Abstract
TFIIA is an important positive regulator of TFIID, the primary promoter recognition factor of the basal RNA polymerase II transcription machinery. TFIIA antagonises negative TFIID regulators such as negative cofactor 2 (NC2), promotes specific binding of the TBP subunit of TFIID to TATA core promoter sequence elements and stimulates the interaction of TBP-associated factors (TAFs) in the TFIID complex with core promoter elements located downstream of TATA, such as the initiator element (INR). Metazoan TFIIA consists of 3 subunits, TFIIAα (35 kDa), β (19 kDa) and γ (12 kDa). TFIIAα and β subunits are encoded by a single gene and result from site-specific cleavage of a 55 kDa TFIIA(α/β) precursor protein by the protease Taspase1. Metazoan cells have been shown to contain variable amounts of TFIIA (55/12 kDa) and Taspase1-processed TFIIA (35/19/12 kDa) depending on cell type, suggesting distinct gene-specific roles of unprocessed and Taspase1-processed TFIIA. How precisely Taspase1 processing affects TFIIA functions is not understood. Here we report that Taspase1 processing alters TFIIA interactions with TFIID and the conformation of TFIID/TFIIA promoter complexes. We further show that Taspase1 processing induces increased sensitivity of TFIID/TFIIA complexes to the repressor NC2, which is counteracted by the presence of an INR core promoter element. Our results provide first evidence that Taspase1 processing affects TFIIA regulation of TFIID and suggest that Taspase1 processing of TFIIA is required to establish INR-selective core promoter activity in the presence of NC2.
Abbreviations
| CPE | = | core promoter element |
| INR | = | initiator core promoter element |
| NC2 | = | negative cofactor 2 |
| RNAP II | = | RNA polymerase II |
| TBP | = | TATA-binding protein |
| TFII (TFIIA | = | -D), general transcription factor for RNA polymerase II |
Introduction
The TFIID multi-protein complex plays a central role in the regulation of transcription initiation by RNA polymerase II (RNAP II) as the primary core promoter recognition factor that initiates assembly of the basal transcription machinery composed of RNAP II and general transcription factors (GTFs) TFIIA, -B, -E, -F, -H.Citation1 TFIID consists of the TATA-binding protein TBP and 13 TBP-associated factors (TAFs).Citation2 TBP specifically recognizes the TATA element,Citation1 while TFIID TAFs have been implicated in the recognition of core promoter elements (CPEs) located downstream of the TATA box region, including the INR element encompassing the transcription start site (TSS; TAF1/2), and the DCE (TAF1), MTE and DPE (TAF6/9) located downstream of the TSS.Citation1,3,4 Core promoters in metazoan genomes are structurally highly diverse and typically contain only a small subset of known CPEs, which determine the position of the primary transcription start site, basal promoter strength and the response of the promoter to transcription regulators.Citation3,5-7 How precisely the readout of core promoter sequence information by TFIID is integrated with regulatory input from transcription activators and repressors to regulate overall transcription output of individual genes is not fully understood.
TATA and INR elements are the best characterized CPEs and are unique in that they are able to direct activator-independent basal RNAP II transcription initiation in the absence of other CPEs. Moreover, when separated by 25–30bp TATA and INR can function in concert in a synergistic manner, resulting in a dramatic increase of promoter activity compared with either TATA or INR alone.Citation3 Consistent with the modular nature of TFIID promoter interactions, INR-directed transcription initiation and synergy between TATA and INR elements requires TFIID TAFs,Citation8-11 whereas the TBP TATA-binding function is largely dispensable for INR-directed transcription initiation in the absence of a TATA box.12 Moreover, TAF interactions with the INR element enhance the promoter binding activity of isolated TFIID at TATA-containing promoters in a TFIIA-dependent manner.Citation13 These observations clearly establish an important role of TAFs in the functional readout of the INR element. However, TAF/promoter interactions are not sufficient to mediate the strong functional synergy of TATA and INR elements observed in crude nuclear extracts.Citation14,15 A strong response of TATA-containing core promoters to the presence of an INR element requires positive and negative TAF- and INR-dependent cofactors (TICs),Citation14 some of which were recently identified.Citation15,16 We previously demonstrated that positive-acting TICs counteract repression of TATA-dependent basal transcription by the ubiquitous repressor NC2 in the presence of a correctly spaced INR element in a TFIID TAF-dependent manner.Citation15 More recently, the chromatin architectural protein HMGA1 and the Mediator coactivator complex were identified as positive-acting TICs that selectively stimulate basal TATA-dependent transcription in the presence of an INR and, at the same time, antagonize NC2 repression.Citation16
NC2 represses RNAP II transcription initiation complex assembly by binding to TATA-bound TBP and preventing recruitment of TFIIB.Citation1 Furthermore, binding of NC2 has been shown to affect dynamic conformational changes within the TBP/TATA nucleoprotein complex, which lead to a loss of TATA specificity and mobilization of the TBP/DNA complex away from the TBP TATA-binding site.Citation17,18
The general RNAP II transcription factor TFIIA is a positive regulator of TFIID function and counteracts NC2 repression by competing with NC2 for binding to TBP.Citation1 TFIIA further modulates the promoter-binding activity of TFIID by stabilizing TBP/TATA interactionsCitation1 and by promoting interactions between TFIID TAFs and DNA sequences downstream of TATA, such as the INR element.Citation13,19,20 Indeed, TFIIA has been shown to be absolutely required for INR-mediated and TAF-dependent transcription initiation in the absence of a TATA box as well as for TAF-dependent INR-mediated stimulation of basal transcription from TATA-containing promoters.Citation14 TFIIA appears to modulate interactions of TAFs with specific core promoter DNA sequences by stabilizing specific TFIID complex conformations.Citation13,20 Recent single particle electron studies of TFIID revealed that TFIID can exist in at least 2 structurally distinct states, a canonical state observed predominantly in the absence of DNA and a ‘re-arranged’ state observed in the presence of promoter DNA.Citation21 Interestingly, these studies suggest a dual role of TFIIA in maintaining both the canonical state of TFIID in the absence of DNA and in stabilizing the re-arranged state of TFIID when promoter-bound.Citation21
In metazoan cells TFIIA exists predominantly as a heterotrimeric complex composed of TFIIAα (35 kDa), TFIIAβ (19 kDa) and TFIIAγ (12 kDa) subunits.Citation1,22 TFIIAα and TFIIAβ originate from a single gene and are produced by site-specific cleavage of a 55 kDa TFIIAα/β precursor protein by the protease Taspase1.Citation23 The relative abundance of unprocessed TFIIA and Taspase1-processed TFIIA in mammalian cells varies significantly between cell types,Citation22 suggesting that Taspase1 processing of TFIIA is regulated in a cell-type specific manner. Unprocessed TFIIA has been shown to associate with free TBP in the absence of DNA to form a transcriptionally active TBP-TFIIA-containing complex (TAC) lacking TAFs.Citation24 In contrast, TFIID complexes isolated at low stringency conditions were found to be associated only with the processed form of TFIIA.Citation24 Interestingly, the processed form of TFIIA was also found to associate in human cells with constitutively expressed TBP-related factor TRF2 Citation25 and TFIIA processing was recently shown to be required for TRF2-mediated transcription initiation at spermatogenic promoters during mammalian spermatogenesis.Citation26 These observations suggest that TFIIA processing plays a role in regulating association of TFIIA into functionally distinct complexes with free TBP, the TFIID complex and TBP-related factors. To what extend Taspase1 processing affects TFIIA functions as regulator of TFIID activity has so far not been investigated.
Here we report that Taspase1-processed heterotrimeric TFIIA (35/19/12 kDa) and unprocessed heterodimeric TFIIA (55/12 kDa) differentially affect TFIID topology and function and provide evidence that TFIIA processing by Taspase1 is required for optimal TFIIA activity in mediating TFIID TAF-dependent resistance to NC2 repression and INR-selective basal promoter activity in the presence of NC2. Our results suggest that TFIIA regulation through Taspase1 contributes to gene-specific regulation of transcription depending on core promoter architecture.
Results
A subpopulation within purified TFIID mediates INR-specific resistance to NC2 repression
We used a previously established two-template transcription assay to further investigate core promoter-dependent NC2 repression of basal transcription in a purified system. We reconstituted in vitro transcription with immunoaffinity purified FLAG:epitope-tagged human TFIID complex (f:TFIID), recombinant TFIIB, TFIIE and TFIIF and highly purified native TFIIH and RNAP II (). In this assay, the transcription activity of two variants of the murine TdT model core promoter, containing either only a consensus TATA box (mTdT-TATA) or both TATA and INR core promoter elements (mTdT-TATA/INR), is directly compared by primer extension analysis using the same radiolabelled primer (). As reported previously,Citation15 and consistent with the absence of positive-acting TAF- and INR-dependent cofactor (TIC) activities in our reconstituted system, we observe comparable levels of basal transcription from mTdT-TATA and mTdT-TATA/INR promoters (, lanes 1–6). However, f:TFIID titration experiments in the presence of recombinant NC2 (rNC2) gave unanticipated results. At low f:TFIID concentrations rNC2 efficiently repressed transcription from both mTdT-TATA and mTdT-TATA/INR promoters (, compare lanes 1–3 with lanes 7–9), consistent with previous observations.Citation15 However, with increasing concentrations of f:TFIID, mTdT-TATA/INR transcription became partially resistant to NC2 repression, whereas mTdT-TATA transcription remained completely repressed (, compare lanes 4 and 10). Furthermore, at saturating f:TFIID concentrations, when further addition of f:TFIID did not further increase basal transcription in our system, rNC2 repressed transcription from the mTdT-TATA promoter but, at the same time, stimulated transcription from the mTdT-TATA/INR template (, compare lanes 5, 6 with lanes 11, 12). Thus, under conditions when f:TFIID concentrations are exceeding saturating levels in our reconstituted system, we observe INR-selective basal transcription activity in the presence of NC2. This observation suggested that a small subpopulation of f:TFIID complexes present in our highly purified f:TFIID preparation is resistant to NC2 repression in the presence of an INR core promoter element.
Figure 1. INR-selective basal promoter activity in a purified system containing f:TFIID complex and rNC2. (A) SDS PAGE analysis of purified human GTFs used for in vitro transcription. rTFIIA is shown in . (B) Two-template in vitro transcription assay with mTdT promoter variants containing only TATA (TATA) or TATA and INR elements (TATA/INR). The mTdT-TATA promoter template contains additional 26-bp polylinker sequence downstream of the mTdT core promoter region (−41/+52), which does not affect core promoter activity and allows the analysis of transcripts originating from both TATA and TATA/INR variants in parallel by primer extension using the same radioactive primer. Transcription reactions (20 µl) were carried out at 30°C for 1 h and contained 50 fmol of each promoter template, 10 ng 6His:TFIIB, 10 ng rTFIIE, 10 ng rTFIIF, 0.5 μl TFIIH, 0.2 μl RNAP II, f:TFIID corresponding to 1, 2, 5, 10, 20, 30 ng TBP (lanes 1–6 and 7–12) and 5 ng rNC2 (lanes 7–12). 32P-labeled primer extension products originating from mTdT-TATA and mTdT-TATA/INR transcripts were resolved by 6% denaturing PAGE and visualized and quantified by PhosphorImager analysis.
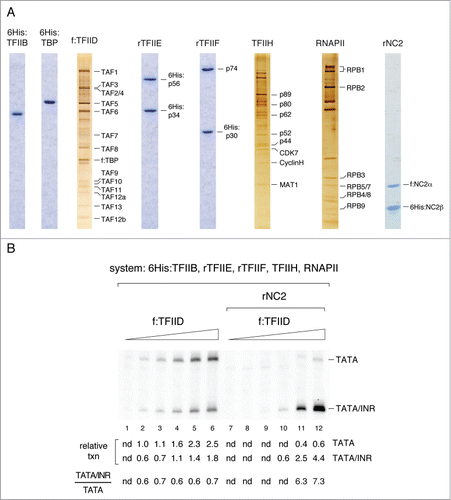
Figure 2. INR element interactions counteract disruption of TFIIA-associated TFIID/promoter complexes by NC2. (A) Immunoblot analysis of two independent f:TFIID preparations A and B, containing low and high amounts of associated TFIIA, respectively. (B) Schematic of the mTdT promoter variants used in Mg2+ agarose EMSA assays. (C–F) Mg2+ agarose EMSA assays. Binding reactions (10 µl) contained 5 fmol DNA template, 2.5 ng 6His:TBP or an equivalent amount of f:TFIID, 2 ng rTFIIA (rIIA) or 2 ng rNC2 as indicated. (C) TATA-dependent core promoter binding by 6His:TBP, but not immunoaffinity purified f:TFIID complex (preparation B), requires TFIIA. (D) f:TFIID/promoter complexes are supershifted with anti-TFIIA antibody. (E) Disruption of TFIIA-associated f:TFIID promoter complexes by NC2. (F) The INR element stabilizes TFIIA-associated f:TFIID promoter complexes in the presence of NC2.
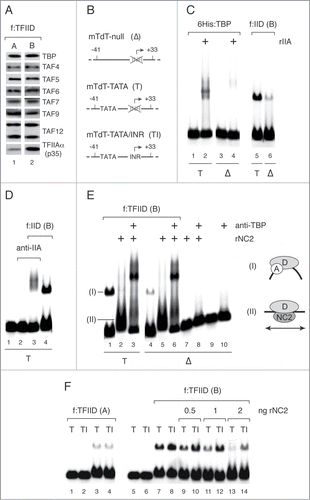
Figure 3. Reconstitution of INR-selective basal promoter activity in the presence of NC2 requires Taspase1 processing of TFIIA. (A) SDS PAGE analysis of purified recombinant and natural TFIIA preparations. rTFIIA was reconstituted from bacterially expressed unprocessed TFIIAα/β (p55) and 6His:TFIIAγ (6His:p12); rTFIIA was processed with recombinant Taspase1 (xTASP1) to yield rTFIIA (p35/p19/6His:p12); natural TFIIA (p35/p19/p12) was purified from the TFIIA (0.1 M KCl) HeLa nuclear extract fraction. (B) Two-template in vitro transcription assays with mTdT promoter variants containing only TATA (TATA) or TATA and INR elements (TATA/INR) were carried out and analyzed as described in the legend to and contained 20 ng rNC2, 5 ng, 10 ng and 20 ng of rTFIIA, rTFIIAxTASP1 or nTFIIA as indicated (lanes 2–11).
A subpopulation of immunoaffinity purified f:TFIID contains stably associated TFIIA
We noticed variable amounts of TFIIA within our highly purified f:TFIID preparations (). f:TFIID is purified by immunoaffinity chromatography from the HeLa nuclear extract phosphocellulose (P11) 0.85 M KCl/ DE-52 0.3 M KCl TFIID fraction.Citation27 Given the stringent conditions of the purification protocol, TFIIA in f:TFIID preparations must be tightly associated with the TFIID complex. Because TFIIA is required for synergistic binding of TFIID to TATA and INR core promoter elementsCitation13 and for INR-dependent basal transcription in vitro,Citation14 we speculated that the subpopulation of TFIID complexes in our f:TFIID preparations that is resistant to NC2 repression in the presence of an INR element is stably associated with TFIIA. To further investigate this possibility, we carried out immunoblot analyses and identified two independent f:TFIID preparations (A) and (B), that differ significantly in TFIIA content. As shown in , f:TFIID preparation (A) contains significantly lower amounts of TFIIA compared to f:TFIID preparation (B). Based on titration of rTFIIA in immunoblot experiments and comparison with TFIIA signals in f:TFIID we estimate that less than 10% of TFIID complexes are stably associated with TFIIA in the f:TFIID (B) preparation. Thus, while TFIIA content can vary considerably between individual f:TFIID preparations, the majority of TFIID complexes in f:TFIID is not associated with TFIIA. Of note is further, that we could only detect Taspase1-processed 35 kDa TFIIAα, but not unprocessed 55 kDa TFIIAα/β, in our f:TFIID preparations. Thus, TFIID in HeLa nuclear extracts appears to be exclusively associated with Taspase1-processed TFIIA.
TATA-dependent promoter binding of f:TFIID in Mg2+ agarose EMSA assays requires TFIIA
Next we used Mg2+ agarose EMSA assaysCitation28 to compare the binding of f:TFIID preparations containing lower (A) or higher (B) amounts of associated TFIIA () to mTdT promoter derivatives lacking an INR element and containing a consensus TATA box (T) or lacking a TATA box or any other known core promoter element (Δ) (). TATA-mediated promoter binding of human TFIID in Mg2+ agarose EMSA assays was previously shown to be strongly stimulated by TFIIA.Citation19 Consistent with this, human recombinant TFIIA reconstituted from unprocessed 55 kDa TFIIAα/β and 6His:epitope-tagged 12 kDa TFIIAγ subunits efficiently stimulated binding of 6His:TBP (, compare lanes 1 and 2). In contrast, strong binding of immunoaffinity purified f:TFIID (B), containing associated TFIIA, could be observed in the absence of added recombinant TFIIA (, lane 5). Binding of both 6His:TBP and f:TFIID to the promoter construct was almost completely abolished in the absence of TATA (Δ; , compare lane 2 with lane 4 and lane 5 with lane 6). Thus, binding of 6His:TBP and binding of f:TFIID preparation B to the mTDT core promoter were dependent on the TATA-box. Weak binding of 6His:TBP and f:TFIID to mTdT lacking a consensus TATA box (Δ; , lanes 4 and 6) likely reflects the presence of an additional low-affinity TATA sequence in our 260 bp mTdT promoter DNA template.
Importantly, f:TFIID (B) showed significantly higher core promoter binding activity than f:TFIID (A) (, compare lanes 3 and 4 with lanes 7 and 8, respectively), consistent with a higher TFIIA content in f:TFIID (B) (). Indeed, pre-formed f:TFIID/promoter complexes were quantitatively supershifted with anti-TFIIA antibody (). Taken together, these findings suggest that only TFIID complexes stably associated with TFIIA within our f:TFIID preparations are capable of forming stable complexes with the TATA-containing promoter DNA probe in Mg2+ agarose EMSAs.
Increased resistance of TFIIA-associated TFIID/promoter complexes to disruption by NC2 in the presence of an INR element
We next investigated whether the subpopulation of f:TFIID associated with TFIIA could mediate INR-dependent resistance to NC2 repression, observed in in vitro transcription assays (), at the level of stable TFIID/promoter complex formation. NC2 has been shown to bind to the concave underside of the TBP/TATA complex, thereby forming a ring-like protein structure with TBPCitation29 that prevents recruitment of TFIIA or TFIIB.Citation30,31 Furthermore, NC2 binding induces dynamic conformational changes into the TBP/DNA nucleo-complex, which lead to a loss of TBP-induced DNA bending and to mobilization of the TBP/DNA/NC2 complex away from the TATA site.Citation18
Consistent with the results of previous studies that examined the effect of NC2 on isolated TBP, rNC2 completely disrupted TATA-dependent f:TFIID/TFIIA promoter complexes (, compare lanes 1 and 2). At the same time, rNC2 stimulated quantitative binding of free DNA template into a different, fast migrating f:TFIID/DNA complex (complex II, , lane 2). This rNC2-induced binding of f:TFIID to DNA was independent of a TATA box element (, compare lanes 2 and 5). Anti-TBP antibody supershifts confirmed the presence of TBP in fast-migrating f:TFIID/NC2/DNA complexes (, lanes 3, 6). No DNA binding was observed in the presence of rNC2 alone (, lanes 7, 8).
We interpret these results as follows. First, the strong reduction in the mobility of DNA complexes formed with TFIID in EMSAs compared to free DNA is primarily caused by TBP DNA bending.Citation32-34 Second, in the presence of Mg2+ ions, the retention of (unbend) protein/DNA complexes is greatly reduced in agarose EMSA gels compared to classical acrylamide EMSAs.Citation28 Slow migrating f:TFIID EMSA complexes formed in the absence of rNC2 are therefore likely to contain the cognate bend TFIIA/TBP/TATA complex (, complex (I)), whereas fast migrating TATA-independent f:TFIID EMSA complexes formed in the presence of rNC2 represent an unbend/mobile conformational state (, complex (II)). Taken together, these results suggest that NC2 is able to compete with TFIID-associated TFIIA and to efficiently bind to TFIID lacking TFIIA to form TFIID/DNA/NC2 complexes. As previously reported for NC2 interactions with isolated TBP, NC2-induced conformational changes alter TBP/DNA interactions within TFIID, resulting in a stabilization of TATA-independent DNA interactions.Citation17 This results in a loss of TBP-induced DNA bending and presumably in mobilization of the TFIID/NC2 nucleoprotein complex (,Citation18).
Finally, we tested whether the presence of an INR element would influence the stability of TFIID/TFIIA complexes when challenged with rNC2. We therefore titrated rNC2 into binding reactions containing f:TFIID (B) () and a mTdT promoter derivative containing only a TATA box (T) or both a TATA box and an INR element (TI). As shown in , addition of rNC2 disrupted f:TFIID complexes formed both with mTdT-T and mTdT-TI promoter constructs. However, f:TFIID/TFIIA complexes formed on mTdT-TI promoter were more resilient to NC2 disruption than f:TFIID/TFIIA complexes formed on the mTdT-T promoter lacking an INR element (, lanes 7–14). These data support the idea that INR-mediated resistance to NC2 repression is established, at least to a certain degree, at the level of TFIID/TFIIA promoter complex formation.
Taspase1-processed TFIIA counteracts NC2 repression of basal transcription in an INR-dependent manner
The results presented above suggested that TFIID-associated TFIIA plays a role in INR-mediated resistance to NC2 repression. This was interesting, since previously we did not observe INR-dependent resistance to NC2 repression using our reconstituted in vitro transcription system containing bacterially expressed recombinant human TFIIA, reconstituted from unprocessed recombinant 55 kDa TFIIAα/β and 6His:epitope-tagged 12 kDa TFIIAγ.Citation15 Because f:TFIID associated exclusively with Taspase1-processed TFIIA composed of 35 kDa TFIIAα, 19 kDa TFIIAβ and 12 kDa TFIIAγ subunits, we hypothesized that Taspase1 cleavage of TFIIAα/β might be important for TFIIA to mediate INR-dependent resistance of TFIID/promoter complexes to NC2.
To investigate this possibility we digested our rTFIIA (p55/p12) preparation with highly purified recombinant Taspase1Citation23,35 and re-purified processed rTFIIA (p35/p19/p12; ) by ion exchange chromatography on RESOURCE Q and phosphocellulose (P11) resins. In addition, we purified natural Taspase1-processed human TFIIA (p35/p19/p12) from the classical 0.1 M KCl phosphocellulose (P11) HeLa nuclear extract fraction ().
Next, we used two-template in vitro transcription assays to compare the ability of unprocessed recombinant TFIIA(p55/p12), Taspase1-processed recombinant TFIIA (p35/p19/p12) and natural Taspase1-processed TFIIA (p35/p19/p12) to antagonize NC2 repression at mTdT-T and mTdT-TATA/INR promoters in our purified system containing high amounts of f:TFIID preparation (A), that contains less TFIIA (). In the absence of rNC2, we observed slightly preferential transcription from mTdT-TATA compared to the mTdT-TATA/INR promoter (TI/T ratio: 0.6, , lane 1; see also ). We then added sufficient amounts of rNC2 to completely repress transcription from both mTdT-TATA and mTdT-TATA/INR promoters (, lane 2) and titrated increasing amounts of TFIIA in order to counteract NC2 repression and to recover transcription activity (, lanes 3–11).
Recombinant unprocessed rTFIIA (p55/6His:p12; ; lanes 3–5) reversed NC2 repression at the mTdT-TATA/INR promoter with slightly higher efficiency than at mTdT-TATA (TI/T ratio 1.6 – 2.6; , compare lanes 1 with lanes 3–5). This relatively modest preference of rTFIIA for stimulating NC2 repressed basal transcription in the presence of an INR decreases with increasing rTFIIA concentrations, suggesting that the presence of an INR element enhances the affinity of rTFIIA for f:TFIID.
Taspase1 processing had no detectable effect on rTFIIA-mediated reversal of NC2 repression at the mTdT-TATA/INR promoter but severely reduced the ability of rTFIIA to counteract NC2 repression in the absence of an INR element (, lanes 6–8). As shown in , addition of 10 ng rTFIIA or rTFIIA processed with Taspase1 (rTFIIAxTASP) fully restored mTdT-TATA/INR transcription to levels observed in the absence of NC2 (, compare lane 1 with lanes 4 and 7). At the same time, unprocessed rTFIIA stimulated mTdT-TATA transcription to about 1/2 of mTdT-TATA/INR transcription levels (TI/T ratio: 2; , lane 4), whereas the same amount of rTFIIAxTASP1 stimulated mTdT-TATA transcription only to about 1/16 of mTdT-TATA/INR transcription levels (TI/T ratio: 16; , lane 7). Importantly, the presence of an INR element enhanced the activity of natural Taspase1-processed TFIIA purified from human nuclear extracts in a very similar manner to Taspase1-processed rTFIIA (TI/T ratio: 16; , lane 11).
These observations suggest that TFIIA needs to be processed by Taspase1 in order to support TFIID TAF-dependent resistance of an INR-containing promoter to NC2. Furthermore, our results suggest that, in the context of our reconstituted in vitro transcription system, (i) Taspase1 processing weakens TFIIA interactions with TATA-bound TFIID that are competitive with NC2 binding and (ii) that the interaction between TFIID and Taspase1 processed TFIIA on promoters is stabilized by the presence of an INR core promoter element.
Taspase1 processing increases stimulation of TFIID promoter binding by rTFIIA and changes TFIIA/TFIID/promoter complex properties
To further investigate the effect of Taspase1 processing of TFIIA on functional TFIIA interactions with TFIID we examined TFIIA/TFIID/promoter complex assembly with unprocessed and Taspase1-cleaved TFIIA in Mg2+ agarose EMSA assays. Titration experiments revealed both quantitative and qualitative effects of Taspase1 processing on TFIIA activity. First, Taspase1 processing significantly increased the ability of rTFIIA to stimulate TFIID promoter binding (). Second, Taspase1-processed rTFIIA as well as purified natural processed TFIIA () formed TFIID/TFIIA/promoter complexes with electrophoretic mobility indistinguishable from promoter complexes formed by TFIIA-associated f:TFIID (, compare lane 5 with lanes 1–4; , compare lanes 1,7 to lanes 5, 6, 11, 12; , compare lanes 1,2 to lanes 3,4). In contrast, association of unprocessed rTFIIA resulted in 2 different TFIID promoter complexes, with increased and with reduced electrophoretic mobility compared to complexes formed with natural TFIIA-associated TFIID (). These observations suggest that Taspase1 processing affects both the ability of TFIIA to interact with TFIID and the structure of resulting TFIIA/TFIID promoter complexes.
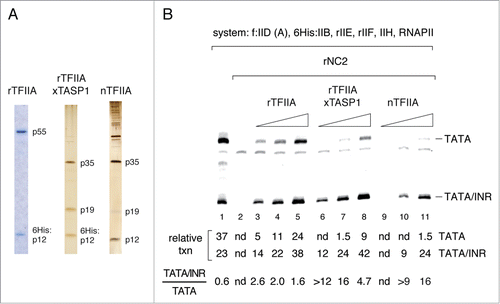
Figure 4. Taspase1 processing increases rTFIIA activity in stimulating TFIID promoter binding and changes TFIIA/TFIID/promoter complex properties. Mg2+ agarose EMSA assays. Binding reactions (10 µl) contained 5 fmol DNA template and 2.5 ng TBP equivalent of f:TFIID (preparation A, see ). (A) Stimulation of f:TFIID promoter binding by rTFIIA and rTFIIA processed by Taspase1 (rTFIIAxTASP1). (B) TFIIA stimulation of f:TFIID binding to mTdT promoter derivatives containing only a TATA box or TATA and INR elements. Binding reactions contained 2 ng rTFIIAxTASP1 or natural TFIIA (nTFIIA) or 2, 5, 10 ng unprocessed rTFIIA. (C) INR-mediated resistance to disruption of f:TFIID promoter complexes by NC2 is mediated by processed natural TFIIA but not by unprocessed rTFIIA. Binding reactions contained 2 ng nTFIIA or rTFIIA and 2 ng rNC2 as indicated.
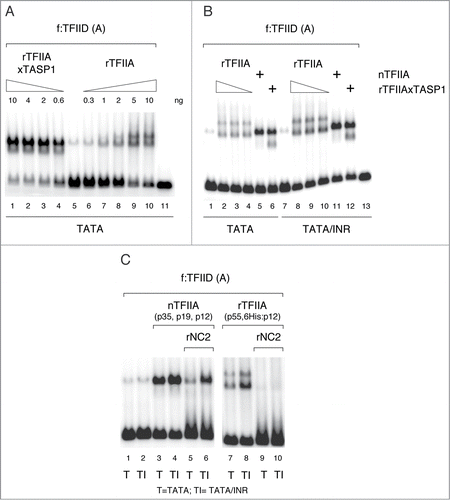
The presence of an INR element modestly increased TFIID promoter binding both in the presence of unprocessed and Taspase1-processed TFIIA (). Thus, Taspase1 processing appears not to specifically affect the ability of TFIIA to stimulate TFIID promoter binding in an INR-dependent manner. Finally, we asked whether NC2 differentially affects TFIID promoter complexes formed with either processed, natural TFIIA or with unprocessed recombinant TFIIA formed on promoters lacking or containing an INR element. For this purpose we challenged TFIID complexes with an amount of rNC2 that was sufficient to disrupt TFIID/TFIIA/promoter complexes and to promote quantitative assembly of the free promoter DNA probe into fast migrating TFIID/NC2 complexes (). Under these conditions, rNC2 completely disrupted TFIID complexes formed with unprocessed rTFIIA, regardless of the presence of an INR element ( lanes 7–10). In contrast, rNC2 only partially disrupted TFIID complexes formed with an equivalent amount of natural processed TFIIA complex (, lanes 3–6). Moreover, TFIID complexes formed with processed TFIIA were more resilient to disruption by rNC2 when formed on promoter DNA containing an INR element in addition to a TATA box (; compare lanes 3, 4 with lanes 5, 6). These results closely resemble those observed with the subpopulation of TFIID tightly associated with processed TFIIA present in f:TFIID preparations (, lanes 7–14).
Taken together, these findings demonstrate that processed and unprocessed TFIIA form TFIID/TFIIA promoter complexes with distinct properties. Firstly, the different electrophoretic mobilities of TFIID complexes formed with unprocessed and processed TFIIA indicate differences in TFIID/TFIIA nucleoprotein complex topology, presumably a different degree of overall DNA bending. Secondly, a stabilization of TFIID promoter complexes by the INR core promoter element, when challenged with NC2, could only be observed in the presence of Taspase1 processed TFIIA, but not with unprocessed TFIIA. Thus Taspase1 processing of TFIIA is required to establish INR-dependent stabilization of TFIID/promoter complexes in the presence of NC2 (see also ).
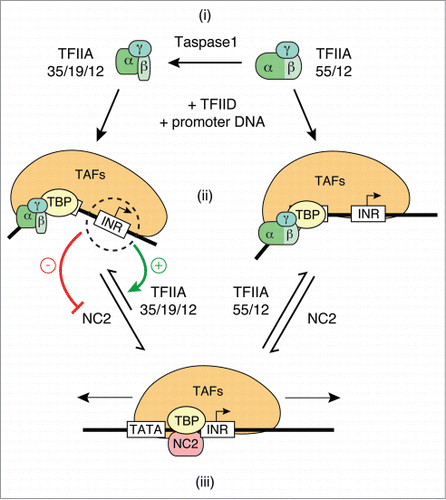
Figure 5. Role of TFIIA processing by Taspase1 in INR-specific transcription. (i) Unprocessed heterodimeric TFIIA is composed of 55 kDa TFIIAα/β and 12 kDa TFIIAγ subunits. Taspase1 cleavage results in heterotrimeric TFIIA complex composed of 35 kDa TFIIAα, 19 kDa TFIIAβ and 12 kDa TFIIAγ. (ii) TFIID promoter complexes formed with unprocessed TFIIA or Taspase1-processed TFIIA have distinct properties. Different mobilities of TFIID complexes formed with unprocessed and processed TFIIA in Mg2+ EMSAs () indicate differences in nucleoprotein complex topology. Both unprocessed and Taspase1-processed TFIIA compete with NC2 for binding to TBP in TFIID. In vitro transcription experiments () suggest that processing of TFIIA by Taspase1 weakens TFIIA interactions with TATA-bound TFIID that are competitive with NC2 binding in a manner that is compensated by the presence of an INR element. As a result, stabilization of TFIID promoter complexes by the INR core promoter element to counteract binding by NC2 is only observed with Taspase1-processed TFIIA (35/19/12; ). (iii) NC2 binding disrupts TFIID/DNA interactions, eliminates DNA bending and results in mobilization of TFIID away from TATA box ().
Discussion
Previous work showed that strong stimulation of TATA-dependent basal promoter activity by the INR core promoter element seen in crude nuclear extracts involves simultaneous action of negative and positive cofactors in addition to GTFs, RNAP II and TFIID TAFs.Citation14,15 Positive cofactors for INR function include the Mediator complex and the architectural transcription factor HMGA1, which stimulate the activity of the basal transcription machinery in the presence of an INR element in a TFIID TAF-dependent manner and, at the same time, counteract repression of TATA-dependent transcription initiation by NC2.Citation15,16
Starting point of the work presented here was the surprising observation that our reconstituted in vitro transcription system composed of highly purified human GTFs and RNAP II can support significant levels of INR-selective core promoter activity in the presence of NC2 when supplied with excess amounts of highly purified f:TFIID (). Importantly, excess amounts of f:TFIID did not support INR-specific basal transcription in the absence of NC2 (), confirming that our reconstituted system, including f:TFIID, is free of TAF-and INR-dependent cofactors that mediate INR-dependent stimulation of transcription independent of the presence of NC2.Citation16 Rather, our results suggested that a subpopulation of TFIID in f:TFIID is associated with an activity distinct from HMGA1 or Mediator that can mediate INR-dependent resistance to NC2 repression. Further investigations revealed variable amounts of TFIIA stably associated with a small proportion (≤ 10%) of TFIID in our f:TFIID preparations. TFIIA acts as an important positive cofactor for TFIID function.Citation1 TFIIA stabilizes TBP/TATA complexes through interactions with TBP opposite of the binding site for TFIIB, and with promoter DNA upstream of TATACitation20,36-39 and counteracts NC2 repression by competing for binding to TBP.Citation1 Importantly, TFIIA also promotes interactions between TFIID TAFs and core promoter DNA sequences downstream of the TATA regionCitation13,19,20 and has been shown to be an essential cofactor for TAF-dependent INR function both in the absence and presence of a TATA element.Citation14
Consistent with these observations, results of site-specific DNA crosslinking studies provided first evidence that TFIIA affects the positioning of individual TFIID TAFs relative to downstream promoter DNA within TFIID nucleoprotein complexes.Citation20 Recent high-resolution single particle electron microscopy studies revealed 2 distinct TFIID conformations.Citation21 Free TFIID exists predominantly in a ‘canonical’ state whereas TFIID bound to DNA adopts a ‘re-arranged’ state. Interestingly, TFIIA was found to stabilize both the canonical TFIID conformation in the absence of DNA as well as the re-arranged state of promoter-bound TFIID.Citation21
Whether and to what extent processing by Taspase1 affects TFIIA functions, in particular the ability of TFIIA to modulate TFIID (TBP) activity has been a longstanding unresolved question. Original studies reported that recombinant human TFIIA reconstituted from purified bacterially expressed TFIIAα/β precursor and TFIIAγ subunits can substitute for natural processed TFIIA isolated from human cells in promoter binding experiments with recombinant TBP and in vitro transcription experimentsCitation40-43 and unprocessed bacterially expressed recombinant human TFIIA has since be widely used in biochemical studies designed to investigate the function of TFIIA in early RNAP II transcription pre-initiation complex (PIC) assembly and in the regulation of both free TBP and the TFIID complex.
The key findings in this study are (i) that Taspase1 processing of TFIIA affects the conformation of TFIID/TFIIA promoter complexes and (ii) that Taspase1 processed TFIIA (35/19/12 kDa), but not unprocessed TFIIA (55/12 kDa), supports significant levels of INR-mediated resistance to NC2, and hence INR-selective core promoter activity in a highly purified in vitro transcription system lacking TAF- and INR-dependent cofactors Mediator and HMGA1.
We detect only the Taspase1-processed form of TFIIA in our highly purified f:TFIID preparations, in agreement with earlier studies.Citation24 We show further that Taspase1 processing increases binding of recombinant TFIIA to form stable TFIID/TFIIA promoter complexes in Mg2+ agarose EMSAs (), suggesting that Taspase1 processing enhances TFIIA interactions with TFIID. We observe clearly distinct electrophoretic mobilities of TATA-dependent TFIID/TFIIA/promoter complexes formed with unprocessed and Taspase1-processed TFIIA, indicative of significant differences in overall TFIID/DNA complex topology (). Importantly, the functional properties of recombinant TFIIA processed with Taspase1 in vitro and natural Taspase1-processed TFIIA isolated from HeLa nuclear extracts were indistinguishable both in DNA binding and in in vitro transcription assays (), confirming that in vitro Taspase1-processed recombinant TFIIA reconstituted with unprocessed TFIIAα/β and natural processed TFIIA (35, 19, 12 kDa) complex have comparable functional properties.
Altered electrophoretic mobility of TFIID/DNA complexes in Mg2+ EMSAs could reflect differences in TBP DNA bending, which contributes significantly to the reduced mobility of TBP or TFIID complexes in EMSAs.Citation28,32 Indeed, results of DNA-binding studies using quantitative fluorescence energy transfer (FRET) suggest that TFIIA binding reduces DNA bending of TATA DNA by TBP.Citation44 Thus altered TBP/TFIIA interactions caused by TFIIA processing, could directly affect the trajectory of the DNA within TFIID/promoter complexes, and thus alter interactions of individual TFIID TAFs (and of the remaining GTFs and RNAP II) with promoter DNA downstream of the TATA box region.
Previous studies demonstrated that TFIIA-induced TAF interactions with the INR core promoter element stabilize TFIID/promoter complexes in Mg2+ agarose EMSAs.Citation13 Consistent with these results, we observe enhanced resilience of promoter complexes formed with f:TFIID containing stably associated TFIIA to NC2 in the presence of an INR element. Importantly, this INR-dependent stabilization of TFIID promoter complexes in the presence of NC2 could only be observed with Taspase1-processed TFIIA but not with unprocessed TFIIA (). Disruption of TFIID/TFIIA complexes with increasing concentrations of NC2 resulted in a loss of TBP-induced DNA bending, indicated by a dramatic increase in electrophoretic mobility of TFIID/DNA complexes, and the formation of stable NC2/TFIID/DNA complexes lacking TATA-specificity (). These observations extend the results of earlier studies that investigated NC2 interaction with isolated TBPCitation17,18 and first demonstrated a loss of TATA-specificity and mobilisation of TBP complexes away from the TATA box upon NC2 binding.
Finally, we observed significant levels of INR-selective transcription initiation in the presence of NC2 when we supplemented our purified transcription system with either natural Taspase1-processed TFIIA isolated from HeLa nuclear extract or with Taspase1-processed recombinant TFIIA, but not with unprocessed recombinant TFIIA reconstituted with the 55 kDa TFIIAα/β precursor and TFIIAγ (). However, in the context of our in vitro transcription system, Taspase1 processing does not enhance TFIIA reversal of NC2 repression in the presence of an INR but instead diminishes the ability of TFIIA to counteract NC2 repression in the absence of an INR element (). Thus Taspase1 processing appears on one hand to weaken specific TFIIA interactions with TBP that are competitive with NC2 and, on the other hand, to stabilize conformational changes within the TFIID nucleoprotein complex that promote TAF/INR interactions and, in turn, assembly of a functional RNAP II transcription initiation complex.
Taken together, our results extend earlier studies suggesting a role of Taspase1 processing in regulating TFIIA association with TFIID, isolated TBP, and TBP-related factorsCitation24-26 and provide first evidence that Taspase1-processed heterotrimeric TFIIA (35/19/12 kDa) and unprocessed heterodimeric TFIIA (55/12 kDa) differentially affect TFIID topology and core promoter-specific TFIID functions. Thus regulation of TFIIA activity by Taspase1 processing adds yet another layer of complexity to the intricate functional interplay between positive and negative factors involved in the functional readout of core promoter sequence elements and the gene-specific regulation of transcription by cis-acting transcription activators and repressors.Citation6,15,16,45
How might Taspase1 cleavage of the TFIIAα/β precursor protein affect TFIIA interactions with TFIID? The Taspase1 cleavage site (269-LVLQVD|GTGDT)Citation23 is located in a functionally poorly defined non-conserved linker region in the TFIIAα/β precursor protein, that connects evolutionary conserved N- and C-termini, which are absolutely required for TFIIA functionCitation46 1. X-ray structure analysis of ternary TBP/TFIIA/TATA complexes, containing a minimal TFIIA complex composed only of the conserved TFIIAα/β N- and C-termini, revealed a boot-shaped TFIIA structure composed of 2 distinct domains orientated approximately 120°C from each other, in which all 3 TFIIA subunits are in intimate contact.Citation36,37,39 The top of the boot interacts with TBP and DNA upstream TATA and is a 12 strand β-barrel structure formed by interaction between the C-terminus of TFIIAα/β (TFIIAβ) with the C-terminal half of TFIIA. The foot of the boot is located away from TBP and TATA and consists of a 4-helix bundle composed of the N-terminus of TFIIAα/β (TFIIAα) and the N-terminal half of TFIIAγ. It is important to note that the non-conserved linker region of TFIIA is missing in these structures. It seems obvious that this region could have a significantly impact on the relative orientation of the 2 conserved TFIIA domains and, at the same time, greatly contribute to TFIIA/TAF interactions in the context of TFIID. Cleavage by Taspase1 within this region might affect the conformational flexibility of the entire TFIIA structure and, consequently, TFIIA/TBP and/or TFIIA/TAF interactions, the relative orientation of TBP and TAF subunits within TFIID, and the ability of TFIID to interact with specific core promoter elements downstream of TATA. Further work is needed to address these important questions.
Materials & Methods
Promoter templates
The TATA/INR model promoter template pPGTdT(TATA/+59) is a pGEM7Zf (Promega) derivative containing within an ApaI/ BamHI insert mTdT core promoter sequences from −41 to +59 with a consensus TATA box at position +30Citation10,15 in front of 5 GAL4 binding sites. The corresponding TATA-only model promoter template pPGTdT(TATAΔINR/−59)-L is a derivative of pPGTdT(TATA/−59) containing 6 point mutations which eliminate INR function without affecting start site selection and a 26-bp polylinker sequence downstream of the mTdT core promoter region (−41/+52), which does not affect core promoter activity.Citation10,15 EMSA probes TI, T, Δ were generated by PCR using plasmids pTOG5TdT(−41TATA/+33), pVC5GTdT-TATAΔINR and pVC5GTdTΔINR, respectively. pTOG5TdT(−41TATA/+33) is a pGEM7Zf (Promega) derivative containing, within a PstI/ BamHI insert, mTdT core promoter sequences from −41 to +33 with a consensus TATA box at position +30Citation10 in front of 5 GAL4 binding sites and serves as TATA/INR model promoter (TI; ). pVC5GTdT-TATAΔINR derives from pTOG5TdT(−41TATA/+33) and contains 6 point mutations identical to pPGTdT(TATAΔINR/−59)-L, which eliminate the INR, and serves as TATA-only (TI; ) promoter template. pVC5GTdTΔINR derives from pVC5GTdT-TATAΔINR and contains the natural TATA-less mTdT promoter from −41 to +33 with the same 6 point mutations to eliminate the INR and serves as ‘null’ promoter template lacking any known core promoter elements (Δ; ).
In vitro RNA polymerase II transcription system
All proteins were stored at −70°C in BC-buffer (20 mM Tris·Cl pH7.9 @4°C, 20% glycerol, 0.2 mM EDTA pH 8.0, 10 mM β-mercaptoethnanol) containing 100 mM KCl (BC-100) unless indicated otherwise. The preparation of recombinant RNA polymerase II general transcription factors (RNAP II GTFs; rTBP, rTFIIA, -IIB, -IIE, -IIF) and FLAG:epitope-tagged TFIID (f:TFIID) was described previously.Citation15,20,27 TFIIH was purified from the HeLa nuclear extract phosphocellulose (P11, Whatman) 0.85 M KCl fractionCitation47 as follows. The eluate was dialised in BC-100 and passed through DEAE FF (GE Healthcare Life Sciences). The flow-through was then loaded onto a Resource S (GE Healthcare Life Sciences) column and eluted with a 20 column volumes linear gradient from 0.1 to 0.3 M KCl in BC buffer. TFIIH-containing fractions (170–240 mM KCl) were identified by immunoblotting, pooled and precipitated with (NH4)2SO4 at 46% saturation. After ultracentrifugation, the protein pellet was resuspended in BC buffer. The sample was adjusted to 1.4 M (NH4)2SO4 and, after ultracentrifugation to remove any insoluble material, further purified on a Source 15 ISO (isopropyl; GE Healthcare Life Sciences) column developed with a linear gradient from 1.4 to 0 M (NH4)2SO4 in BC buffer. Finally, TFIIH-containing fractions (1.2 to 1 M (NH4)2SO4) were pooled, concentrated on NanoSep spin columns (Pall; cut-off 10 kDa) and further purified by size exclusion chromatography on Superdex 200 (GE Healthcare Life Sciences) in BC-buffer containing 400 mM KCl.
RNA polymerase II (RNAP II) was purified from HeLa nuclear pellet as previously describedCitation48 with the following modifications. All chromatography steps were carried out in TGEED buffer (50 mM Tris·Cl pH 7.2 @ 21°C, 20% glycerol, 1 mM EDTA, 1 mM EGTA, 2 mM DTT, 1 mM NaF, 10 mM NaPPi) in the presence of protease inhibitors (Sigma P8430). After precipitation with (NH4)2SO4 samples were adjusted to 70 mM (NH4)2SO4 by dialysis and further purified on DEAE FF (GE Healthcare Life Sciences) developed with a 10 column volume linear gradient from 80 to 600 mM (NH4)2SO4 followed by chromatography on a Heparin POROS column (Life Technologies) developed with a 5 column volumes linear gradient from 80 to 600 mM (NH4)2SO4 and finally by chromatography on a TSKgel DEAE 5PW column (Tosoh Bioscience) developed with a 20 column volumes linear gradient from 80 to 600 mM (NH4)2SO4. Fractions containing highly purified RNAP II were identified by immunoblot analysis and SDS PAGE and silver staining. Purified RNAP II was dialyzed in BC-100, snap-frozen in liquid N2 and stored at −70°C.
Natural TFIIA (nTFIIA 35/19/12 kDa, see ) was purified from the HeLa nuclear extract 0.1 M KCl phosphocellulose (P11, Whatman) fractionCitation47 by ion exchange chromatography on Mono Q resin (GE Healthcare Life Sciences) followed by affinity chromatography on Ni2+ NTA resin (Qiagen) and, finally, TBP-affinity chromatography using 6His:epitope-tagged TBP from Saccaromyces cerevisae (6His:scTBP) coupled to Affi-Gel 10 resin BioRad). TFIIA was eluted from the 6His:scTBP affinity column with BC buffer containing 700 mM KCl (BC-700), snap-frozen in liquid N2 and stored at −70°C.
Taspase1 processing of recombinant bacterially expressed TFIIA (55/12 kDa) was carried out as follows: 50 µg rTFIIA (;Citation15) was cleaved with 50 µg recombinant 6His:epitope-tagged Taspase1 enzymeCitation23,35 for 1 h at 37°C in 1.4 ml BC buffer containing 100 mM KCl, 1.5 mM MgCl2, 0.1 mg/ml insulin and 2 mM dithiothreitol (DTT). Following protease cleavage, Taspase1-processed rTFIIA (35/19/12 kDa) was separated from Taspase1 by ion exchange chromatography on Resource Q (GE Healthcare Life Sciences) and phosphocellulose (P11; Whatman) resins.
Antibodies
Polyclonal antibodies for human TBP, TAFs 4, 5, 6, 7, 9, 12 and human TFIIA, raised against recombinant TFIIAα/β precursor, () were a kind gift from Robert G Roeder.
In vitro transcription
Two-template in vitro transcription assays and primer extension to analyze in parallel basal transcription from mTdT promoter variants containing only TATA (TATA) or TATA and INR elements (TATA/INR) have been described previously.Citation15
Mg2+ agarose electrophoretic mobility shift assays
Mg2+ EMSAs were carried out as described.Citation28 Briefly, 5fmol 32P-labeled 255 bp promoter template was incubated with purified proteins as described in the figure legends for 45 min at 30°C under transcription conditions in the absence of NTPs. Protein/DNA complexes were resolved at 4°C in 1.4% or 1.5% agarose gels in 1x TBE buffer containing 5mM MgCl2. After electrophoresis, gels were dried on DE81 chromatography paper (Whatman) and signals were visualized by autoradiography.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Petra Gross for help in the initial stages of the study. We are also grateful to Robert G Roeder for providing reagents used in this study.
Funding
This work was supported by core funding from Marie Curie Cancer Care and by Incentive Funding for Rated Researchers by the National Research Foundation of South Africa (CPRR13080225272, Grant No. 81120) to TO.
References
- Thomas MC, Chiang C-M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105-78; PMID:16858867; http://dx.doi.org/10.1080/10409230600648736
- Papai G, Weil PA, Schultz P. New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev 2011; 21:219-24; PMID:21420851; http://dx.doi.org/10.1016/j.gde.2011.01.009
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem 2003; 72:449-79; PMID:12651739; http://dx.doi.org/10.1146/annurev.biochem.72.121801.161520
- Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol 2008; 20:253-9; PMID:18436437; http://dx.doi.org/10.1016/j.ceb.2008.03.003
- Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev 2001; 15:2503-8; PMID:11581155; http://dx.doi.org/10.1101/gad.937701
- Gross P, Oelgeschläger T. Core promoter-selective RNA polymerase II transcription. Biochem Soc Symp 2006:225-36; PMID:16626302
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Developmental Biology 2010; 339:225-9; PMID:19682982; http://dx.doi.org/10.1016/j.ydbio.2009.08.009
- Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A 1990; 87:4509-13; PMID:2141169; http://dx.doi.org/10.1073/pnas.87.12.4509
- Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev 1994; 8:821-9; PMID:7926770; http://dx.doi.org/10.1101/gad.8.7.821
- Martinez E, Chiang C-M, Ge H, Roeder RG. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J 1994; 13:3115-26; PMID:7518774
- Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 1995; 81:1115-25; PMID:7600579; http://dx.doi.org/10.1016/S0092-8674(05)80016-9
- Martinez E, Zhou Q, L'Etoile ND, Oelgeschläger T, Berk AJ, Roeder RG. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci U S A 1995; 92:11864-8; PMID:8524864; http://dx.doi.org/10.1073/pnas.92.25.11864
- Emami KH, Jain A, Smale ST. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev 1997; 11:3007-19; PMID:9367983; http://dx.doi.org/10.1101/gad.11.22.3007
- Martinez E, Ge H, Tao Y, Yuan CX, Palhan V, Roeder RG. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol 1998; 18:6571-83; PMID:9774672
- Malecová B, Gross P, Boyer-Guittaut M, Yavuz S, Oelgeschläger T. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem 2007; 282:24767-76; PMID:17584739; http://dx.doi.org/10.1074/jbc.M702776200
- Xu M, Sharma P, Pan S, Malik S, Roeder RG, Martinez E. Core promoter-selective function of HMGA1 and Mediator in Initiator-dependent transcription. Genes Dev 2011; 25:2513-24; PMID:22156211; http://dx.doi.org/10.1101/gad.177360.111
- Gilfillan S, Stelzer G, Piaia E, Hofmann MG, Meisterernst M. Efficient binding of NC2.TATA-binding protein to DNA in the absence of TATA. J Biol Chem 2005; 280:6222-30; PMID:15574413; http://dx.doi.org/10.1074/jbc.M406343200
- Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol 2007; 14:1196-201; PMID:17994103; http://dx.doi.org/10.1038/nsmb1328
- Lieberman PM, Berk AJ. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes Dev 1994; 8:995-1006; PMID:7926793; http://dx.doi.org/10.1101/gad.8.9.995
- Oelgeschläger T, Chiang C-M, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature 1996; 382:735-8; PMID:8751448; http://dx.doi.org/10.1038/382735a0
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID Binds to Core Promoter DNA in a Reorganized Structural State. Cell 2013; 152:120-31; PMID:23332750; http://dx.doi.org/10.1016/j.cell.2012.12.005
- Høiby T, Zhou H, Mitsiou DJ, Stunnenberg HG. A facelift for the general transcription factor TFIIA. Biochim Biophys Acta 2007; 1769:429-36; PMID:17560669; http://dx.doi.org/10.1016/j.bbaexp.2007.04.008
- Zhou H, Spicuglia S, Hsieh JJ-D, Mitsiou DJ, Høiby T, Veenstra GJC, Korsmeyer SJ, Stunnenberg HG. Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol Cell Biol 2006; 26:2728-35; PMID:16537915; http://dx.doi.org/10.1128/MCB.26.7.2728-2735.2006
- Mitsiou DJ, Stunnenberg HG. TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAalphabeta precursor and the TFIIAgamma subunit. Mol Cell 2000; 6:527-37; PMID:11030333; http://dx.doi.org/10.1016/S1097-2765(00)00052-6
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci U S A 1999; 96:13720-5; PMID:10570139; http://dx.doi.org/10.1073/pnas.96.24.13720
- Oyama T, Sasagawa S, Takeda S, Hess RA, Lieberman PM, Cheng EH, Hsieh JJ. Cleavage of TFIIA by taspase1 activates TRF2-specified mammalian male germ cell programs. Dev Cell 2013; 27:188-200; PMID:24176642; http://dx.doi.org/10.1016/j.devcel.2013.09.025
- Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder RG. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J 1993; 12:2749-62; PMID:7687540
- Zerby D, Lieberman PM. Functional analysis of TFIID-activator interaction by magnesium-agarose gel electrophoresis. Methods 1997; 12:217-23; PMID:9237166; http://dx.doi.org/10.1006/meth.1997.0474
- Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, Meisterernst M, Burley SK. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 2001; 106:71-81; PMID:11461703; http://dx.doi.org/10.1016/S0092-8674(01)00417-2
- Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J 1996; 15:3105-16; PMID:8670811
- Mermelstein F, Yeung K, Cao J, Inostroza JA, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev 1996; 10:1033-48; PMID:8608938; http://dx.doi.org/10.1101/gad.10.8.1033
- Horikoshi M, Bertuccioli C, Takada R, Wang J, Yamamoto T, Roeder RG. Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc Natl Acad Sci U S A 1992; 89:1060-4; PMID:1736286; http://dx.doi.org/10.1073/pnas.89.3.1060
- Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993; 365:520-7; PMID:8413605; http://dx.doi.org/10.1038/365520a0
- Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993; 365:512-20; PMID:8413604; http://dx.doi.org/10.1038/365512a0
- Hsieh JJ-D, Cheng EH-Y, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 2003; 115:293-303; PMID:14636557; http://dx.doi.org/10.1016/S0092-8674(03)00816-X
- Geiger JH, Hahn S, Lee S, Sigler PB. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 1996; 272:830-6; PMID:8629014; http://dx.doi.org/10.1126/science.272.5263.830
- Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 1996; 381:127-51; PMID:8610010; http://dx.doi.org/10.1038/381127a0
- Lagrange T, Kim TK, Orphanides G, Ebright YW, Ebright RH, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci U S A 1996; 93:10620-5; PMID:8855228; http://dx.doi.org/10.1073/pnas.93.20.10620
- Bleichenbacher M, Tan S, Richmond TJ. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J Mol Biol 2003; 332:783-93; PMID:12972251; http://dx.doi.org/10.1016/S0022-2836(03)00887-8
- DeJong J, Roeder RG. A single cDNA, hTFIIA/alpha, encodes both the p35 and p19 subunits of human TFIIA. Genes Dev 1993; 7:2220-34; PMID:8224848; http://dx.doi.org/10.1101/gad.7.11.2220
- Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev 1994; 8:2336-48; PMID:7958900; http://dx.doi.org/10.1101/gad.8.19.2336
- Ozer J, Moore PA, Bolden AH, Lee A, Rosen CA, Lieberman PM. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev 1994; 8:2324-35; PMID:7958899; http://dx.doi.org/10.1101/gad.8.19.2324
- DeJong J, Bernstein R, Roeder RG. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci U S A 1995; 92:3313-7; PMID:7724559; http://dx.doi.org/10.1073/pnas.92.8.3313
- Hieb AR, Halsey WA, Betterton MD, Perkins TT, Kugel JF, Goodrich JA. TFIIA changes the conformation of the DNA in TBP/TATA complexes and increases their kinetic stability. J Mol Biol 2007; 372:619-32; PMID:17681538; http://dx.doi.org/10.1016/j.jmb.2007.06.061
- Martinez E. Core promoter-selective coregulators of transcription by RNA polymerase II. Transcription 2012; 3; PMID:23117823; http://dx.doi.org/10.4161/trns.21846
- Kang JJ, Auble DT, Ranish JA, Hahn S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol 1995; 15:1234-43; PMID:7862117
- Reinberg D, Horikoshi M, Roeder RG. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem 1987; 262:3322-30; PMID:3818643
- Castaño E, Gross P, Wang Z, Roeder RG, Oelgeschläger T. The C-terminal domain-phosphorylated IIO form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc Natl Acad Sci U S A 2000; 97:7184-9; PMID:10852970; http://dx.doi.org/10.1073/pnas.140202297
