7(1):12485. doi:10.1038/ncomms12485
- Gams W. 1971. Cephalosporium-artige schimmelpilze (Hyphomycetes). Stuttgart (Germany): Gustav Fischer Verlag.
- González CD, Chaverri P. 2017. Corinectria, a new genus to accommodate Neonectria fuckeliana and C. constricta sp. nov. from Pinus radiata in Chile. Mycol Prog. 16(11–12):1015–1027. doi:10.1007/s11557-017-1343-8.
- Gräfenhan T, Schroers HJ, Nirenberg HI, Seifert KA. 2011. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol. 68:79–113. doi:10.3114/sim.2011.68.04.
- Höhnel von F. 1915. Fragmente zur Mykologie (XVII. Mitteilung, Nr. 876 bis 943). Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften Math.-naturw. Klasse Abt I. 124:49–159.
- Höhnel von F. 1920. Fungi imperfecti. Beiträge zur Kenntnis derselben Hedwigia. 60:56–89.
- Huang SK, Jeewon R, Hyde KD, Bhat DJ, Wen TC. 2018. Novel Taxa within Nectriaceae: cosmosporella gen. nov. and Aquanectria sp. nov. from Freshwater Habitats in China. Crypto Mycol. 39(2):169–192. doi:10.7872/crym/v39.iss2.2018.169.
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: bayesian inference of phylogeny. Bioinformatics. 17(8):754–755. doi:10.1093/bioinformatics/17.8.754.
- Hyde KD. 2020. Refined families of Sordariomycetes. Mycosphere. 11(1):305–1059. doi:10.5943/mycosphere/11/1/7.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. In: 3rd edn. London: Eyre Methuen.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Lechat C, Fournier J. 2015. Varicosporella, a new aquatic genus in Nectriaceae from France. Ascomyceteorg. 7:1–8.
- Lechat C, Fournier J. 2016. Varicosporellopsis, a new aquatic genus from South of France. Ascomyceteorg. 8:96–100.
- Lechat C, Fournier J, Nordén B. 2015. Stylonectria norvegica (Nectriaceae), a new species from Norway. Ascomyceteorg. 7:220–224.
- Lin YW, Weng RQ. 2011. Rhynchophorus ferrugineus (Oliver) was intercepted from inbound parcels at Fujian Port. Plant Quar. 25:24.
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 16(12):1799–1808. doi:10.1093/oxfordjournals.molbev.a026092.
- Lombard L, Sandoval-Denis M, Lamprecht SC, Crous PW. 2019. Epitypification of Fusarium oxysporum – clearing the taxonomic chaos. Persoonia. 43(1):1–47. doi:10.3767/persoonia.2019.43.01.
- Lombard L, van der Merwe NA, Groenewald JZ, Crous PW. 2015. Generic concepts in Nectriaceae. Stud Mycol. 80(1):189–245. doi:10.1016/j.simyco.2014.12.002.
- Luo J, Zhuang WY. 2008. Two new species of Cosmospora (Nectriaceae, Hypocreales) from China. Fungal Divers. 31:83–93.
- Lynch SC, Twizeyimana M, Mayorquin JS, Wang DH, Na F, Kayim M, Kasson MT, Thu PQ, Bateman C, Rugman-Jones P, et al. 2016. Identification, pathogenicity and abundance of Paracremonium pembeum sp. nov. and Graphium euwallaceae sp. nov.—two newly discovered mycangial associates of the polyphagous shot hole borer (Euwallacea sp.) in California. Mycologia. 108(2):313–329. doi:10.3852/15-063
- Lyu Y, Duan WJ. 2021. The current status and suggestions of interception of quarantine fungi at China ports from 2017 to 2019. Plant Quar. 35:70–76.
- Maryani N, Lombard L, Poerba YS, Subandiyah S, Crous PW, Kema GHJ. 2019. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud Mycol. 92:155–194.
- Ma GD, Zhang MZ, Chen WJ. 2017. First interception of Diaporthe phaseolorum at national postal inspection ports. Plant Quar. 21:38.
- Medeiros Araújo MB, Moreira GM, Nascimento LV, GdA N, SRdC N, Pfenning LH, Ambrósio M. 2020. Fusarium rot of melon is caused by several Fusarium species. Plant Pathol. 70(3):712–721. doi:10.1111/ppa.13328.
- Miller MA, Pfeiffer W, Schwartz T 2012. The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: proceedings of the 1st conference of the extreme science and engineering discovery environment: bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA: 1–8.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre. Sweden: Uppsala University.
- O’Donnell K, Cigelnik E. 1997. Two Divergent Intragenomic rDNA ITS2 Types within a Monophyletic Lineage of the FungusFusariumAre Nonorthologous. Mol Phylogenet Evol. 7(1):103–116. doi:10.1006/mpev.1996.0376.
- O’Donnell K, Kistlerr HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 95(5):2044–2049. doi:10.1073/pnas.95.5.2044.
- O’Donnell K, Kistler HC, Tacke BK, Casper HH. 2000a. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA. 97(14):7905–7910. doi:10.1073/pnas.130193297.
- O’Donnell K, Nirenberg H, Aoki T, Cigelnik E. 2000b. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 41(1):61–78. doi:10.1007/BF02464387.
- O’Donnell K, Sutton DA, Rinaldi MG, Sarver BA, Balajee SA, Schroers HJ, Summerbell RC, Robert VA, Crous PW, Zhang N, et al. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol. 48(10):3708–3718. doi:10.1128/JCM.00989-10
- Petrak F. 1924. Mykologische Notizen. VII Ann Mycol. 22:1–182.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. doi:10.1007/BF02772108.
- Pyšek P, Hulme PE, Simberloff D, Bacher S, Blackburn TM, Carlton JT, Dawson W, Essl F, Foxcroft LC, Genovesi P, et al. 2020. Scientists’ warning on invasive alien species. Biol Rev Camb Philos Soc. 95:1511−1534.
- Qiu MH, Zhang RF, Xue C, Zhang SS, Li SQ, Zhang N, Shen QR. 2012. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fertil Soils. 48:807−816.
- Reeb V, Lutzoni F, Roux C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the Euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol Phylogenet Evol. 32(3):1036–1060. doi:10.1016/j.ympev.2004.04.012.
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 98(6):625–634. doi:10.1016/S0953-7562(09)80409-7.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Rossman A. 1996. Morphological and molecular perspectives on systematics of the Hypocreales. Mycologia. 88(1):1–19. doi:10.1080/00275514.1996.12026620.
- Roux J, Steenkamp ET, Marasas WFO, Wingfield MJ, Wingfield BD. 2001. Characterization of Fusarium graminearum from Acacia and Eucalyptus using β-tubulin and histone gene sequences. Mycologia. 93(4):704–711. doi:10.1080/00275514.2001.12063201.
- Samuels GJ. 1988. Species of Nectria (Ascomycetes, Hypocreales) having orange perithecia and colorless, striate ascospores. Brittonia. 40:306–331.
- Sandoval-Denis M, Lombard L, Crous PW. 2019. Back to the roots: a reappraisal of Neocosmospora. Persoonia. 43(1):90–185. doi:10.3767/persoonia.2019.43.04.
- Smith N. 2015. Phytelephas macrocarpa. In: palms and people in the Amazon. Geobotany studies. doi:10.1007/978-3-319-05509-1_53
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312−1313.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172(8):4238–4246. doi:10.1128/jb.172.8.4238-4246.1990.
- Wang F, Wang K, Cai L, Zhao M, Kirk PM, Fan G, Sun Q, Li B, Wang S, Yu Z, et al. 2023. Fungal names: a comprehensive nomenclatural repository and knowledge base for fungal taxonomy. Nucleic Acids Res. 51(D1):708–716. doi:10.1093/nar/gkac926
- Wei WC. 2019. First interception of live cockroach with pyomonas aeruginosa nucleic acid in Nanning. China Inspection and Quarantine. Z1:13.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 18:315–322.
- Wingfield MJ, De Beer ZW, Slippers B, Wingfield BD, Groenewald JZ, Lombard L, Crous PW. 2012. One fungus, one name promotes progressive plant pathology. Mol Plant Pathol. 13:604−613.
- Xu F, Liu W, Song Y, Zhou Y, Xu X, Yang G, Wang J, Zhang J, Liu L. 2021. The distribution of Fusarium graminearum and Fusarium asiaticum causing fusarium head blight of wheat in relation to climate and cropping system. Plant Dis. 105:2830−2835.
- Yin LP, Ma GY 2004. Strengthen the firewall for entry quarantine of international postal parcels. International Business. 007. https://kns.cnki.net/kcms/detail/detail.aspx?FileName=GJSB200412270074&DbName=CCND2004.
- Zhang K, Su YY, Cai L. 2013. An optimized protocol of single spore isolation for fungi. Crypto Mycol. 34:349−356.
- Zhang ZF, Zhou SY, Eurwilaichitr L, Ingsriswang S, Raza M, Chen Q, Zhao P, Liu F, Cai L. 2020. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers. 106:29−136.
- Zhao P, Duan WJ, Liu F, Zhou X, Fan GM, Ma ZY, Cai L. 2021. Construction and application of reference databases for plant quarantine fungi in China. Acta Microbiol Sinica. 61:3806−3819.