 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
During transcription, RNA polymerase moves downstream along the DNA template and maintains a transcription bubble. Several recent structural studies of transcription complexes with a complete transcription bubble provide new insights into how RNAP couples the nucleotide addition reaction to its directional movement.
During transcription, the DNA-dependent RNA polymerase (RNAP) moves downstream along template DNA and synthesizes RNA molecules composed of hundreds to thousands of nucleotides without the need of a helicase to unwind nucleic acid duplexes. The net effect of each RNA extension step during transcription is that RNAP takes one nucleoside triphosphate, extends the nascent RNA by one nucleotide, and generates one pyrophosphate ion (PPi). This is an energetically favorable reaction with free energy change of about −5.6 kcal/mol under standard conditions.Citation1,2 It remains obscure how the RNAP couples this chemical energy output to its directional movement along the DNA template.
RNAP and the transcription bubble
Cellular RNAP are large multi-subunit protein complexes that display a crab-claw-shaped architecture with a central cleft and two pincers, one of which, the “clamp,” is mobile.Citation3,4 The DNA template binds in the central cleft, and NTP substrates enter the active center through one channel, while the RNA product exits the active center through another channel. The NTP entrance channel (secondary channel) is separated from the central cleft (primary channel) by the long bridge helix (BH) that traverses the central cleft. The RNAP active center is located deep inside on the floor of the central cleft, and next to the secondary channel.
While moving along the DNA during transcription, RNAP maintains a transcription bubble with single-stranded non-template DNA and a segment of hybrid formed by the template-strand DNA and the nascent RNA. Several conserved structural elements lining the inner surface of the RNAP channels play critical roles in maintaining the downstream and upstream edges of the transcription bubble ().
Figure 1. (A) The mammalian transcription elongation complex (TEC)Citation18 and (B) the E. coli sS-transcription initiation complex (sS-TIC)Citation17 with a complete bubble. The nucleic acids are shown as colored spheres: nontemplate strand, blue; template strand, purple; RNA, green. The RNAP core enzymes are shown as grey surface diagrams and the structure elements critical for bubble maintenance are shown as colored tubes or ribbons: lid, orange; rudder, red; FL1, wheat; FL2, yellow; BH, red; TL, cyan. (C) and (D) are schematic representations of a post-translocated TEC and a pre-translocated TIC respectively. The TEC contains a 10-nt bubble with no extra unpaired DNA residues at both edges of the bubble.
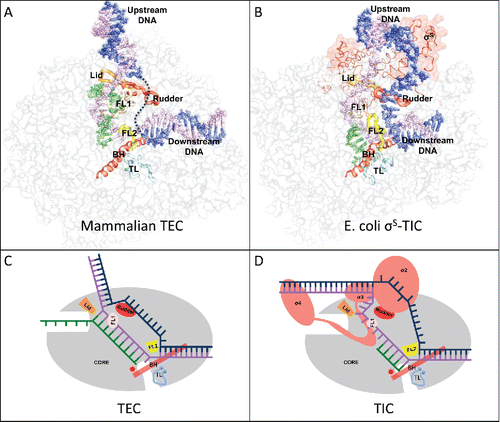
The downstream edge of the transcription bubble is essentially the same for both transcription initiation and elongation. The downstream DNA separation is maintained by the conserved structural element called the fork loop 2 (FL2); and blocking at the downstream end of the DNA–RNA hybrid are the bridge helix and a conserved mobile element called trigger loop (TL). Conformational changes of the bridge helix and the trigger loop remodel the active site and modulates the enzymatic activity of cellular RNAP.Citation3-10
The bacterial FL2 is rigid and well-ordered even in the absence of downstream DNA. Various structural studies of bacterial transcription complexes show that the downstream edge of the DNA bubble is well-defined and maintained by the rigid FL2 of the bacterial RNAP, and there are no single stranded residues bridging the downstream DNA duplex and the templating DNA residue at the active site. The eukaryotic FL2 appears to be flexible, and there remains some uncertainty about the downstream edge of a eukaryotic transcription bubble.
RNAP makes sequence-specific interactions with the upstream promoter DNA during transcription initiation. In bacteria, the σ factor of the RNAP holoenzyme recognizes the consensus −35 and −10 hexamers and forms base-specific interactions with the −11A and −7T residues of the promoter DNA. The sequence-specific interactions help separate the DNA strands starting from the promoter −11 position, and the DNA bubble expands as RNAP moves downstream for RNA synthesis. Bacterial RNA polymerase preferentially initiates RNA synthesis seven or eight nucleotides (nt) downstream from the −10 hexamer,Citation11,12 suggesting that an initiation bubble of about 13 nt is likely the energetically most favorable state. Structural studies show that RNAP comfortably accommodates both the non-template and template DNA strands of a 13-nt bubble.Citation13-15 When the initiation bubble expands to 14 nt or larger, the non-template strand DNA starts to loop out and the template strand DNA presses onto the σ3 domain (DNA scrunching), and the transcription complex becomes stressed.Citation16,17 An initiation bubble of 16 nt or larger would start causing the σ3 domain to separate from the RNAP core. It is likely that the DNA bubble never expands significantly larger than 16 nt during transcription initiation before σ3–σ4 domains separate from the core for transition into the elongation phase.
There remains no consensus about the upstream edge of an elongation bubble. A recent Cryo-EM study of the mammalian transcription elongation complexes with a complete DNA bubble suggests that the template strand DNA base pairs with the non-template DNA immediately after DNA–RNA separation by the “lid” element.Citation18 The DNA separation is maintained by the universally conserved “rudder” element and the fork loop 1 (FL1). This DNA annealing immediately after DNA–RNA separation was evident for the single-subunit RNAP,Citation19 and may also be true in the crystal structure of a yeast RNAP II transcription elongation complex.Citation20 It was shown that bacterial transcription elongation complexes contain a 9- or 10-bp hybrid depending on the translocation status.Citation5,21 If there are no single-stranded DNA residues bridging the upstream DNA duplex and the hybrid, the transcription bubble might be maintained at a size as small as 10 nt by bacterial RNAPs during processive transcription elongation. Since the free energy levels associated with different translocation states of a transcription complex would be profoundly affected by the making and breaking of nucleic acid base stacking at both edges of the transcription bubble, this lack of extra unpaired DNA residues at both edges of an elongation bubble could be of critical importance in coordinating transcription translocation by the RNAPs.
Nucleotide addition cycle
RNA synthesis in both transcription initiation and elongation involves nucleotide addition cycles (NAC) comprising four fundamental steps: translocation, NTP binding, catalysis, and PPi release (). At the end of the previous NAC, the nascent RNA 3′-end still occupies the nucleotide addition site. After RNAP translocates along the DNA template to move the RNA 3′ residue upstream and bring the next template DNA base into the active site, NTP is allowed to enter the active site and base pair with the DNA template to extend the RNA by one nucleotide, generating a PPi and putting the new RNA 3′-end at the nucleotide addition site. Release of PPi completes the cycle and resets the stage for the next round of nucleotide addition. The reverse reaction, called pyrophosphate exchange or pyrophosphorolysis, also happens in the presence of excessive PPi and leads to formation of NTP and cleavage of the terminal nucleotides.Citation22-24
Figure 2. The steps of the nucleotide addition cycle are related to six basic states E1 to E6 with regard to translocation, NTP/PPi association and active site opening. These six states include three quickly equilibrating groups QE1 (pre-translocated E1 and post-translocated E2), QE2 (NTP-associated E3 and E4) and QE3 (PPi-associated E5 and E6), which are connected by slower diffusion-controlled and chemical reaction steps. E1 and E2 states are also likely equilibrating quickly with others states such as active site closed E0, backtracked E7 and hypertranslocated E8 states. Both E7 and E8 represent ensembles of translocation states, however, it is likely that only the 1-nt backtracking and 1-nt hyper forward translocation states are of significance in many cases. Solid arrows represent rapid processes; dashed arrows represent potentially rate-limiting processes. Some states (such as the active site-open E1, E2, E7 and E8 states) are more ratchetable and could become ratcheted/clamp-opened (E9 states). The off-pathway ratcheting processes (rate constants kr's) are expected to be slower than the on-pathway processes. TL/open, active site-open trigger loop conformation; TH/close, active site-closed trigger helices conformation.
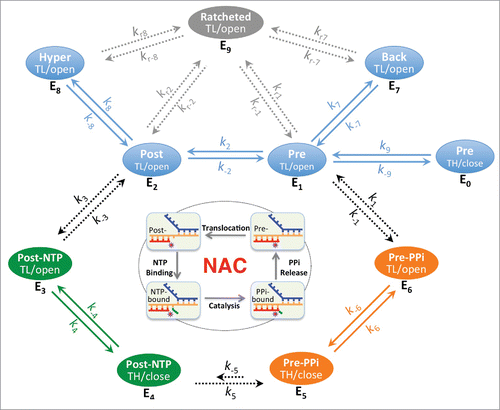
NTP binds to the post-translocated RNAP with an open active site. In T7 RNA polymerase, it was proposed that PPi release is tightly coupled with active site opening and RNAP translocation.Citation25 In E. coli RNA polymerase, this tight coupling was not observed, but it was suggested that the PPi release occurs shortly before or concurrently with the translocation,Citation26 and that transcription translocation might require an unfolded TL.Citation27 TL unfolding allows access to the active site through the secondary channel. Previous structural studies of cellular RNAP complexes with nucleic acids displayed an overwhelming preference for the post-translocated state with an open active site,Citation17 and thus gave an impression that both post-translocation and RNAP active site opening are energetically favorable. However, recent studies showed a pre-translocated complex with a closed RNAP active site in both the presence and absence of a bound PPi, suggesting that PPi release, TL unfolding, and transcript translocation are separable events during transcription.Citation16,17 Since the active site opening of a closed complex appears to be too small for a PPi to freely pass through,Citation17 TL unfolding might be necessary before PPi can dissociate from the RNAP active site. The cycling process of nucleotide addition might be divided into six basic states (E1 to E6 in ). NTP binds only to the E2 state and would shift the equilibrium of transcription to the post-translocated states as was shown previously,Citation28 whereas PPi favors binding to the pre-translocated E1 state and would shift the equilibrium in the opposite direction.
The movement of RNAP along the nucleic acid scaffold might be visualized in terms of thermally controlled transitions between free energy minima associated with translocation states along the template. This thermally controlled transition is expected to be a rapid event that happens in microseconds time scale. In addition to the quick motion between the pre- and post-translocation states, it is known that translocation can go further in both directions, forming the backtracked E7 and the hyper forward translocated E8 states. It is likely that, after each nucleotide addition, enzyme states E0, E1, E2, E7, and E8 equilibrate quickly (QE1, ). The enzyme distribution among these equilibrating states is determined by the relative free energy levels, which might be estimated using the nearest-neighbor stacking parameters of nucleic acids.Citation29,30 Quick changes in equilibrium are also expected between states E3 and E4 (QE2) and between states E5 and E6 (QE3) as they involve only local conformational changes in RNAP. Two diffusion-controlled processes (between E2 and E3 and between E6 and E1) and the chemical reaction step (between E4 and E5) connect these quickly equilibrating groups to form the nucleotide addition cycle.
A kinetic model of the nucleotide addition cycle
According to the diagram shown in , the net rate of nucleotide addition might be represented as(1)
(1) where En represents the concentration of individual enzyme species En, and the kn's are the reaction constants as shown in . The following analyses are based on the assumption that the chemical reaction (E4 to E5) is the rate-limiting step with quick changes in equilibrium among species of each QE group and between E2 and E3 after each nucleotide addition. Under assay conditions with very low NTP concentrations, the diffusion-controlled E2 to E3 step could become the rate-limiting step, and similar analyses might also be made but are not included here.
(1). When the concentration of pyrophosphate is low and E5 is negligible, then(2)
(2)
By applying the equilibrating processes, we can get a Michaelis–Menten equation(3)
(3) with
(4)
(4)
(5)
(5) where ETotal is the total concentration of all the En species actively involved in the nucleotide addition cycle; S is the concentration of substrate NTP, KdS is the NTP dissociation constant,
(6)
(6)
Keq4 is the equilibrium constant between QE2 species,(7)
(7) and fE2 is the fraction of E2 in QE1 species,
(8)
(8)
Based on this kinetic model, Km increases significantly if the post-translocated state is energetically unfavorable, and vmax is strongly affected by the QE2 equilibrium. Non-cognate substrates likely disfavor active site closure and would display reduced vmax values.
(2). When pyrophosphate is present in a significant amount, such as in vivo, PPi-containing species become non-negligible, then(9)
(9) where P is the concentration of pyrophosphate, and KdP is the pyrophosphate dissociation constant,
(10)
(10)
Keq6 is the equilibrium constant between QE3 species,(11)
(11) and fE1 is the fraction of E1 in QE1 species,
(12)
(12)
Since , the reverse pyrophosphorolysis reaction is negligible under normal assay conditions, and equation (Equation9
(9)
(9) ) could be simplified to a Michaelis–Menten equation-like form with the same vmax as shown in equation (Equation4
(4)
(4) ) and a Km of the form
(13)
(13)
When the PPi-containing species become non-negligible (), the Km for the substrate NTP would be significantly affected by the presence of pyrophosphate. When (
) and pyrophosphorolysis remains negligible, the nucleotide addition rate might become relatively slow and proportional to fE2/fE1:
(14)
(14)
Since fE1 and fE2 are expected to be strongly sequence-dependent, this might be the mechanistic basis for the sequence-dependent frequent short pauses during transcription. By applying nearest-neighbor stacking analysesCitation29,31 to the transcription bubble models shown in , it can be seen that some sequences strongly prefer the pre-translocated E1 state over the post-translocated E2 state (). This kinetic model makes predictions consistent with the observed pauses during transcription elongationCitation32,33 (unpublished analyses).
On the other hand, this model also suggests that the presence of the NTP that is complementary to the next template residue would promote PPi dissociation, which could be used to explain the effects of the incoming NTP in the measured rates of PPi releaseCitation24 and nucleotide additionCitation34 without involving a secondary NTP-binding site in the RNAP. Structural studies of transcription complexes showed no evidence for a secondary NTP-binding site in the RNAP.
In an extreme case, when the reverse pyrophosphorolysis reaction becomes non-negligible, the chemical reaction might reach equilibrium or even be reversed. According to equation (Equation9(9)
(9) ), an equilibrium of nucleotide addition (
) can be reached by increasing the [PPi]/[NTP] ratio so that
(15)
(15)
Although k5 is expected to be about 104 faster than k−5, when (), the chemical reaction could easily reach equilibrium in an in vitro assay as PPi accumulates with each nucleotide addition, which is probably what we observed in the σS-TIC crystals.Citation17
Regulation of transcription
As shown in , transcription can be regulated at various steps of the nucleotide addition cycle. Some factors, such as NTP or PPi analogs, modulate the formation or distribution of QE2 or QE3 species, and thus might act as competitive regulators; whereas many other factors act at the translocation step and affect the distribution of QE1 species. It is known that upstream RNA hairpin formation could also be coupled to RNAP forward translocation, and similarly, DNA or RNA translocases could interact directly with the nucleic acid and power either forward or backward translocation by ATP hydrolysis.Citation35 Some DNA sequences favor the pre-translocation state and cause transcriptional pausesCitation32,33; whereas sequence-specific interactions between the RNAP and the DNA could also alter the translocation preference and play a role in regulating transcription efficiency.Citation33,36
As shown by Sekine et al.,Citation37 transcription complexes at some states are more ratchetable, and thus could become ratcheted. A ratcheted complex contains a wide-open central cleft caused by the swing movement of the clamp and a ratcheting movement between the core and shelf modules that form the active center.Citation38 Ratchetable and ratcheted states are expected to be inter-convertible, presumably at a slow rate. A known slow process similar to the ratcheting movement is the rate-limiting I1 to I2 conversion, which likely involves RNAP clamp opening, during the formation of an RNAP-promoter open complex.Citation39 Some of the ratcheted complexes, such as those with extensive backtracking or an RNA hairpin in the RNA exit channel, are long-lived. Since ratcheting involves BH bending, it is likely that only those states with an unfolded TL, such as states E1, E2, E7, and E8, are ratchetable. Extended pauses at some ratchetable states would increase the chance to form long-lived ratcheted states. Apparently, formation of the off-pathway ratcheted complexes would decrease the effective total amount of complexes in active transcription (ETotal in equations (Equation4(4)
(4) ) and (Equation9
(9)
(9) )).
The universally conserved NusG-family proteins bind the clamp helices of the RNAP near the upstream edge of the transcription bubble, and make bridging interactions between the two pincers of the RNAP central cleft. NusG bridging of the two pincers could potentially prevent RNAP from ratcheting open. This inhibition of RNAP ratcheting is likely what makes NusG essential for E. coli viability. Ratcheted or more ratchetable complexes, such as those with backtracked residues, are frequently targeted by Gre factors from the secondary channel for cleavage of backtracked RNA residues. ppGpp binds on the outer surface of RNAPCitation40 and thus could increase the ratchetability of RNAP for its cooperative role with DksA, another RNAP secondary channel-binding protein.Citation41 It is known that RapA binds at the RNA exit channel and could potentially suppress RNA hairpin formation and promote backtranslocation.Citation35 It remains to be revealed how other factors, such as NusA, interact with the exiting RNA to modulate transcription termination or antitermination.
Abbreviations
| BH | = | bridge helix |
| NTP | = | nucleoside triphosphate |
| PPi | = | pyrophosphate |
| RNAP | = | RNA polymerase |
| TL | = | trigger loop |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH grant GM22778 (to T.A.S.). T.A.S. is an investigator of the Howard Hughes Medical Institute.
References
- Frey PA, Arabshahi A. Standard free energy change for the hydrolysis of the α, β-phosphoanhydride bridge in ATP. Biochemistry 1995; 34:11307-11310; PMID:7547856; http://dx.doi.org/10.1021/bi00036a001
- Dickson KS, Burns CM, Richardson JP. Determination of the free-energy change for repair of a DNA phosphodiester bond. J Biol Chem 2000; 275:15828-15831; PMID:10748184; http://dx.doi.org/10.1074/jbc.M910044199
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 2003; 13:31-39; PMID:12581657; http://dx.doi.org/10.1016/S0959-440X(02)00005-2
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol 2002; 12:89-97.; PMID:11839495; http://dx.doi.org/10.1016/S0959-440X(02)00294-4
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature 2007; 448:163-168; PMID:17581591; http://dx.doi.org/10.1038/nature05931
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 2006; 127:941-954; PMID:17129781; http://dx.doi.org/10.1016/j.cell.2006.11.023
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell 2007; 27:406-419; PMID:17679091; http://dx.doi.org/10.1016/j.molcel.2007.06.008
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 2001; 292:1876-1882; PMID:11313499; http://dx.doi.org/10.1126/science.1059495
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 2002; 417:712-719; PMID:12000971; http://dx.doi.org/10.1038/nature752
- Mejia YX, Nudler E, Bustamante C. Trigger loop folding determines transcription rate of Escherichia coli's RNA polymerase. Proc Natl Acad Sci USA 2015; 112:743-748; PMID:25552559; http://dx.doi.org/10.1073/pnas.1421067112
- Vvedenskaya IO, Zhang Y, Goldman SR, Valenti A, Visone V, Taylor DM, Ebright RH, Nickels BE. Massively Systematic Transcript End Readout, “MASTER:” Transcription Start Site Selection, Transcriptional Slippage, and Transcript Yields. Mol Cell 2015; 60:953-965; PMID:26626484; http://dx.doi.org/10.1016/j.molcel.2015.10.029
- Shimada T, Yamazaki Y, Tanaka K, Ishihama A. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One 2014; 9:e90447; PMID:24603758; http://dx.doi.org/10.1371/journal.pone.0090447
- Feng Y, Zhang Y, Ebright RH. Structural basis of transcription activation. Science 2016; 352:1330-1333; PMID:27284196; http://dx.doi.org/10.1126/science.aaf4417
- Bae B, Chen J, Davis E, Leon K, Darst SA, Campbell EA. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife 2015; 4:e08505; PMID:26349034; http://dx.doi.org/10.7554/eLife.08505
- Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife 2015; 4:e08504; PMID:26349032; http://dx.doi.org/10.7554/eLife.08504
- Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell 2015; 58:534-540; PMID:25866247; http://dx.doi.org/10.1016/j.molcel.2015.03.010
- Liu B, Zuo Y, Steitz TA. Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci U S A 2016; 113:4051-4056; PMID:27035955; http://dx.doi.org/10.1073/pnas.1520555113
- Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature 2016; 529:551-554; PMID:26789250; http://dx.doi.org/10.1038/nature16482
- Schwinghammer K, Cheung AC, Morozov YI, Agaronyan K, Temiakov D, Cramer P. Structure of human mitochondrial RNA polymerase elongation complex. Nat Struct Mol Biol 2013; 20:1298-1303; PMID:24096365; http://dx.doi.org/10.1038/nsmb.2683
- Barnes CO, Calero M, Malik I, Graham BW, Spahr H, Lin G, Cohen AE, Brown IS, Zhang Q, Pullara F, et al. Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble. Mol Cell 2015; 59:258-269; PMID:26186291; http://dx.doi.org/10.1016/j.molcel.2015.06.034
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 2007; 448:157-162; PMID:17581590; http://dx.doi.org/10.1038/nature05932
- Rozovskaya TA, Chenchik AA, Beabealashvilli R. Processive pyrophosphorolysis of RNA by Escherichia coli RNA polymerase. FEBS Lett 1982; 137:100-104; PMID:6175533; http://dx.doi.org/10.1016/0014-5793(82)80323-2
- Maitra U, Hurwitz J. The role of deoxyribonucleic acid in ribonucleic acid synthesis. 13. Modified purification procedure and additional properties of ribonucleic acid polymerase from Escherichia coli W. J Biol Chem 1967; 242:4897-4907; PMID:4293779
- Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol 2008; 381:1106-1113; PMID:18638485; http://dx.doi.org/10.1016/j.jmb.2008.06.089
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 2004; 116:393-404; PMID:15016374; http://dx.doi.org/10.1016/S0092-8674(04)00120-5
- Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res 2012; 40:7442-7451; PMID:22570421; http://dx.doi.org/10.1093/nar/gks383
- Brueckner F, Cramer P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 2008; 15:811-818; PMID:18552824; http://dx.doi.org/10.1038/nsmb.1458
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell 2005; 120:183-193; PMID:15680325; http://dx.doi.org/10.1016/j.cell.2004.11.045
- SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 2004; 33:415-440; PMID:15139820; http://dx.doi.org/10.1146/annurev.biophys.32.110601.141800
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 1995; 34:11211-11216; PMID:7545436; http://dx.doi.org/10.1021/bi00035a029
- Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 2010; 38:D280-282; PMID:19880381; http://dx.doi.org/10.1093/nar/gkp892
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 2014; 344:1042-1047; PMID:24789973; http://dx.doi.org/10.1126/science.1251871
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 2014; 344:1285-1289; PMID:24926020; http://dx.doi.org/10.1126/science.1253458
- Holmes SF, Erie DA. Downstream DNA sequence effects on transcription elongation. Allosteric binding of nucleoside triphosphates facilitates translocation via a ratchet motion. J Biol Chem 2003; 278:35597-35608; PMID:12813036; http://dx.doi.org/10.1074/jbc.M304496200
- Liu B, Zuo Y, Steitz TA. Structural basis for transcription reactivation by RapA. Proc Natl Acad Sci U S A 2015; 112:2006-2010; PMID:25646438; http://dx.doi.org/10.1073/pnas.1417152112
- Strobel EJ, Roberts JW. Two transcription pause elements underlie a sigma70-dependent pause cycle. Proc Natl Acad Sci U S A 2015; 112:E4374-4380; PMID:26216999; http://dx.doi.org/10.1073/pnas.1512986112
- Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell 2015; 57:408-421; PMID:25601758; http://dx.doi.org/10.1016/j.molcel.2014.12.014
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 2010; 468:978-982; PMID:21124318; http://dx.doi.org/10.1038/nature09573
- Gries TJ, Kontur WS, Capp MW, Saecker RM, Record MT, Jr. One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proc Natl Acad Sci U S A 2010; 107:10418-10423; PMID:20483995; http://dx.doi.org/10.1073/pnas.1000967107
- Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 2013; 50:430-436; PMID:23623685; http://dx.doi.org/10.1016/j.molcel.2013.03.020
- Ross W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL, Gourse RL. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell 2016; 62:811-823; PMID:27237053; http://dx.doi.org/10.1016/j.molcel.2016.04.029
