ABSTRACT
This study aimed to identify important mycotoxigenic fungi and accurate detection of mycotoxin in stored maize grains using molecular methods. The current study also optimised the real-time PCR (RT-PCR) assay. The melting curve was established to identify isolated fungal species of Aspergillus (4), Fusarium (3), Penicillium (3), and Alternaria (one). A multiplex polymerase chain reaction (mPCR) technique was developed for the detection and characterisation of mycotoxin producing fungi, mycotoxin metabolic pathway genes, and the determination of eleven mycotoxins in stored maize grains using high-performance liquid chromatography (HPLC). The mPCR results indicated positive signals for potentially mycotoxigenic fungal species tested of Aspergillus, Fusarium, Penicillium, and Alternaria. A protocol for multiplex reverse transcription-polymerase chain reaction (mRT-PCR) was tested to distinguish between free and contaminated, stored maize with aflatoxin B1 (AFB1). The expression pattern of four aflatoxin biosynthetic pathway genes, AFB1 (aflQ, aflP, aflO, and aflD), was a good marker for contaminated, stored maize grains. HPLC analysis showed that maize grain samples were contaminated with mycotoxins, and the concentration was above the detection level. The results indicate that the polyphasic approach might provide a sensitive, rapid, and accurate method for detecting and identifying mycotoxigenic fungal species and mycotoxins in stored maize grains.
1. Introduction
The maize crop is a major agricultural product in human diets and one of the world’s most important sources of staple food and animal feed (Dorn et al. Citation2011; Choi et al. Citation2018). The maize crop is the most susceptible to mycotoxigenic fungi and mycotoxins contamination among cereals (Pleadin et al. Citation2013; Kirincic et al. Citation2015; Tima et al. Citation2016). Mycotoxins are the secondary metabolites biosynthesised by various fungal strains that significantly affect the quality of food products, thus causing severe problems for humans and animals (IARC Citation1993). Traditionally, mycotoxigenic fungal strains in affected crops were classified into two different classes: “field fungi”, which generally invade maize crops before harvest, such as Fusarium spp., and “storage fungi” or (saprophytic), which contaminate maize grains after harvest, like Aspergillus and Penicillium genera, and become a severe problem. However, pathogenic strains from Aspergillus flavus affected maize grains in the agricultural field and in stored grains (Miller Citation1995; Šimerda Citation1996). The most significant and economically relevant mycotoxigenic fungi and their mycotoxins of stored maize grains are Aspergillus spp. [aflatoxins (AF)], Fusarium spp. [fumonisins (FUM), trichothecenes (TCT), zearalenone (ZEA), deoxynivalenol (DON)], Penicillium spp. [ochratoxin A (OTA), patulin (PAT), and citrinin (CIT)], and Alternaria spp. [alternariol (AOH)] (Richard Citation2007). These mycotoxigenic fungi and mycotoxins must be detected, and identification is essential for assessing food quality and developing control strategies for food safety (Eskola et al. Citation2020). Morphologically based methods have long been employed to detect and identify mycotoxigenic fungi contamination in food and feed materials. These previous methods have several drawbacks, such as not being accurate, time-consuming, and less sensitive (Ramirez et al. Citation2009; Kim et al. Citation2020). Molecular diagnostics have been developed during the last two decades by real-time PCR (RT-PCR) to detect mycotoxigenic fungi belonging to the four major mycotoxin-producing genera, namely, Aspergillus (Sardiñas et al. Citation2011; Ghafori et al. Citation2021), Fusarium, Penicillium (Tannous et al. Citation2015; Susca et al. Citation2020), and Alternaria (Kordalewska et al. Citation2015), in various food and feed matrices. The detection and characterisation of mycotoxigenic fungus using multiplex PCR (mPCR), which targets the genes responsible for the biosynthesis of mycotoxins, is rapid, inexpensive, and reliable (Priyanka et al. Citation2015). Compared to monoplex PCR, mPCR allows for the simultaneous amplification of more than two genomic DNA regions in a single PCR reaction (Yli-Mattila et al. Citation2017). The mPCR assay with competitive interior amplification control to eliminate false, negative findings was developed by utilising specific primers for each of the fungi species and optimising and validating them using standard isolates (Priyanka et al. Citation2013; Neera and Murali Citation2021).
Chromatographic techniques for detecting and quantifying mycotoxins have been constantly developed and improved (Eshelli et al. Citation2018). The chromatographic method of analysis is widely used for the determination of mycotoxins from food sources (Sadhasivam et al. Citation2017). In the current work, we develop a sensitive and specific assay to detect eleven species belonging to mycotoxigenic fungi by RT-PCR and mPCR, mPCR assay to detect mycotoxin metabolic pathway genes simultaneously, high-performance liquid chromatography (HPLC) technique for detection and accurate analysis of eleven mycotoxins in stored maize grains obtained from storage warehouses in Riyadh, Saudi Arabia.
2. Materials and methods
2.1. Maize grain samples
During this study, samples were collected from six maize grain storage warehouses in Riyadh 6 months after harvest. We divided the warehouse into five sections, and from each section, we collected 30 samples (1,000 g) of maize grain stored for human consumption. The grain sample temperature and moisture contents were 25–33 °C, with a humidity range of 10.2%–13.5%. The obtained grains were packed in an airtight container and transported to the laboratory for further analysis. Aliquots of grains (100 g) were packed in containers and lyophilised. Samples were mechanically powdered and stored at −10 °C for further experiments.
2.2. DNA extraction
We used a modified version of the CTAB method (Murray and Thompson Citation1980). A maize grain (10 g) was ground with a laboratory blender. Standard and isolated mycotoxigenic fungi were grown for 48 h at 25 °C, peptone yeast glucose media (PYG) (Table S1). In berif, the maize grain powder (300 mg) or the lyophilised mycelium (200 mg) was pulverised in a mortar with a small amount of glass sand and transferred to a 50 mL centrifugation tube containing TES buffer, proteinase K, 5 mol/L NaCl, cetyltrimethylammonium bromide, chloroform-isoamyl alcohol and isopropanol. The lysis mixture was incubated at 45 °C for 45 min and mixed by turning the tubes every 10 min. The upper phase was transferred to another centrifugation tube and spun for 20 min at 4,000 g. The aqueous phase was transferred to a new tube containing isopropanol and mixed thoroughly. The supernatant was decanted and the pellet was rinsed with 70% (v/v) ethanol, dried, and dissolved in TE. The process took longer than the traditional method of extracting DNA. Undissolved material was removed by centrifugation. DNA was concentrated by ethanol 96% (v/v) precipitation and dissolved in TE. DNA was extracted using a cetyltrimethylammonium bromide (CTAB)-based protocol (Brandfass and Karlovsky Citation2008). The amount of extracted DNA from the fungal strains was assayed using the NanoDrop (ND-1000) spectrophotometer (NanoDrop Technologies, Wilmington, USA).
2.3. Real-time PCR of mycotoxigenic fungi
RT-PCR used specific primers to amplify target DNA to detect eleven species of mycotoxigenic fungi. RTi-PCR reactions were carried out using a PCR machine (real time analysis system, Rotor-Gene 6000, Qiagen, Hilden, Germany). All details of used primer sequences, conditions, and parameters of RTi-PCR were available in (Table S2, S5).
2.4. Multiplex PCR for mycotoxigenic fungi and genes of mycotoxin biosynthetic
Multiplex PCR analysis was utilised in this study to enable multiple detections of a broad spectrum of mycotoxigenic fungi in maize grains storage warehouses in Saudi Arabia. Species-specific primers were used for the determination of fungi in a single PCR reaction and the used primers were tabulated (Table S3, S4). The PCR reaction was optimised by analysing various concentrations of primers, annealing temperature, and the amount of template DNA. For multiplex PCR, various primers were tested, with different annealing temperature, and amplified product was finally verified. All mPCR parameters, conditions, and primers sequences are shown in Tables S3, S4, and S5. PCR products were electrophoresed with 1% (w/v) agarose containing ethidium bromide, using TAE buffer. The 1× TAE buffer was prepared with 1.0 mmol/L EDTA and 40 mmol/L Tris-acetate. A DNA marker (a 100 bp DNA ladder) and samples were run for 1 h at 96 V.
2.5. Multiplex PCR analysis to determine fungal strains from the contaminated grains
Multiplex PCR analysis was validated and assessed the presence of various fungal strains using a single PCR analysis from the artificially contaminated grains with fungal spores. The fungal spore suspensions (104, 105, 106 spores/g) of Alternaria spp.; Penicillium spp., Fusarium spp., and Aspergillus spp. were inoculated into 2 g sterilised maize grains. The control sample was free of spores. They were incubated for 1 week for mycelium growth, and the extracted DNA was used for the characterisation of fungal strains in an mPCR assay.
2.6. Preparation of mycotoxin standard solutions
Mycotoxin standards such as aflatoxins (AFs), fumonisins (FB1, FB2), trichothecenes (TCT), zearalenone (ZEA), ochratoxin A (OTA), patulin (PAT), alternariol (AOH) were prepared at 1 mg/mL concentration using HPLC grade methanol. These mycotoxin stock solutions were prepared at various concentrations within the working concentration range using methanol as a diluent. The diluted stock solutions were stored at – 20 °C for further analysis.
2.7. Mycotoxins assays
2.7.1. AFs
Detecting and determining AFs production were performed according to the method described previously (Christian Citation1990). Aflatoxins were extracted by blending maize grains (5 g) and extracted with methanol and water (80:20, v:v) for 2 min. and filtered through Whatman’s filter paper. The extracted solvent was removed by evaporation at 35 °C under vacuum conditions. The extracted aflatoxin was dried and dissolved in acetic acid, methanol, and water (20:20:60, v:v:v, respectively), and these solvent systems were used for the determination of aflatoxin. The flow rate was adjusted to 1 mL/min and the sample was run for 35 min.
2.7.2. Fusarium toxins
Fusarium toxins from the grains were extracted by blending 5 g of the ground sample with a solvent comprising sodium chloride (5 g), methanol (100 mL), and water (20 mL) at an 80:20 ratio. It was spun for 60 s and finally filtered using a micro-fibre filter (1 μm). Ten millilitres of the sample was mixed with wash buffer (40 mL) and filtered using a 1 μm filter. Assays for fusarium toxins, including fumonisin, trichothecenes, and zearalenone amounts, were performed on HPLC (Mazzani et al. Citation2001).
2.7.3. Penicillium toxins
Penicillium toxins were extracted by blending maize grains (5 g) with a 20 mL liquid mobile phase solution of a mixture of acetonitrile: water (5:95, v:v) for 2 min and then filtering through Whatman’s filter paper. The amount of Penicillium toxins, ochratoxin A, and patulin were determined as described earlier (Christian Citation1990).
2.7.4. Alternaria toxins
Alternaria toxin was extracted by blending ground maize grain (5 g) with 20 mL of methanol for 2 min and filtering through Whatman’s filter paper (No. 1). The filtrate was measured and further clarified using ammonium sulphate (60 mL, 20%) and the clear filtrate was obtained after repeated extraction with chloroform. For analysis using HPLC, the solvent phases were collected, pooled, and dried. It was dissolved in a minimum volume of methanol (2 mL) and a toxin assay was performed as described previously (Li et al. Citation2001).
3. Results
3.1. Specificity of the RT-PCR assays
The melting-curve analysis of RT-PCR (Figure S1) showed the presence of single peaks for 10 species, which are members of four potentially mycotoxigenic fungi genera, and were associated with maize grain samples. Aspergillus spp. (Figure S1a–d), Fusarium spp. (Figure S1e–g), Penicillium spp. (Figure S1h–j), and A. alternate (Figure S1k). Melting temperatures are used to detect potentially mycotoxigenic fungal genera found on stored maize grains (Table S1). Aspergillus spp., including A. flavus, A. niger, and A. ochraceus also, the Fusarium genus: F. proliferatum, F. oxysporum, and F. verticillioides. Penicillium spp. such as P. chrysogenum, P. expansum, P. oxalicum, and A. alternate were also identified. All earlier fungal species were present except A. parasiticus.
3.2. Standard curve
The linearity, efficiency, and the limit of quantification assay of detection were estimated from the standard curves with mycotoxigenic fungi (10 isolates) genomic DNA ranging from 0.001 to 1 ng (Figure S2). The cycle threshold (Ct) values from the standard curves showed a linear dynamic relationship between Ct values and mycotoxigenic fungi DNA concentration (ng). The lower detection limit (0.001 ng) of the mycotoxigenic fungi corresponds to a Ct range of 29 to 34 cycles, and the higher (1 ng) corresponds to a Ct range of 17 to 23 cycles.
The standard curve was established for slope, and R2 values ranging from −3.2657 to −3.5471 and 0.9573 to 0.9933, respectively, and the amplification efficiency [Efficiency, E (%)] was calculated ().
Table 1. The cycle threshold, E, slop, and R2 values of mycotoxigenic fungi based on the standard curve.
3.3. Multiplex PCR specificity and sensitivity
A total of four Aspergillus spp., three Fusarium spp., three Penicillium spp., and one Alternaria spp. were identified (Table S6). The selected primers showed district specificity and were used for the detection of fungal strains. A total of four Aspergillus spp., three Fusarium spp., three Penicillium spp., and one species of Alternaria spp. were characterised. The utilised primers amplified target food pathogens. Also, each primer displayed remarkable potential and enabled the detection of mycotoxin-producing fungi and molecular characterisation up to strain level. To validate the multiplex PCR approach, the genomic DNA of the common mycotoxin-producing standard fungi was amplified using specific primers (Figure S3). mPCR sensitivity was examined using maize grain powder inoculated with fungal spores of mycotoxin-producing organism. Using 103, 104 and 105 spores/g with 8, 16, 24, and 48 h, the results were depicted in Figure S4. Mycotoxin-producing fungal DNA was characterised after 8 h inoculation with 104 spores inoculums dose, providing an excellent tool in a short time for maize grain powder samples. On the other hand, the inoculums at 106 spores dosage improved target DNA detection after 48 h. In general, improved target DNA detected was achieved at increasing incubation time (Figure S4).
3.4. Multiplex PCR for mycotoxigenic fungi isolated from stored maize grain naturally contaminated
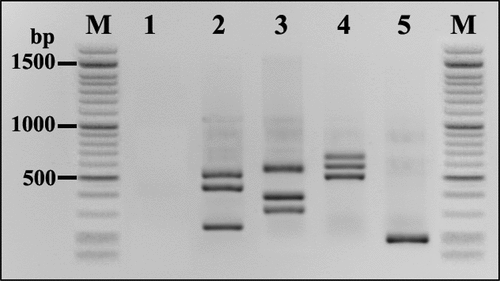
Amplification products were clearly visual, indicating the presence of four potentially mycotoxigenic fungi, A. flavus (500 bp), A. niger (245 bp) and A. ochraceus 430 bp (lane 2), F. oxysporum (340 bp), F. proliferatum (585 bp), and F. verticillioides (370 bp) (lane 3), P. chrysogenum (585 bp), P. expansum (533 bp) and P. oxalicum (611 bp) (lane 4), finally A. alternata (184 bp) (lane 5) ().
3.5. Distribution of mycotoxin-producing fungi in maize sample
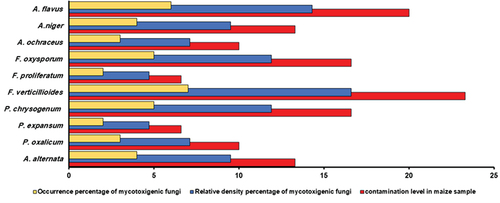
The 30 storage maize samples revealed the presence of ten mycotoxigenic fungi species: Aspergillus spp. included A. flavus (6 isolates), A. niger (4 isolates), A. ochraceus (4 isolates); Fusarium spp. included F. oxysporum (5 isolates), F. proliferatum (2 isolates), and F. verticillioides (7 isolates); Penicillium spp. had P. chrysogenum (5 isolates), P. expansum (2 isolates), P. oxalicum (3 isolates); and A. alternate (6 isolates) (Table S7). shows one example of detecting mycotoxigenic fungi in naturally contaminated, stored maize grain samples. Among the 30 stored maize grain samples, 11 exhibited positive signals for potentially mycotoxigenic fungi based on PCR of species-specific primers (Table S6). The results demonstrated the dominance of Fusarium spp. in all the samples analysed, with a percentage of 33.3%. Regarding F. verticillioides, the isolation percentage found was 16.6%, followed by F. oxysporum, at 11.9% of the total fungi. Aspergillus spp. came in second in terms of dominance, ahead of Penicillium spp. and Alternaria spp. The same trend was observed in the contamination level of storage maize samples ().
3.6. Analysis of genes involved in mycotoxin biosynthetic pathway
In this study, the amplification of mRT-PCR products indicated the presence of two aflatoxin producing A. flavus strains and the absence of aflatoxin-producing A. niger and A. ochraceus from the contaminated grains (). The expression patterns of four aflatoxins (AFs) genes (aflD, aflO, aflP, and aflQ) can be utilised to conclude the AF-synthesising ability of A. flavus isolates.
Figure 3. mPCR of stored maize grain. (a) Lane: M: 100 bp DNA ladder; Lanes: 1–2: Maize grain naturally contaminated by aflatoxigenic A. flavus, (using RNA, positive sample for four genes aflD, aflO, aflP, and aflQ), 3: Non-toxigenic isolates of A. flavus; 4: Non-toxigenic isolates of A. Niger; 5: Non-toxigenic isolates of A. ochraceus, and (b) Lane 1: Maize grain naturally contaminated by toxigenic F. verticillioides (using DNA, positive sample for two genes fum6, fum8); 2: Non-toxigenic F. verticillioides; 3–4: Non-toxigenic F. proliferatum; 5: Non-toxigenic F. oxysporum.
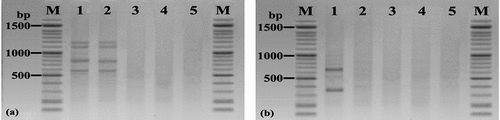
The mPCR protocol was validated based on fum6, and fum8 genes involved in the biosynthesis of fumonisin by screening 30 stored maize samples, illustrating one toxigenic F. verticillioides isolates and the absence of toxigenic species related to Fusarium spp. (). The general PCR and RT-PCR analysis results are described in . The gene expression was analysed by using four genes (aflD, aflO, aflP and aflQ) obtained from four aflatoxigenic A. flavus isolates (samples 11,12, 29, 30), two non-aflatoxigenic isolates. PCR analyses used for the detection fum6 and fum8 were essential genes for fumonisin showed five toxigenic F. verticillioides isolates (samples 8, 9, 11,13 and 14) and two non-toxigenic isolates. All species of Penicillium and Alternaria were non-toxigenic isolates.
Table 2. Detection of mycotoxin biosynthetic pathway genes by different PCR methods in stored maize grain samples.
3.7. Quantification of mycotoxin in stored maize grain
Using HPLC, aflatoxins AFB1 and AFG2 were detected in four analysed samples (11, 12, 29 and 30), at concentrations ranging from 17.93 to 1.18 µg/kg for AFB1 and 19.48 to 13.24 µg/kg for AFG2. Fumonisins FB1 and FB2 contaminated five stored maize samples (8,9,11,13,14) at levels ranging from 0.4 to 9.1 µg/kg. Sample No. 11 was contaminated with ABF1, AFG2, FB1, and FB2 (). An elevated level of AFB1 (>2 µg/kg) was detected in one post-harvest maize sample and was not suitable for consumption. On the other hand, FB1 and FB2 levels were below the EU regulatory limit of 200–4,000 µg/kg. Twenty-two samples of post-harvest maize were free from mycotoxins.
Molecular analysis of the maize sample No. 9 revealed high expression of genes, such as aflQ, aflP, aflO, and aflD ( lane 1), which support the aflatoxin (AF) biosynthetic pathway. PCR-analysis detected the presence of fum5 and fum6 genes revealed in Fusarium mycotoxin in the five maize samples (, lanes 1; samples 14). The results of the different PCR assays were found to be in good agreement with those discovered by HPLC analysis of 30 stored maize grain samples (). Overall, 8 maize samples tested were positive for PCR reaction and detection of mycotoxins, such as samples no. 8, 9, 11, 12, 29 and 30. All these samples tested positive for mycotoxin genes. All positive mycotoxins genes appeared in 30 stored maize grain samples () . 22 samples were free of mycotoxigenic fungi and mycotoxin. 30 stored maize grain samples were grouped into four groups based on genotype and chemotype profiles (). Four groups have been proposed: (1) Category I included three samples (10%) that exhibited a complete amplified pattern of four AFs genes related to AFB1 and AFG2 production and found A. flavus isolates; (2) Category II contained four samples (13.33%) that showed an amplified pattern of two fum6, fum8 genes related to FB1 and FB2 production and found F. verticillioides, F. proliferatum isolates; (3) Category III retained one sample (3.33%) that displayed an amplified pattern of four AFs genes and two fum6, fum8 genes related to AFB1, AFG2, FB1 and FB2 production and discovered A. flavus, F. verticillioides, and F. proliferatum, and finally; (4) Category IV included 22 samples (73.33%) that exhibited a negative pattern for all mycotoxins genes amplicons and non-producers of mycotoxins.
Table 3. Molecular characterisation and different mycotoxin contamination in stored maize grain samples.
Table 4. Molecular characterisation and aflatoxigenicity of 30 stored maize grain samples.
Table 5. Genotype and chemotype patterns of 30 stored maize grain samples based on mycotoxins genes and mycotoxins production.
4. Discussion
The contamination of stored maize grains by mycotoxigenic fungi is a serious concern where various reports are revealing the existence of these pathogenic fungi in many countries, such as China (Qin et al. Citation2020), Finland (Hietaniemi et al. Citation2016), Italy (Giorni et al. Citation2019), north-east Russia (Grünig et al. Citation2020), South Africa (Biemond et al. Citation2021), Spain (Luz et al. Citation2021), and United States (Lane et al. Citation2018).
In the field, several environmental factors play significant roles in mycotoxigenic fungi invasion, colonisation, growth, spread, development, distribution, frequency, and survival, and their subsequent accumulation of mycotoxins. These factors include temperature, relative humidity, and water activity (Richard et al. Citation2003; Smith et al. Citation2016). Further, stress factors, including mechanical injury, insect damage, high crop densities, weed competition, poor fertilisation, and drought, can reduce the natural defensive mechanisms of plants and, as a consequence, promote fungal colonisation and mycotoxin production (Awuchi et al. Citation2020).
Global warming and other climate change increase the risk of maize contamination by mycotoxigenic fungi in north-western Europe, which might change their geographic distribution and lead to a greater presence of the mycotoxins in new geographic areas (Miedaner and Juroszek Citation2021). The mycotoxins quantity produced will depend on environmental factors, chemical factors (oxygen, carbon dioxide, composition of substrate, fungicides, and pesticides), and biological factors such as genetic factors, susceptible plants, insects, and initial spore load (Bryden Citation2012). We think it is a complex matrix of many factors that lead to mycotoxigenic fungi growth and the production of mycotoxins. We need good agronomic, hygiene, storage, and manufacturing practices. The culture-based method is suitable for an initial overview of the entire fungal community, even if it does not provide information reliably covering all species. Furthermore, this method is strenuous, time-consuming, and requires qualified microbiologists with comprehensive knowledge of fungal taxonomy for lower-level species identification. Also, growth rates among fungal isolates are not the same; they are either prevalent, fast-growing or vigorously spread isolates (Bretträger et al. Citation2022).
To avoid the drawbacks of conventional detection strategies, molecular diagnostic techniques such as RT-PCR are recent technologies that have been used in the detection of mycotoxigenic fungi by amplification of a specific DNA sequence in the fungal genome and are a rapid, accurate, and specific alternative to conventional detection methods.
In this study, particular protocols were used to detect A. flavus, A. parasiticus, A. ochraceus, A. niger, F. oxysporum, F. proliferatum, F. verticillioides, and P. chrysogenum, P. expansum, P. oxalicum and A. alternata contamination stored in maize grains by RT-PCR. Optimising a RT-PCR analysis is crucial, when using SYBR green dye. Our protocols have excellent efficiency (94%), and the maximum R2 coefficient is considered a potential indicator of the robustness of our protocols.
The power of RT-PCR for the past two decades has substantially improved the specificity and sensitivity of the characterisation of mycotoxigenic fungi (Tsang et al. Citation2018). Detection of mycotoxin-producing fungi in an early stage is crucial for controlling mycotoxins from entering the food chain. So far, a significant RTi-PCR has proven to be a valuable tool for improving the diagnosis of Aspergillus spp. (Sardiñas et al. Citation2011; Ghafori et al. Citation2021), Fusarium spp. (Jiménez-Fernández et al. Citation2010), Penicillium spp. (Lignell et al. Citation2008; Sardiñas et al. Citation2011) and Alternaria spp. (Kordalewska et al. Citation2015).
The ITS regions have been recommended as the main fungal barcode by the International Fungal Barcoding Consortium in 2012 (Schoch et al. Citation2012). Moreover, the ITS regions do not helpful for the detection of fungi from the genus Fusarium, and this leads to the analysis of EF1-α genes for DNA barcode analysis (Al-Hatmi et al. Citation2016). In this study, we developed a multiplex PCR method to analyse potential mycotoxigenic fungi belonging to various genera. Also, the mycotoxigenic isolates of Alternaria spp., Penicillium spp., Fusarium spp., and Aspergillus spp. demonstrated amplification during mPCR analysis and showed positive results for all fungal genera based on the eleven-primer pairs cocktail. The developed mPCR assay is accurate, rapid, easy, and cost-effective and could be a valuable alternative to traditional methods for (1) very early detection, (2) rapid screening tool, and (3) high-level sensitivity and specificity for mycotoxin-producing fungi in contaminated grains. Mycotoxigenic fungi are a severe threat to food and feed safety and security. It is urgently needed to be sure that food and feed matrices are safe and clear of mycotoxigenic fungi (Priyanka et al. Citation2013; Rahman et al. Citation2020).
The selected primer pair revealed the remarkable potential and enabled mycotoxin-in-producing fungal pathogens to detect and identify the strains. By analysing unique specific primer pairs, the PCR-based method was useful for detecting mycotoxins and fungi (Gu et al. Citation2017). It has been previously reported that the ITS regions of A. flavus and A. ochraceus rDNA were highly sensitive than other methods (Gil-Serna et al. Citation2009). Species-specific primers have been used for the determination of A. niger and the traditional method using ITS region amplifications (Mule et al. Citation2006; Gil-Serna et al. Citation2009). The ribosomal ITS1, ITS2 and a portion of the calmodulin gene were used to design a species-specific primer for F. proliferatum. These pairs of primers produced PCR products of 585 bp for these fungi. The developed PCR assay should supply a powerful tool for detecting F. proliferatum in maize grains (Mule et al. Citation2004).
In this study, DNA was used from A. flavus isolates as a template for mRT-PCR, revealing DNA failed to produce any correlation between the aflatoxin gene and expression. For this reason, we depend on RNA to detect aflatoxigenic A. flavus. Four A. flavus isolates successfully generated positive correlations on genes such as aflD, aflO, aflP and aflQ. Fifteen aflatoxigenic isolates of A. flavus were obtained from dairy feeds in Zimbabwe. The five targeted AF cluster genes aflD, aflR, aflS, aflM, and aflP were used for examining these isolates in this study. The five genes were present as follows: aflD (100%) and aflS (100%), aflR (66.6%), aflM (60%), and aflP (80%). Two genes, aflD and aflS, were significantly associated with aflatoxigenic A. flavus isolates (Nleya et al. Citation2021).
In the mPCR-based molecular bioassay, various primers were mixed in a single reaction for the determination of various genes involved in the mycotoxin biosynthesis pathway (Rahman et al. Citation2020). The occurrence of mycotoxin contamination in stored grains of wheat was assessed by mRT-PCR. Specific primers were used in multiplex PCR to detect aflatoxins genes, Fusarium toxin genes, and Penicillium toxin genes. These specific primers are useful for the determination of aflatoxin and fumonisin produced by A. flavus and F. verticillioides in wheat grains (Sadhasivam et al. Citation2017).
In this study, mPCR assay was used for the determination of aflatoxin, fumonisin, trichothecene, zearalenone, and ochratoxin A synthesised Aspergillus spp., Fusarium spp. and Penicillium spp. (Priyanka et al. Citation2013). Sixteen AFB1-producing fungal isolates have been isolated from peanut kernels and produced AFB1-producing genes were determined and differentiated non-aflatoxigenic isolates and aflatoxigenic strains (Mahmoud Citation2015). Aflatoxin-producing Aspergillus flavus was determined using the analysis of aflO and aflQ genes (Jamali et al. Citation2013). Five genes including, aflR, aflS, aflD, aflO, and aflQ were determined from various fungal strains. This protocol shows a good correlation between the AF genes’ expression patterns, analysed by mRT-PCR, and aflatoxin production (Degola et al. Citation2007, Citation2009). HPLC method has been used for the determination of various toxins from feed and food sources (Pascale Citation2009; Al-Wadai et al. Citation2013; Mahmoud Citation2015; ARM et al. Citation2021). AFB1 production has been used for the determination of pathogenic A. flavus (Al-Wadai et al. Citation2013).
5. Conclusions
Among the 30 stored maize grain samples, 11 exhibited positive signals for potentially mycotoxigenic fungi based on PCR of species-specific primers. The test samples revealed ten species from four different fungal genera. Aspergillus spp. included A. flavus (6 isolates), A. niger (4 isolates), A. ochraceus (4 isolates), Fusarium spp. including F. oxysporum (5 isolates), F. proliferatum (2 isolates), and F. verticillioides (7 isolates). Also, Penicillium spp. were P. chrysogenum (5 isolates), P. expansum (2 isolates), P. oxalicum (3 isolates), and A. alternate (6 isolates). All previous isolates were detected by RT-PCR and mPCR containing various species-specific primers. The amplification of mRT-PCR products indicated the presence of two aflatoxigenic A. flavus isolates and the absence of aflatoxigenic A. niger and A. ochraceus. The mPCR protocol based on two genes of fumonisin (fum6, fum8) has been verified by displaying only one toxigenic F. verticillioides isolate and the absence of toxigenic species related to Fusarium spp. Penicillium spp. and Alternaria spp. isolates were tested for toxigenicity by an mPCR assay, and the results revealed that all isolates were toxigenic. Thirty stored maize samples were analysed for 11 different mycotoxins using an HPLC method. The results revealed that eight contaminated stored grain samples were contaminated with AFB1, AFG2, FB1, and FB2.
Supplemental Material
Download MS Word (2.2 MB)Acknowledgements
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (No. PNURSP2023R84), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21501203.2023.2213704.
Additional information
Funding
References
- Al-Hatmi AM, Van Den Ende AG, Stielow JB, Van Diepeningen AD, Seifert KA, McCormick W, Assabgui R, Gräfenhan T, De Hoog GS, Levesque CA. 2016. Evaluation of two novel barcodes for species recognition of opportunistic pathogens in Fusarium. Fungal Biol. 1202(2):231–245. doi:10.1016/j.funbio.2015.08.006.
- Al-Wadai AS, Al-Othman MR, Mahmoud MA. 2013. Molecular characterization of Aspergillus flavus and aflatoxin contamination of wheat grains from Saudi Arabia. Genet Mol Res. 12:3335–3352.
- ARM AE-A, Shehata SHM, Hisham SM, Alobathani AA. 2021. Molecular profile of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus flavus isolated from stored maize. Saudi J Biol Sci. 28(2):1383–1391. doi:10.1016/j.sjbs.2020.11.073.
- Awuchi CG, Ukpe AE, Asoegwu CR, Uyo CN, Ngoka KE. 2020. Environmental impacts of food and agricultural production: a systematic review. Eur Acad Res. 8:1120–1135.
- Biemond PC, Stomph TJ, Kumar PL, Struik PC. 2021. How maize seed systems can contribute to the control of mycotoxigenic fungal infection: a perspective. Agronomy. 11(11):2168. doi:10.3390/agronomy11112168.
- Brandfass C, Karlovsky P. 2008. Upscaled CTAB-based DNA extraction and real-time PCR assays for Fusarium culmorum and F. graminearum DNA in plant material with reduced sampling error. Int J Mol Sci. 9(11):2306–2321. doi:10.3390/ijms9112306.
- Bretträger M, Becker T, Gastl M. 2022. Screening of mycotoxigenic fungi in barley and barley malt (Hordeum vulgare L.) using Real-Time PCR-a comparison between molecular diagnostic and culture technique. Foods. 11(8):1149–1161. doi:10.3390/foods11081149.
- Bryden WL. 2012. Mycotoxin contamination of the feed supply chain: implications for animal productivity and feed security. Anim Feed Sci Technol. 173(1–2):134–158. doi:10.1016/j.anifeedsci.2011.12.014.
- Choi JH, Lee S, Nah JY, Kim HK, Paek JS, Lee S, Ham H, Hong SK, Yun SH, Lee T. 2018. Species composition of and fumonisin production by the Fusarium fujikuroi species complex isolated from Korean cereals. Int J Food Microbiol. 267:62–69. doi:10.1016/j.ijfoodmicro.2017.12.006.
- Christian G. 1990. HPLC tips and tricks. Oxford (London): Great Britain at the Iden Press; p. 608.
- Degola F, Berni E, Dall’Asta C, Spotti ER, Marchelli I, Ferrero C, FM R. 2007. A multiplex RT-PCR approach to detect aflatoxigenic strains of Aspergillus flavus. J Appl Microbiol. 103(2):409–417. doi:10.1111/j.1365-2672.2006.03256.x.
- Degola F, Berni E, Spotti E, Ferrero I, Restivo FM. 2009. Facing the problem of “false positives”: re-assessment and improvement of a multiplex RT-PCR procedure for the diagnosis of A. flavus mycotoxin producers. Int J Food Microbiol. 129(3):300–305. doi:10.1016/j.ijfoodmicro.2008.12.016.
- Dorn B, Forrer HR, Jenny E, Wettstein FE, Bucheli TD, Vogelgsang S. 2011. Fusarium species complex and mycotoxins in grain maize from maize hybrid trials and from grower’s fields. J Appl Microbiol. 111:693–706.
- Eshelli M, Qader M, Jambi E, Hursthouse A, Rateb K. 2018. Current status and future opportunities of omics tools in mycotoxin research. Toxins. 10(11):433–445. doi:10.3390/toxins10110433.
- Eskola M, Kos G, Elliott CT, Hajšlová J, Mayar S, Krska R. 2020. Worldwide contamination of food-crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25. Crit Rev Food Sci Nutr. 60:2773–2789.
- Faria CB, Abe CAL, da Silva CN, Tessmann DJ, Barbosa-Tessmann IP. 2012. New PCR assays for the identification of Fusarium verticillioides, Fusarium subglutinans, and other species of the Gibberella fujikuroi complex. Int J Mol Sci. 13(1):115–132. doi:10.3390/ijms13010115.
- Ghafori S, Habibipour R, Bayat S. 2021. Optimization of a real-time PCR assay for identification of Aspergillus fumigatus and Aspergillus Niger from flour samples: comparison of phenotypic and genotypic methods. Gene Reports. 22:100993–101005.
- Gil-Serna J, Vázquez C, Sardiñas N, González-Jaén MT, Patiño B. 2009. Discrimination of the main Ochratoxin A-producing species in Aspergillus section Circumdati by specific PCR assays. Int J Food Microbiol. 136(1):83–87. doi:10.1016/j.ijfoodmicro.2009.09.018.
- Giorni P, Bertuzzi T, Battilani P. 2019. Impact of Fungi Co-occurrence on Mycotoxin Contamination in Maize During the Growing Season. null. 10:1265. doi:10.3389/fmicb.2019.01265.
- Gonzalez-Salgado A, Gonzalez-Jaen T, Vazquez C, Patino B. 2008. Highly sensitive PCR-based detection method specific for Aspergillus flavus in wheat flour. Food Addit Contam Part. 25(6):758–764. doi:10.1080/02652030701765715.
- Grünig M, Mazzi D, Calanca P, Karger DN, Pellissier L. 2020. Crop and forest pest metawebs shift towards increased linkage and suitability overlap under climate change. Commun Biol. 3:233–249.
- Gu Q, Yang ZH, Zhao DM, Zhang D, Wang Q, Ma LS, Zhu JH. 2017. Development of a semi-nested PCR-based method for specific and rapid detection of Alternaria solani causing potato early blight in soil. Curr Microbiol. 74(9):1083–1088. doi:10.1007/s00284-017-1284-0.
- Hietaniemi V, Sari R, Yli-Mattila Peltonen T, Sari JM, Kartio M, Sieviläinen E, Koivisto T, Parikka P. 2016. Updated survey of the Fusarium species and toxins in Finnish cereal grains. Food Addit Contam Part A. 33(5):831–848.
- IARC. Some naturally occurring aromatic amines. Mycotoxins; International Agency for Research on Cancer: Lyon (France), 1993.
- Jamali M, Karimipour M, Shams-Ghahfarokhi M, Amani A, Razzaghi-Abyaneh M. 2013. Expression of aflatoxin genes aflO (omtB) and aflQ (ordA) differentiates levels of aflatoxin production by Aspergillus flavus strains from soils of pistachio orchards. Res Microbiol. 164(4):293–299. doi:10.1016/j.resmic.2012.12.008.
- Jiménez-Fernández D, Montes-Borregob M, Navas-Cortésb JA, Jiménez-Díaz RM, Landa BB. 2010. Identification and quantification of Fusarium oxysporum in planta and soil by means of an improved specific and quantitative PCR assay. Appl Soil Ecol. 46:372–382.
- Kim WB, Park C, Cho SY, Chun HS, Lee DG, Kalendar R. 2020. Development of multiplex real-time PCR for rapid identification and quantitative analysis of Aspergillus species. PLoS One. 15(3):e0229561. doi:10.1371/journal.pone.0229561.
- Kirincic S, Škrjanc B, Kos N, Kozolc B, Pirnat N, Tavˇcar-Kalcher G. 2015. Mycotoxins in cereals and cereal products in Slovenia-Official control of foods in the years 2008–2012. Food Control. 50:157–165. doi:10.1016/j.foodcont.2014.08.034.
- Kordalewska M, Brillowska-Dąbrowska A, Jagielski T, Dworecka-Kaszak B. 2015. PCR and real-time PCR assays to detect fungi of Alternaria alternata species. Acta Biochim Pol. 62(4):707–712. doi:10.18388/abp.2015_1112.
- Lane B, Sharma S, Niu C, Maina AW, Wagacha JM, Bluhm BH, Woloshuk CP. 2018. Changes in the fungal microbiome of maize during hermetic storage in the United States and Kenya. Front Microbiol. 9:2336–2351. doi:10.3389/fmicb.2018.02336.
- Lignell U, Meklin T, Rintala H, Hyvarinen A, Vepsalainen A, Pekkanen J, Nevalainen A. 2008. Evaluation of quantitative PCR and culture methods for detection of house dust fungi and streptomycetes in relation to moisture damage of the house. Lett Appl Microbiol. 47(4):303–308. doi:10.1111/j.1472-765X.2008.02431.x.
- Li F, Toyazaki N, Yoshizawa T. 2001. Production of Alternaria Mycotoxins by Alternaria alternata Isolated from Weather-Damaged Wheat. J Food Prot. 64(4):567–571. doi:10.4315/0362-028X-64.4.567.
- Luque MI, Andrade MJ, Rodríguez A. 2013. Development of a multiplex PCR method for the detection of patulin-, ochratoxin A- and aflatoxin-producing moulds in foods. Food Anal Methods. 6(4):1113–1121. doi:10.1007/s12161-012-9516-1.
- Luz C, Carbonell R, Quiles JM, Torrijos R, de Melo Nazareth T, Mañes J, Meca G. 2021. Antifungal activity of peracetic acid against toxigenic fungal contaminants of maize and barley at the postharvest stage. Lwt. 148:111754.
- Mahmoud MA. 2015. Detection of Aspergillus flavus in stored peanuts using Real-Time PCR and the expression of aflatoxin genes in toxigenic and atoxigenic A. flavus isolates. Foodborne Pathog Dis. 12(4):289–296. doi:10.1089/fpd.2014.1854.
- Mazzani C, Borges O, Luzon O, Barrientos V, Quijada P. 2001. Occurrence of Fusarium moniliforme and fumonisins in kernels of maize hybrids in Venezuela. Braz J Microbiol. 32(4):345–349. doi:10.1590/S1517-83822001000400018.
- Miedaner T, Juroszek P. 2021. Global warming and increasing maize cultivation demand comprehensive efforts in disease and insect resistance breeding in north-Western Europe. Plant Pathol. 70:1032–1046.
- Miller JD. 1995. Fungi and mycotoxins in grain-implications for stored-product research. J Stored Prod Res. 31:1–16.
- Mule G, Susca A, Logrieco A, Stea G, Visconti A. 2006. Development of quantitative real-time PCR assay for the detection of Aspergillus carbonarius in grapes. Int J Food Microbiol. 111:S28–S34.
- Mule G, Susca A, Stea G, Moretti A. 2004. A species-specific PCR assay based on the calmodulin partial gene for identification of Fusarium verticillioides, F. proliferatum and F. subglutinans. Eur J Plant Pathol. 110:495–502.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325.
- Neera A, Murali HS. 2021. Development and evaluation of multiplex PCR for detection of T-2 and zearalenone producing Fusarium spp. Lett Appl Microbiol. 73:363–371.
- Nguyen HN, Nguyen TMT, Nguyen TH. 2015. Developing a PCR assay to detect Aspergillus parasiticus. Sci Technol Dev. 18(T1):43–49.
- Nleya N, Ngoma L, Adetunji MC, Mwanza M. 2021. Biodiversity of aflatoxigenic Aspergillus species in dairy feeds in Bulawayo, Zimbabwe. Front Microbiol. 11:599605.
- Nutz S, Döll K, Karlovsky P. 2011. Determination of the LOQ in real-time PCR by receiver operating characteristic curve analysis: application to qPCR assays for Fusarium verticillioides and F. proliferatum. Anal Bioanal Chem. 401:717–726.
- Pascale M. 2009. Detection methods for mycotoxins in cereal grains and cereal products. Proc Nat Sci Matica Srpska Novi Sad. 117:15–25.
- Pleadin J, Vahcic N, Perši N, Ševelj D, Markov K, Frece J. 2013. Fusarium mycotoxins’ occurrence in cereals harvested from Croatian fields. Food Control. 32:49–54.
- Prashant K, Mishra A, Roland TVF, Alastair C. 2003. Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusaria. FEMS Microbiol Lett. 218:329–332.
- Priyanka SR, Venkataramana M, Balakrishna K, Murali HS, Batra HV. 2013. Development and evaluation of a multiplex PCR assay for simultaneous detection of major mycotoxigenic fungi from cereals. J Food Sci Technol. 52:486–492.
- Priyanka SR, Venkataramana M, Balakrishna K, Murali HS, Batra HV. 2015. Development and evaluation of a multiplex PCR assay for simultaneous detection of major mycotoxigenic fungi from cereals. J Food Sci Technol. 52:486–492.
- Qin PW, Xu J, Jiang Y, Hu L, van der Lee T, Waalwijk C, Zhang WM, Xu XD. 2020. Survey for toxigenic Fusarium species on maize kernels in China. World Mycotoxin J. 13:213–223.
- Rahman HU, Xiaofeng Y, Xianfeng R, Wen Z, Qi Z, Peiwu L. 2020. Multiplex PCR assay to detect Aspergillus, Penicillium and Fusarium species simultaneously. Food Addit Contam Part A. 37:1939–1950.
- Ramirez M, Castro C, Palomares JC, Torres MJ, Aller AI, Ruiz M, Aznar J, Martin-Mazuelos E. 2009. Molecular detection and identification of Aspergillus spp. from Clinical Samples Using real-time PCR. Mycoses. 52:129–134.
- Richard JL. 2007. Some major mycotoxins and their mycotoxicoses-An overview. Int J Food Microbiol. 119:3–10.
- Richard JL, Payne GA, Desjardins AE. 2003. Mycotoxins: risks in plant, animal and human systems. CAST Task Force Rep. 139:101–103.
- Sadhasivam S, Britzi M, Zakin V, Kostyukovsky M, Trostanetsky A, Quinn E, Sionov E. 2017. Rapid detection and identification of mycotoxigenic fungi and mycotoxins in stored wheat grain. Toxins. 9:302.
- Sardiñas N, Vázquez C, Gil-Serna J, González-Jaén MT, Patiño B. 2011. Specific detection and quantification of Aspergillus flavus and Aspergillus parasiticus in wheat flour by SYBR Green quantitative PCR. Int J Food Microbiol. 145:121–125.
- Scherm B, Palomba M, Serra D, Marcello A, Migheli Q. 2005. Detection of transcripts of the aflatoxin genes aflD, aflO, and aflP by reverse-transcription polymerase chain reaction allows differentiation of aflatoxin-producing isolates of Aspergillus flavus and Aspergillus parasiticus. Int J Food Microbiol. 98:201–210.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci. 109:6241–6246.
- Šimerda B. Moulds and mycotoxins. Náš. chov. 1996, 8, Czech journal. 18–19.
- Smith MC, Madec S, Coton E. 2016. Natural co-occurrence of mycotoxins in foods and feeds and their in vitro combined toxicological effects. Toxins. 8:94–106.
- Susca A, Stea G, Mule G, Perrone G. 2007. Polymerase chain reaction (PCR) identification of Aspergillus Niger and Aspergillus tubingensis based on the calmodulin gene. Food Addit Contam Part. 24:1154–1160.
- Susca A, Villani A, Moretti A, Stea G, Logrieco A. 2020. Identification of toxigenic fungal species associated with maize ear rot: calmodulin as single informative gene. Int J Food Microbiol. 319:108491–108504.
- Tannous J, Atoui A, El Khoury A, Kantar S, Chdid N, Oswald IP, Puel O, Lteif R. 2015. Development of a real-time PCR assay for Penicillium expansum quantification and patulin estimation in apples. Food Microbiol. 50:28–37.
- Tima H, Brückner A, Mohácsi-Farkas C, Kiskó G. 2016. Fusarium mycotoxins in cereals harvested from Hungarian fields. Food Addit Contam Part B. 9:127–131.
- Tsang CC, Tang JY, Lau SK, Woo PC. 2018. Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era–past, present and future. Comput Struct Biotechnol J. 16:197–210.
- Yli-Mattila T, Nayaka SC, Venkataramana M, Yörük E. 2017. Multiplex detection of Fusarium species. In: Dr WJ, editor. Mycotoxigenic fungi: methods and protocols. New York (NY): Springer; p. 269–291.