ABSTRACT
Candida albicans is able to switch between two epigenetic cell types, namely white and opaque. Multiple conserved signalling pathways control the switch between white and opaque cell types in response to environmental changes. Here, we report the regulatory roles of the endosomal Rab family GTPase Vps21 and associated key components of the Vps21 signalling pathway in white-opaque switching and mating in C. albicans. Deletion of VPS21 promoted a switch from the white to the opaque phenotype in the presence of N-acetyl-glucosamine (GlcNAc). Consistently, inactivation of the guanine nucleotide exchange factor of Vps21 (Vps9) and downstream components in the Vps21 pathway (Vps3, Vac1, and Pep12) had similar promoting effects on phenotypic switching. The mating efficiency of opaque cells is much higher than that of white cells under standard laboratory culture conditions. However, compared to the wildtype strain, the vps21/vps21, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains exhibited dramatically reduced mating efficiencies. Quantitative RT-PCR assays demonstrated that inactivation of the Vps21 signalling pathway led to downregulation of pheromone expression and mating response pathway associated genes. Taken together, our findings indicate that the conserved Vps21 signalling pathway plays critical roles in the regulation of cell-type switching and mating in C. albicans.
1. Introduction
The opportunistic yeast pathogen Candida albicans can cause cutaneous diseases as well as life-threatening systemic infections in humans (Brown et al. Citation2012). An important characteristic of C. albicans is its ability to undergo morphological transitions between several phenotypes (Whiteway and Bachewich Citation2007; Huang Citation2012). This ability is associated with several biological processes including its rapid adaptation to environmental changes, virulence, and sexual mating (Soll Citation2009; Sudbery Citation2011; Huang Citation2012; Wang et al. Citation2023). For example, opaque cells are better at colonising superficial niches, whereas white cells are more virulent in systemic infection models (Soll Citation2009; Sudbery Citation2011; Huang Citation2012).
The bistable and heritable white-opaque switch is a typical phenotypic switching system in C. albicans (Slutsky et al. Citation1987; Soll Citation2009; Huang Citation2012). White and opaque cells exhibit significant differences in colony and cellular appearances, virulence, and mating competency (Slutsky et al. Citation1987; Miller and Johnson Citation2002; Soll Citation2009; Huang Citation2012). Under standard laboratory culture conditions, opaque cells mate one million times more efficiently than white cells (Miller and Johnson Citation2002). However, we recently reported that C. albicans white cells can mate efficiently under glucose starvation conditions (Guan et al. Citation2023). These findings provide new insights into the adaptive capabilities of C. albicans and its potential mating strategies in various environmental contexts.
Many environmental cues and signalling pathways are involved in the regulation of white-opaque switching and mating in C. albicans (Slutsky et al. Citation1987; Soll Citation2009; Huang Citation2012). For example, high levels of CO2, acidic pH, and N-acetyl-glucosamine (GlcNAc) promote white-to-opaque switching, while high temperatures and basic pH favour the white cell development (Huang et al. Citation2009, Citation2010; Lohse and Johnson Citation2009; Soll Citation2009; Huang Citation2012; Sun et al. Citation2015). Multiple signalling pathways are involved in the regulation of this environmental cues-induced phenotypic switch. For example, the cAMP-dependent PKA signalling pathway, the Ste11-Hst7-Cek1/Cek2-mediated MAPK cascade pathway, and the osmotic sensing pathway play critical roles in the regulation of white-opaque switching and mating in C. albicans (Huang et al. Citation2010; Ramirez-Zavala et al. Citation2013; Liang et al. Citation2014). In activation of the cAMP-dependent PKA signalling pathway reduced the efficiency of white-to-opaque switching, while overexpression of the key components of the Ste11-Hst7-Cek1/Cek2-mediated MAPK pathway promotes the development of opaque cells (Huang et al. Citation2010; Ramirez-Zavala et al. Citation2013; Liang et al. Citation2014).
The endosomal GTPase Vps21-mediated signalling pathway is conserved in fungi and involved in the regulation of a number of important biological processes including vacuolar biogenesis and trafficking, autophagy, and morphological transitions (Rieder et al. Citation1996; Gerrard et al. Citation2000; Palmer et al. Citation2003; Chen et al. Citation2014). Vps9 is a guanine nucleotide exchange factor (GEF) for Vps21 and facilitates the exchange of GTP for GDP on the guanine nucleotide binding site, leading to GTPase activation (Johnston et al. Citation2013; Chen et al. Citation2014; Borchers et al. Citation2021). The CORVET tethering complex components Vps3, Vac1, and Pep12 (the soluble N-ethylmaleimide-sensitive factor attachment protein receptor, t-SNARE) are major downstream factors in the Vps21 signalling pathway (Johnston et al. Citation2013; Chen et al. Citation2014; Borchers et al. Citation2021). Large vacuoles are commonly observed in C. albicans opaque cells and shmoos, the elongated morphology yeast cells adopt during mating (Anderson et al. Citation1989; Liang et al. Citation2020). In this study, we investigated whether the Vps21 signalling pathway plays a role in the regulation of white-opaque switching and mating in C. albicans. Our findings show that inactivation of the Vps21 signalling pathway promotes the white-to-opaque switch but represses mating in C. albicans. Furthermore, disruption of the Vps21 signalling pathway leads to downregulation of mating-associated gene expression, which could directly affect the mating response in C. albicans.
2. Materials and methods
2.1. Strains and culture conditions
YPD medium (1% yeast extract, 2% peptone, 2% glucose; 2% agar added for solid medium, w/v) was used for regular growth of C. albicans cells. Modified Lee’s glucose (pH 6.8) and Lee’s GlcNAc (pH 6.8) media containing phloxine B (5 μg/mL) were used for white-opaque switching and quantitative mating assays (Huang et al. Citation2010; Li et al. Citation2023). Synthetic complete [SC, 0.67% yeast nitrogen base with ammonium sulphate and without amino acids (YNB), 2% glucose, and corresponding amino acids] medium was used for selective growth in quantitative mating assays. The strains used in this study are listed in supplementary Table S1.
2.2. Plasmid and strain construction
To delete VPS21, VPS9, VPS3, VAC1, or PEP12 in C. albicans, the fusion PCR recombination strategy was used as described by the Noble group (Noble and Johnson Citation2005). To delete the first copy of each gene, a long fusion PCR fragment of CdHIS1 flanked by ~400 bp upstream and downstream sequences of the target gene was transformed into strain SN152 [MTLa/α, a derivative of SC5314 (Noble and Johnson Citation2005)], generating a heterozygous mutant. A fusion PCR fragment of CmLEU2 or CaARG4 flanked by ~400 bp upstream and downstream sequences of the target gene was then transformed into a heterozygous mutant, generating a homozygous mutant. For white-opaque switching and mating assays, the deletion mutants of VPS21, VPS9, VPS3, VAC1, and PEP12 were converted to MTLa/Δ or MTLΔ/α strains by deleting one allele of the MTL locus. The linearising plasmid L23.14 [a pSFS2A-based MTL locus deletion plasmid (Park and Morschhauser Citation2005; Xie et al. Citation2013)] was used for deletion of the MTL locus.
The method for construction of the reconstituted strains was used as described previously (Li et al. Citation2023). To construct the VPS21 complementation plasmid, a fragment of VPS21-3’-UTR (untranslated region) was amplified from genomic DNA of strain SC5314, digested with two restriction enzymes, XhoI (SalI isocaudamer) and BglII, and subcloned into the SalI/BgIII sites of plasmid pNIM1, generating pNIM1-VPS21-3’-UTR. Then, a fragment containing VPS21-5’-UTR and its ORF was amplified from genomic DNA of strain SC5314, digested with two restriction enzymes, SalI and BamHI (BgIII isocaudamer), and subcloned into the SalI/BgIII sites of plasmid pNIM1-VPS21-3’-UTR, generating the complementation plasmid VPS21p-VPS21. To construct the VPS21Q69L or VPS21S24N complementation plasmid, a fragment containing VPS21Q69L or VPS21S24N-5’-UTR and ORF was obtained by site-directed mutagenesis (Johnston et al. Citation2013) and subcloned into the SalI/BamHI sites of plasmid pNIM1-VPS21-3’-UTR, generating the complementation plasmid VPS21p-VPS21Q69L or VPS21p-VPS21S24N. Similarly, we constructed the complementation plasmid for VPS3 called VPS3p-VPS3 using the same strategy. To construct the VPS9 complementation plasmid, a fragment of VPS9-3’-UTR was amplified and subcloned into the EcoRV/HindIII sites of the plasmid BES116 (Feng et al. Citation1999). The caSAT1 gene (nourseothricin resistance marker adapted for C. albicans) was amplified from the plasmid pSFS2A (Reuss et al. Citation2004) and inserted into the KpnI site of the plasmid BES116-VPS9-3’-UTR. Then, the fragment containing the VPS9-5’-UTR and ORF was amplified from C. albicans genomic DNA and subcloned into the HindIII/KpnI sites of the plasmid BES116-VPS9-3’-UTR-SAT1, generating the complementation plasmid VPS9p-VPS9. To construct the VAC1 complementation plasmid, a fragment of VAC1-3’-UTR was amplified and subcloned into the KpnI/EcoRV site of the plasmid BES116. The fragment containing VAC1-5’-UTR and its ORF was amplified from C. albicans genomic DNA and subcloned into the EcoRV/PstI site of the plasmid BES116-VAC1-3’-UTR. Then, the selectable marker caSAT1 was inserted into the PstI site of the plasmid BES116-VAC1-5’-UTR-ORF-3’-UTR, generating the complementation plasmid VAC1p-VAC1. To construct the PEP12 complementation plasmid, a fragment of PEP12-3’-UTR was amplified and subcloned into the KpnI/HindIII site of the plasmid BES116. The fragment containing PEP12-5’-UTR and its ORF were amplified from genomic DNA and subcloned into the HindIII/PstI sites of the plasmid BES116-PEP12-3’-UTR. Then, the selectable marker caSAT1 was inserted into the PstI site of the plasmid BES116-PEP12-5’-UTR-ORF-3’-UTR, generating the complementation plasmid PEP12p-PEP12. Finally, the constructed complementation plasmids were linearised with restriction enzymes (SalI for pNIM1-based plasmids, HindIII for the plasmid VPS9p-VPS9 and PEP12p-PEP12, and EcoRV for plasmid VAC1p-VAC1) and transformed into the corresponding mutant strains, generating the reconstituted complemented strains (vps21/vps21 + VPS21, vps21/vps21 + VPS21Q69L, vps21/vps21 + VPS21S24N, vps9/vps9 + VPS9, vps3/vps3 + VPS3, vac1/vac1 + VAC1, and pep12/pep12 + PEP12). The primer sequences used for PCR assays are presented in Table S2.
2.3. White-opaque switching assay
White-opaque switching assays were performed as described previously (Huang et al. Citation2010). C. albicans opaque or white cells were initially plated onto Lee’s glucose medium and incubated at 25 °C for 7 days. For quantitative switching assays, homogeneous white or opaque colonies were then selected and replated onto Lee’s glucose or Lee’s GlcNAc medium (pH 6.8) and cultured at 25 °C for 5 days. Three biological repeats were performed. Switching frequency = average percentage of colonies with the alternative phenotype or sectored colonies ± standard deviation (SD).
2.4. Quantitative mating assay
Quantitative mating assays were performed as previously described (Miller and Johnson Citation2002; Xie et al. Citation2013; Li et al. Citation2023). Opaque cells were first grown on Lee’s glucose medium at 25 °C for 5 days. Approximately 2 × 107 cells of MTLa/Δ and MTLΔ/α strains were mixed, spotted onto Lee’s glucose and Lee’s GlcNAc medium (pH 6.8) and incubated at 25 °C for 3 days. The mating mixtures were then resuspended, diluted, and plated onto SC media (without arginine, leucine, or both).
2.5. Quantitative real-time RT-PCR assays
Candida albicans cells (MTLa/Δ or MTLΔ/α) were initially cultured on Lee’s glucose medium at 25 °C for 5 days. For pheromone treatment assays, approximately 2 × 107 opaque cells (MTLa/Δ) were inoculated into liquid Lee’s glucose medium (4 mL) containing 50 μmol/L α-pheromone and incubated at 25 °C for 12 h to examine the relative expression levels of pheromone response associated genes. To examine the relative expression levels of mating associated genes in MTLΔ/α cells, approximately 2 × 107 opaque cells (MTLΔ/α) were incubated in 4 mL of liquid Lee’s glucose medium (pH 6.8) and cultured at 25 °C for 24 h. Fungal cells (MTLa/Δ or MTLΔ/α) were harvested, and total RNA was extracted using RNA purification kit (Thermo Scientific, K0732, Thermo Scientific, Waltham, USA). Total RNA (1 μg per sample) was used to synthesise cDNA with RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific Inc., Shanghai, China). The expression level of the C. albicans ACT1 gene was used for normalisation.
3. Results
3.1. Vps21 regulates white-opaque switching in C. albicans
Opaque cells of C. albicans often contain a large vacuole (Anderson and Soll Citation1987; Soll Citation2009). Since the Vps21 signalling pathway is associated with vacuolar trafficking and protein sorting in fungi (Stack et al. Citation1995; Gerrard et al. Citation2000; Toshima et al. Citation2014), we set out to investigate whether this pathway plays a role in the regulation of C. albicans white-opaque switching. We generated a vps21/vps21 deletion mutant strain in C. albicans using the fusion PCR homologous recombination strategy (Noble and Johnson Citation2005). White-opaque switching assays demonstrated that compared to the WT strain [(14.6 ± 0.2)%] and reconstituted strain (vps21/vps21 + VPS21) [(10.9 ± 1.0)%], the vps21/vps21 mutant strain [(30.9 ± 2.3)%] exhibited a significantly higher white-to-opaque switching frequency on Lee’s GlcNAc medium ( and ). However, both the WT and vps21/vps21 mutant strains demonstrated low frequencies of white-to-opaque switching on Lee’s glucose medium (). The opaque-to-white switching frequencies of the WT strain, vps21/vps21 mutant strain, and reconstituted strain (vps21/vps21 + VPS21) were comparable on both Lee’s glucose and Lee’s GlcNAc media ().
Figure 1. Function of Vps21 and its variants in the regulation of white-to-opaque switching. White cells of the WT control, vps21/vps21 mutant, and reconstituted strains were initially grown on Lee’s glucose medium (pH 6.8) at 25 °C for 7 days. Candida albicans cells of homogeneous white colonies were replated onto Lee’s GlcNAc medium (pH 6.8) and incubated at 25 °C for 5 days. wh, white; op, opaque; op-s, opaque-sectored (colonies containing both white and opaque cells). Scale bar for cells, 10 μm; Scale bar for colonies, 2 mm. Strains used: WT (FDZF208); vps21/vps21 (FDZF263); vps21/vps21 + VPS21 (FDZF489); vps21/vps21 + VPS21S24N (FDZF475); and vps21/vps21 + VPS21Q69L (FDZF473). The mating type of all strains used was MTLa/Δ. The switching frequencies are presented in .
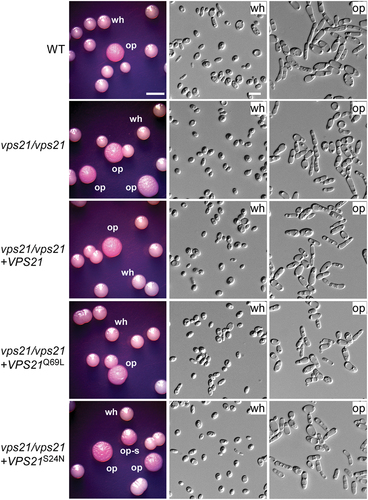
Table 1. White-opaque switching frequencies of the WT control and mutant strains of the Vps21 signalling pathway.
GEF activator, which facilitates the switch between an inactive GDP-bound state and an active GTP-bound state (Chen et al. Citation2014). We next ectopically expressed putative GTP-locked (active, VPS21Q69L) and GDP-locked (inactive, VPS21S24N) states in the vps21/vps21 mutant strain and examined the white-to-opaque switching frequencies. As expected, the VPS21Q69L active strain [(12.4 ± 0.6)%] exhibited a comparable switching frequency to that of the WT control strain [(14.6 ± 0.2)%], whereas the VPS21S24N inactive strain [(30.8 ± 3.0)%] showed an increased frequency on Lee’s GlcNAc medium compared to the WT control strain ( and ). Consistently, we observed that deletion of the GEF activator-encoding gene VPS9 had a similar promoting effect on the induction of the opaque phenotype in C. albicans under the same culture condition ( and ). Taken together, these results suggest that the Vps21 GTPase plays a critical role in regulating white-opaque switching in C. albicans.
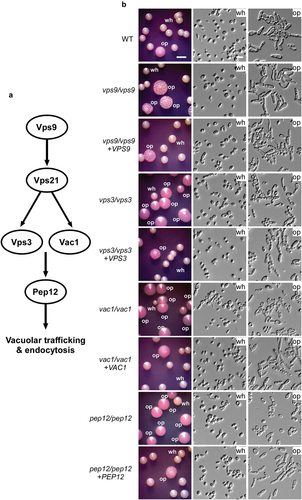
Figure 2. Deletion of VPS9, VPS3, VAC1, and PEP12 promotes white-to-opaque switching. (a) Schematic diagram of the Vps21 signaling pathway. Vps9, a guanine nucleotide exchange factor (GEF) of Vps21; Vps3, a CORVET tethering complex component; Vac1, a vesicle transport protein; and Pep12, a t-SNARE involved in prevacuolar trafficking. (b) White-to-opaque switching assays. White cells of the WT control, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant, and corresponding reconstituted strains were initially grown on Lee’s glucose medium (pH 6.8) for 7 days. Candida albicans cells of homogeneous white colonies were replated onto Lee’s GlcNAc medium (pH 6.8) and incubated at 25 °C for 5 days. Wh, white; op, opaque. Scale bar for cells, 10 μm; Scale bar for colonies, 2 mm. Strains used: WT (FDZF208); vps9/vps9 (FDZF213); vps3/vps3 (FDZF259); vac1/vac1 (FDZF531); pep12/pep12 (FDZF546); vps9/vps9 + VPS9 (FDZF491); vps3/vps3 + VPS3 (FDZF493); vac1/vac1 + VAC1 (FDZF548); and pep12/pep12 + PEP12 (FDZF549). The mating type of all strains used was MTLa/Δ. The switching frequencies are presented in .
3.2. Vps3, Vac1, and Pep12 regulate white-opaque switching
Vps3, Vac1, and Pep12 are downstream factors in the Vps21 signalling pathway (Chen et al. Citation2014). To investigate their roles in white-opaque switching, we generated vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains in C. albicans and performed white-opaque switching assays. As shown in and , all three mutant strains exhibited extremely high white-to-opaque switching frequencies on Lee’s GlcNAc medium [(94.3 ± 3.1)% for the vps3/vps3 mutant strain, (91.7 ± 3.4)% for the vac1/vac1 mutant strain, and (96.9 ± 1.8)% for the pep12/pep12 mutant strain)]. These mutant strains also had relatively high opaque-to-white switching frequencies on Lee’s glucose medium (approximately 30%, ), suggesting that deletion of these genes has a bidirectional effect on promoting phenotypic transitions. Similar to the vps21/vps21 and vps9/vps9 mutant strains, the white phenotypes of the vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains were stable on Lee’s glucose medium (switching frequency < 0.4%), while the opaque phenotypes of these mutant strains were stable on Lee’s GlcNAc medium (switching frequency < 0.4%). As expected, the switching frequencies between white and opaque phenotypes of the VPS3-, VAC1-, and PEP12-reconstituted strains were comparable to those of the WT control strain. Interestingly, we found that the vps3/vps3, vac1/vac1, and pep12/pep12 mutants exhibited a much stronger white-to-opaque switching ability than the vps21/vps21 mutant, indicating that some other signalling pathways or regulators are involved in this regulation. There could be cross-talks and inter-regulations among these pathways in the regulation of phenotypic switching. Taken together, our results suggest that the Vps21 downstream factors Vps3, Vac1, and Pep12 also play roles in the regulation of white-opaque switching in C. albicans.
3.3. The Vps21 signaling pathway regulates sexual mating in C. albicans
Since white-opaque switching regulates mating in C. albicans and closely related species (Pujol et al. Citation2004; Porman et al. Citation2011; Xie et al. Citation2013), we next tested whether the Vps21 signalling pathway plays a role in the regulation of mating. As shown in , quantitative mating assays demonstrated that the inactivation of Vps21 or Vps9 led to an approximately 8- to 193-fold decrease in mating efficiency on Lee’s glucose or Lee’s GlcNAc medium (compared to the WTa × WTα control cross). Notably, the vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains exhibited dramatically decreased mating efficiencies on both media (decreased by three to five orders of magnitude). The mating efficiency of these mutants was particularly low on Lee’s GlcNAc medium (). These results imply that the Vps21 signalling pathway is required for efficient mating in C. albicans.
Table 2. Mating efficiencies of the WT control and mutant strains of the Vps21 signalling pathway.
3.4. Deletion of genes associated with the Vps21 signaling pathway downregulates the expression of mating-related genes
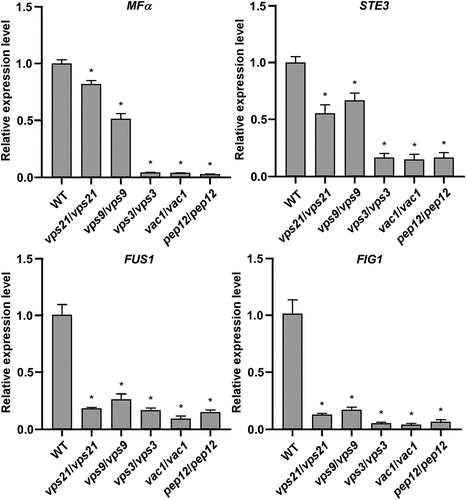
To explore the potential mechanism of the Vps21 signalling pathway in regulating mating in C. albicans, we performed quantitative RT-PCR assays to test the relative expression levels of mating-associated genes. As shown in and S1, the relative expression levels of MFα (encoding the α-pheromone precursor) (Bennett et al. Citation2003; Lockhart et al. Citation2003; Panwar et al. Citation2003), STE3 (encoding the a-pheromone receptor) (Chen et al. Citation2002; Magee et al. Citation2002), FUS1 (encoding a membrane protein required for cell fusion), FIG1 (encoding a membrane protein required for efficient mating and cell fusion), CPH1 (a Saccharomyces cerevisiae STE12 homolog required for mating) (Liu et al. Citation1994), and the mating-specific MAPK pathway-associated genes (including CST20, STE11, HST7, CEK1, and CEK2) (Chen et al. Citation2002; Magee et al. Citation2002) were significantly downregulated in the vps21/vps21, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains (compared to the WT control strain). The changes in relative expression levels of these mating-associated genes largely corresponded to the decreased mating efficiencies in the Vps21 signalling pathway mutant strains.
Figure 3. Relative transcriptional expression of mating associated genes in the WT control and mutant strains of the Vps21 signalling pathway. Opaque cells of the WT control, vps21/vps21, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains were incubated in liquid Lee’s glucose medium (pH 6.8) for 24 hours at 25 °C. Total RNA was extracted for quantitative RT-PCR assays. The expression level of ACT1 was used for normalisation. The average value of the WT control strain for each gene was set as “1”. “*” indicates significant difference between the WT control strain and mutant strain (p < 0.05, two-tailed Student’s t-test). Strains used: WT (FDZF171); vps21/vps21 (FDZF266); vps9/vps9 (FDZF212); vps3/vps3 (FDZF250); vac1/vac1 (FDZF533); and pep12/pep12 (FDZF534). The mating type of all strains used was MTLΔ/α.
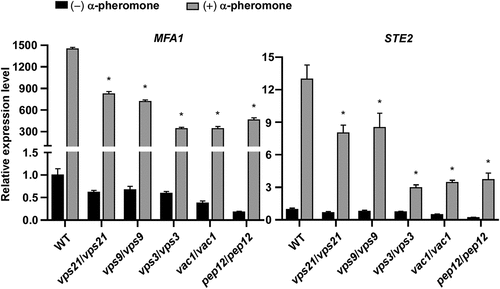
We next examined the induction of mating-associated genes [MFA1 (Dignard et al. Citation2007) and STE2 (Chen et al. Citation2002; Magee et al. Citation2002)] in response to synthetic α-pheromone exposure. Consistently, we observed that inactivation of the Vps21 signalling pathway remarkably suppressed the expression of MFA1 and STE2 in the presence of α-pheromone (). In summary, these findings indicate that the Vps21 signalling pathway is critical for activation of the mating response pathway and is required for efficient mating in C. albicans.
Figure 4. Relative transcriptional expression of MFA1 and STE2 in the absence and presence of α-pheromone in the WT control and mutant strains of the Vps21 signalling pathway. Opaque cells of the WT control, vps21/vps21, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains were incubated in liquid Lee’s glucose medium (pH 6.8) and treated with or without synthetic α-pheromone (50 μg/mL) for 12 hours at 25 °C. Total RNA was extracted for quantitative RT-PCR assays. The expression level of ACT1 was used for normalisation. The average value of the WT control strain for each gene without pheromone treatment was set as “1”. “*” indicates significant difference between the WT control strain and mutant strain (p < 0.05, two-tailed Student’s t-test). Strains used: WT (FDZF208); vps21/vps21 (FDZF263); vps9/vps9 (FDZF213); vps3/vps3 (FDZF259); vac1/vac1 (FDZF531); and pep12/pep12 (FDZF546). The mating type of all strains used was MTLa/Δ.
4. Discussion
To mate efficiently under standard laboratory culture conditions, C. albicans must first undergo a morphological switch from white to opaque cell type (Miller and Johnson Citation2002). Many environmental factors and genes are involved in the regulation of white-opaque switching and mating in C. albicans (Lohse and Johnson Citation2009; Soll Citation2009; Huang Citation2012; Guan et al. Citation2019; Li et al. Citation2023). In this study, we report that the conserved Vps21 signalling pathway regulates both white-opaque switching and sexual mating in C. albicans. Inactivation of the major components of the Vps21 signalling pathway (including Vps21, Vps9, Vps3, Vac1, and Pep12) promoted white-to-opaque switching on Lee’s GlcNAc medium but repressed mating in C. albicans (, ). There is an obvious negative correlation between white-to-opaque switching frequencies and mating efficiencies in the vps21/vps21, vps9/vps9, vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains. For example, the vps3/vps3, vac1/vac1, and pep12/pep12 mutant strains exhibited high white-to-opaque switching frequencies (>90%) but mated poorly (). It remains to be investigated as to why the Vps21 signalling pathway plays opposing roles in the regulation of these two canonically linked biological processes.
A major function of the Vps21 signalling pathway in fungi is in vacuolar biogenesis and trafficking (Stack et al. Citation1995; Rieder et al. Citation1996; Palmer et al. Citation2003; Toshima et al. Citation2014). Previous studies have demonstrated that C. albicans opaque cells and projected opaque cells (“shmoos” formed during mating) often contain large vacuoles (Anderson and Soll Citation1987; Liang et al. Citation2020). Given the global regulatory roles of the Vps21 signalling pathway and vacuoles, it is reasonable that impairment of this pathway or the vacuole itself could affect white-opaque switching and mating in C. albicans. Consistent with this hypothesis, a previous study found that Vps21-mediated vacuolar trafficking controls cellular morphogenesis and is important for hyphal development (Johnston et al. Citation2013). However, the mechanistic association between the Vps21 signalling pathway, vacuoles, phenotypic switching, and mating remains to be established in future studies.
We found that inactivation of genes related to the Vps21 signalling pathway had similar effects on the repression of mating in C. albicans on Lee’s glucose and Lee’s GlcNAc medium (). We observed that the mutant strains of this pathway exhibited distinct white-opaque switching frequencies under different culture conditions (). On Lee’s glucose medium, deletion of these genes (especially VPS3, VAC1, and PEP12) had promoting effects on the induction of the white cell type. However, on Lee’s GlcNAc medium, inactivation of the Vps21 signalling pathway induced the opaque phenotype. These results suggest that the regulatory roles of this pathway in white-opaque switching could be different under different culture conditions. One possible explanation for these observed differences is that GlcNAc is an opaque inducer via the activation of the cAMP/PKA signalling pathway which could have an additive effect on the induction of the opaque phenotype (Huang et al. Citation2010). Quantitative RT-PCR assays demonstrated that deletion of VPS21, VPS9, and their downstream factors led to reduced expression levels of mating-associated genes in C. albicans (). This reduced transcriptional expression could directly repress the mating response ().
In summary, our study indicates that the Vps21 signalling pathway plays a critical role in the control of both white-opaque transitions and sexual mating in Candida albicans. Additionally, the ability of mutant strains of the Vps21 signalling pathway to induce the mating-competent opaque phenotype depends on specific culture conditions. Deletions of the genes associated with this pathway lead to a repressing effect on sexual mating, perhaps through downregulation of expression of mating-associated genes. Our findings shed new light on the biological roles of the Vps21 signalling pathway in the regulation of morphological transitions and sexual mating in C. albicans.
Author contributions
FZ, HD, QZ, JB, LT, CJN, and GH conceived and designed the research. FZ, HD, and GH surveyed the scientific literature. FZ, HD, QZ, JB, LT, CJN, and GH analysed data and wrote the draft manuscript. FZ, HD, QZ, JB, LT, CJN, and GH interpreted the data and reviewed the manuscript. All authors read and approved the manuscript.
Supplemental Material
Download Zip (70.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21501203.2024.2376533.
Additional information
Funding
References
- Anderson J, Cundiff L, Schnars B, Gao MX, Mackenzie I, Soll DR. 1989. Hypha formation in the white-opaque transition of Candida albicans. Infect Immun. 57(2):458–467. doi: 10.1128/iai.57.2.458-467.1989.
- Anderson JM, Soll DR. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol. 169(12):5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987.
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 23(22):8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003.
- Borchers AC, Langemeyer L, Ungermann C. 2021. Who’s in control? Principles of rab GTPase activation in endolysosomal membrane trafficking and beyond. J Cell Biol. 220(9):e202105120. doi: 10.1083/jcb.202105120.
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med. 4(165):165rv113. doi: 10.1126/scitranslmed.3004404.
- Chen J, Lane S, Liu H, Liu H. 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol. 46(5):1335–1344. doi: 10.1046/j.1365-2958.2002.03249.x.
- Chen Y, Zhou F, Zou S, Yu S, Li S, Li D, Song J, Li H, He Z, Hu B, et al. 2014. A Vps21 endocytic module regulates autophagy. Mol Biol Cell. 25(20):3166–3177. doi: 10.1091/mbc.e14-04-0917.
- Dignard D, Al E-N, Logue ME, Butler G, Whiteway M. 2007. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot Cell. 6(3):487–494. doi: 10.1128/EC.00387-06.
- Feng Q, Summers E, Guo B, Fink G. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 181(20):6339–6346. doi: 10.1128/JB.181.20.6339-6346.1999.
- Gerrard SR, Bryant NJ, Stevens TH, Kaiser C. 2000. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 11(2):613–626. doi: 10.1091/mbc.11.2.613.
- Guan G, Tao L, Li C, Xu M, Liu L, Bennett RJ, Huang G. 2023. Glucose depletion enables Candida albicans mating independently of the epigenetic white-opaque switch. Nat Commun. 14(1):2067. doi: 10.1038/s41467-023-37755-8.
- Guan G, Tao L, Yue H, Liang W, Gong J, Bing J, Zheng Q, Veri AO, Fan S, Robbins N, et al. 2019. Environment-induced same-sex mating in the yeast Candida albicans through the Hsf1-Hsp90 pathway. PLoS Biol. 17(3):e2006966. doi: 10.1371/journal.pbio.2006966.
- Huang G. 2012. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 3(3):251–261. doi: 10.4161/viru.20010.
- Huang G, Srikantha T, Sahni N, Yi S, Soll DR. 2009. CO(2) regulates white-to-opaque switching in Candida albicans. Curr Biol. 19(4):330–334. doi: 10.1016/j.cub.2009.01.018.
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR, Hull CM. 2010. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 6(3):e1000806. doi: 10.1371/journal.ppat.1000806.
- Johnston DA, Tapia AL, Eberle KE, Palmer GE. 2013. Three prevacuolar compartment rab GTPases impact Candida albicans hyphal growth. Eukaryot Cell. 12(7):1039–1050. doi: 10.1128/EC.00359-12.
- Li C, Tao L, Guan G, Guan Z, Perry AM, Hu T, Bing J, Xu M, Nobile CJ, Huang G. 2023. Atmospheric humidity regulates same-sex mating in Candida albicans through the trehalose and osmotic signaling pathways. Sci China Life Sci. 66(8):1915–1929. doi: 10.1007/s11427-023-2309-1.
- Liang SH, Cheng JH, Deng FS, Tsai PA, Lin CH. 2014. A novel function for Hog1 stress-activated protein kinase in controlling white-opaque switching and mating in Candida albicans. Eukaryot Cell. 13(12):1557–1566. doi: 10.1128/EC.00235-14.
- Liang W, Guan G, Li C, Nobile CJ, Tao L, Huang G. 2020. Genetic regulation of the development of mating projections in Candida albicans. Emerg Microbes Infect. 9(1):413–426. doi: 10.1080/22221751.2020.1729067.
- Liu H, Kohler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 266(5191):1723–1726. doi: 10.1126/science.7992058.
- Lockhart SR, Zhao R, Daniels KJ, Soll DR. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell. 2(5):847–855. doi: 10.1128/EC.2.5.847-855.2003.
- Lohse MB, Johnson AD. 2009. White-opaque switching in Candida albicans. Curr Opin Microbiol. 12(6):650–654. doi: 10.1016/j.mib.2009.09.010.
- Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol. 46(5):1345–1351. doi: 10.1046/j.1365-2958.2002.03263.x.
- Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 110(3):293–302. doi: 10.1016/S0092-8674(02)00837-1.
- Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 4(2):298–309. doi: 10.1128/EC.4.2.298-309.2005.
- Palmer GE, Cashmore A, Sturtevant J. 2003. Candida albicans VPS11 is required for vacuole biogenesis and germ tube formation. Eukaryot Cell. 2(3):411–421. doi: 10.1128/EC.2.3.411-421.2003.
- Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. 2003. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell. 2(6):1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003.
- Park YN, Morschhauser J. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell. 4(8):1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005.
- Porman AM, Alby K, Hirakawa MP, Bennett RJ. 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci USA. 108(52):21158–21163. doi: 10.1073/pnas.1112076109.
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 3(4):1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004.
- Ramirez-Zavala B, Weyler M, Gildor T, Schmauch C, Kornitzer D, Arkowitz R, Morschhauser J, Mitchell AP. 2013. Activation of the Cph1-dependent MAP kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog. 9(10):e1003696. doi: 10.1371/journal.ppat.1003696.
- Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 341:119–127. doi: 10.1016/j.gene.2004.06.021.
- Rieder SE, Banta LM, Kohrer K, Jm M, Emr SD. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 7(6):985–999. doi: 10.1091/mbc.7.6.985.
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 169(1):189–197. doi: 10.1128/jb.169.1.189-197.1987.
- Soll DR. 2009. Why does Candida albicans switch? FEMS Yeast Res. 9(7):973–989. doi: 10.1111/j.1567-1364.2009.00562.x.
- Stack JH, Horazdovsky B, Emr SD. 1995. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 11(1):1–33. doi: 10.1146/annurev.cb.11.110195.000245.
- Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol. 9(10):737–748. doi: 10.1038/nrmicro2636.
- Sun Y, Cao C, Jia W, Tao L, Guan G, Huang G. 2015. pH regulates white-opaque switching and sexual mating in Candida albicans. Eukaryot Cell. 14(11):1127–1134. doi: 10.1128/EC.00123-15.
- Toshima JY, Nishinoaki S, Sato Y, Yamamoto W, Furukawa D, Siekhaus DE, Sawaguchi A, Toshima J. 2014. Bifurcation of the endocytic pathway into Rab5-dependent and -independent transport to the vacuole. Nat Commun. 5(1):3498. doi: 10.1038/ncomms4498.
- Wang X, Wu SS, Li LM, Yan ZM. 2023. Candida albicans overgrowth disrupts the gut microbiota in mice bearing oral cancer. Mycology. 15(1):57–69. doi: 10.1080/21501203.2023.2256761.
- Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu Rev Microbiol. 61(1):529–553. doi: 10.1146/annurev.micro.61.080706.093341.
- Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, Cao C, Hernday AD, Johnson AD, Zhang L, et al. 2013. White-opaque switching in natural MTLa/α isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 11(3):e1001525. doi: 10.1371/journal.pbio.1001525.
