ABSTRACT
How genes are repressed by steroid hormones remains a matter of debate, and several indirect mechanisms have been proposed. We found that the ligand-activated progesterone receptor recruits to the promoter of downregulated genes a repressor complex composed of HP1γ, the lysine demethylase LSD1, histone deacetylases, coREST, the RNA SRA, and the ATPase BRG1. BRG1 is needed for chromatin remodeling and facilitates the deposition of linker histone variant H1.2, which compacts chromatin and hinders RNA polymerase loading and transcription. Thus, steroid hormone receptors can repress genes in ways reminiscent of those used for gene induction, namely by directly targeting factors that remodel chromatin. But while PR-dependent gene induction in T47D cells is mainly achieved by potentiating enhancer activity, repression acts at the level of gene promoters.
Eukaryotic cells adapt to environmental changes by modifying their gene expression programs via modulation of the rate of transcription of subset of genes. In eukaryotes, regulation of gene transcription is a complex process as genomic DNA is packaged within the nucleus into highly organized and dynamic chromatin, the structure of which determines the rate of RNA polymerase initiation and elongation, and thus provides an opportunity for gene regulation. In fact, many external signals impinging on eukaryotic cells activate signaling pathways converging on enzymes that modulate chromatin structural dynamics. In the case of steroid hormones, their intracellular hormone receptors play a fundamental role acting both as signaling molecules that activated multiple kinase pathways and as hormone activated transcription factors that target chromatin remodeling enzymes to specific binding sites in the genome. We have studied the action of the progesterone analog R5020 on the breast cancer cell line T47D, which express the progesterone receptor (PR). When these cells are incubated with R5020 for 6 hours, thousands of genes exhibit increased binding of PR to their enhancers and consequent increase in transcription.Citation2 However, this is only one side of the coin, as 649 genes are found repressed after hormone exposure.Citation13
Till present, more attention and study has been devoted to gene activation compared to gene repression mediated by hormones. In fact, gene repression is poorly understood, with some authors even considering repression as a secondary effect of activation. It is postulated that downregulation of genes is rather a negligible process and its physiological importance has been questioned. As for the molecular mechanism, it has been proposed that downregulation occurs without interaction between the hormone receptors and the promoter or enhancer sequences of repressed genes, and also that some repressive effects could be explained by squelching of certain components of the transcriptional machinery.Citation21 In the case of progestins (Pg), though the PRB isoform is assumed to function predominantly as an activator of Pg-responsive genes, the PRA isoform has been linked to the repressive actions of Pg.Citation7 In addition, gene expression analysis performed in cell lines that express either PRB or PRA showed that the two PR isoforms largely regulate different subsets of genes with 6 genes uniquely downregulated by PRB, 0 uniquely downregulated by PRA, and 6 downregulated by both receptors.Citation19 Thus, all these findings are controversial and the molecular mechanism of repression is not clear.
Hormone-dependent downregulation requires binding of PR and the kinases ERK and MSK1 to target genes
As mentioned above, 649 genes were downregulated after Pg treatment of T47D breast cancer cells. Gene ontology analysis revealed that these genes are associated with relevant cell functions including intracellular signaling cascades, cell proliferation, cell adhesion, and cell fate commitment. By ChIP-seq experiments, we found that PR binding sites (PRBs) are enriched close to the transcription start site (TSS) of downregulated genes in regions containing sequence motifs similar to those found in PRBs of upregulated genes, discarding the existence of specific negative responsive elements as previously proposed for estrogen receptor (ER).Citation22
The kinetics of PR binding analyzed by ChIP-seq after exposure to hormone for different timesCitation2 showed a more rapid and transient PR binding to downregulated genes compared to upregulated genes. These findings support a direct effect of the hormone on the repression of these genes requiring a transient binding of PR to their promoters.
In the context of gene activation, PR forms a ternary complex with the active form of the kinases ERK and MSK1, which is selectively recruited to the target PRBs.Citation24 We wondered whether these kinases are also recruited to the PRBs associated to downregulated genes. ChIP experiments confirmed the recruitment of ERK1/2 as well as MSK1 to the BCAS1, KRT23, and IGFBP5, three transcriptionally repressed genes exhibiting PRBs in their promoter region. In fact, it is the “active” form of PR phosphorylated in S294 by ERK1/2 associated with the kinases ERK1/2 and MSK1, which is recruited both to activated and repressed genes. These new findings explain our previous results that inhibition of ERK or MSK signaling pathways significantly affected both up and downregulation of hormone-responsive genes.Citation24 During gene activation, MSK1 phosphorylates H3S10phos and promotes the displacement of a repressive complex,Citation24,25 but still the targets of MSK1 during hormonal gene repression remain to be identified.
BRG1 as part of the HP1γ–LSD1.com complex is recruited to repressed genes and needed for hormonal downregulation
We found that the ligand-activated PR recruits to downregulated genes a repressor complex composed of HP1γ, the lysine demethylase LSD1, histone deacetylases, CoREST, and the RNA SRA.Citation13 The same complex is also used by the unliganded PR to keep genes silent in the absence of hormone,Citation25 in a striking case of functional convergence.
Several ATP-dependent remodeling complexes are involved in hormonal gene regulation, including the homologues of SWI/SNF complex, BAF and PBAF, which contain BRG1 and/or BRM ATPase subunits. These complexes have been shown to alter nucleosome structure and to facilitate transcription factor binding and gene activation. In the case of Pg acting on breast cancer cells, BAF has been shown to be needed for gene activation and is recruited to PRBs via an interaction with PR.Citation26 We now found that the ATPase BRG1 but not the whole BAF complex is required during Pg-dependent gene repression. Interestingly, BRG1 is recruited to repressed genes as part of the HP1γ−LSD1 complex and its remodeling activity is critical during this process. The presence of chromatin-remodelers and histone deacetylases was previously described in other repressive complexes, including SMRT, NuRD, and N-CoR, though its functional implication is still uncertain.Citation15
We confirmed the implication of BRG1 in hormone-dependent gene repression in breast cancer cells by RNA-seq experiments. We found that 44% of the down and 26% of the upregulated genes were affected by BRG1 knockdown.Citation13 The repressed genes were significantly more dependent on BRG1 than upregulated genes. Interestingly, knockdown of BRG1 affected hormone-dependent cell proliferation as well as apoptosis, highlighting its key role in these processes. In T47D cells, BRG1 is found at least in two large complexes, one associated with BAF and the other associated with HP1γ, but how the distribution of BRG1 between BAF and the HP1γ complex is controlled is still unknown. As BRG1 is phosphorylated in response to hormone and specific acetylation of BRM, the other ATPase of the BAF complex has been reported,Citation31 one possibility is that post-translational modifications could influence BRG1 interactions.
Chromatin crosstalk and the role of linker histone H1.2 during hormonal gene repression
Post-translational modifications of specific histone tail residues affect transcription by altering DNA–histone interactions, and/or providing specific binding sites for proteins that affect chromatin structure. To repress gene transcription, the LSD1–coREST complex coordinates chromatin changes that are brought about by combinations of distinct chromatin-modifying enzymes. In particular, the H3K9me3 signal is important for the localization of HP1γ protein in the genome.Citation8 In T47D cells, we have found an increase in the H3K9me3 signal in hormone repressed genes and we have identified SUV39H2 as the responsible methyltransferase. Thus, PR recruits the HP1γ–LSD1.com repressor complex with its associated BRG1, but what is the function of the ATPase during repression? It has been proposed that by virtue of its remodeling activity, BRG1 as part of LSD1–coREST complex facilitates access of the complex to chromatin targets.Citation15 We studied the molecular mechanism and found that BRG1 enhanced nucleosome occupancy around the TSS of repressed genes and exhibited hormone-dependent interaction with the linker histone variant H1.2, but not with other H1 variants such as H1.3 and H1.5. BRG1, along with the HP1γ–LSD1 complex and H1.2, co-localizes at the promoter region of repressed genes in ChIP-seq experiments performed in cells exposed to hormone and BRG1 is required for H1.2 loading.Citation13 Histone H1.2 deposition leads to chromatin compaction reducing access of the transcriptional machinery.
Linker histone variants are considered to be widely redundant,Citation4 and we wondered about the reasons for the selective role of H1.2 in hormonal gene repression. The major differences between histone H1.2 and other somatic H1 variants are found in the C-terminal tail, where unique post-translational modifications—including acetylation—has been reported.Citation27 These differences may account for the selective binding of histone H1.2 to the bromodomain of BRG1, but mutational studies will be required to address this possibility. Depletion of H1.2 in T47D cells caused a general decrease in nucleosome spacing and cell cycle arrest in G1.Citation20 Thus, in addition to its selective binding to BRG1 H1.2 exhibits specific properties in T47D cells.
We recently found that nuclear ATP synthesis via the PARP1-PARG-NUDIX5 pathway is required for chromatin remodeling after hormone exposure.Citation28 Blocking nuclear ATP synthesis by depletion of NUDIX5 compromised both hormonal gene activation and gene repression, indicating the deposition of H1.2 by BRG1 may also depend on nuclear ATP generation.
The pioneer factor FOXA1 marks repressed PRBs
A question that remains open is how the PR along with the repressor complex discriminates PRBs associated with the repressed genes. We speculate that these sites are marked by another factor that determines the nature of the PR-associated complex by interacting with one of its subunits. Given that FOXA2 has been reported to interact with BRG1,Citation11 and that FOXA1 is important for steroid receptor dynamics,Citation9 we explored its possible function in repression. We found that prior to hormone exposure, FOXA1 is significantly enriched near the PRBs that mediate hormonal repression as compared with PRBs associated to upregulated genes or to all PRBs. Knockdown of FOXA1 affected a larger proportion of downregulated than of upregulated genes (68.5% vs. 48%, respectively). Moreover, depletion of FOXA1 compromised BRG1 recruitment and prevented hormone-dependent gene repression.Citation13 Thus, one possibility is that FOXA1 contributes to BRG1 targeting, though we cannot exclude additional factors. Interestingly, the activated and repressed genes sharing the HP1γ−LSD1.com repressor complex for hormonal regulation are “marked” at uninduced conditions by the unliganded receptor and FOXA1, respectively.Citation13,25
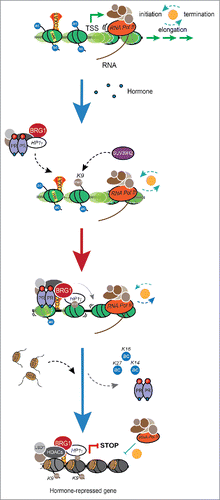
In conclusion, hormonal gene repression requires binding of activated PR to sequences similar to those involved in gene activation, with the distinction that repressive PRBs are near the TSS, whereas activating PRBs are preferentially in distant enhancer regions. In addition, ERK and MSK1 kinases are recruited by PR to both activated and repressed genes, though the phosphorylation target involved in repression has not been identified. The activated PR recruits to downregulated gene promoters a repressive complex composed of HP1γ, histone demethylases, histone deacetylases, the SRA RNA, and the ATPase BRG1. The BRG1 in the repressive complex increases linker histone H1.2 deposition and nucleosome occupancy, leading to chromatin compaction around the TSS that hinders RNA-pol-II loading and maintenance of PR binding ().
Figure 1. Model of gene repression induced by progestins in breast cancer cells. At uninduced conditions, genes are active with RNA pol II and an open chromatin configuration characterized by the presence of the pioneer factor FOXA1 and acetylated histones. Upon hormone exposure, activated PR along with the HP1γ–LSD1.com complex interacts with the ATPase BRG1 and promotes histone deacetylation, demethylation, and chromatin remodeling via BRG1, which increase nucleosome positioning and occupancy. This arrangement of nucleosomes constitutes a suitable platform for histone H1.2 binding, and thus close the target chromatin decreasing RNApol II loading and transcription.
Open questions
But we are still halfway in the study of the mechanism of repression induced by hormones. We were able to understand how a subset of 150 out of 649 genes are repressed but still around 500 genes are downregulated by other mechanisms that remain to be elucidated. This subset of bona fide repressed genes are affected by knockdown of BRG1, HP1γ, and FOXA1, and showed increase in nucleosome positioning along with H1.2 deposition after hormone induction.Citation13 Our studies indicate that the deacetylases HDAC1 and HDAC2 present in other repressor complexes as NURD and Sin3, and acting in concert with Polycomb repressive complexes (PRC1/PRC2), are potential candidates to complete the list of machineries responsible for hormonal repression.Citation16 In fact, phosphorylation of HDAC1, HDAC2, and HDAC3 stimulates enzyme activity and is also required for their incorporation into the Sin3, NuRD, and CoREST corepressor complexes.Citation17,23 This may be one of the functions of the kinases during repression.
What is the scope of this mechanism of gene repression? We can expect that a similar mechanism could operate with the receptors of other steroid hormones. It has been shown that ER, androgen receptor (AR), and glucocorticoid receptor (GR) interact with LSD1, BRG1, and HDACs, respectively, and that depletion of these cofactors affect receptor-dependent transcription.Citation1,6,12,18,29 Moreover, FOXA1 has a relevant role in hormone-receptor binding to chromatin.Citation30 However, whether other components of the repressing mechanism, in particular H1.2 deposition, are used for gene repression by other steroid hormones remains to be established.
Most of our understanding of the molecular mechanisms of ER and PR signaling comes from in vitro studies with hormone-receptor positive cell lines. Estrogens and Pg act in vivo on a subset of mammary epithelial cells and relegate biological functions to paracrine factors as Receptor Activator of Nuclear factor Kappa B Ligand (RANKL) and members of the Wnt family.Citation10 One point certainly worth to analyze is the dynamics of PR binding in normal human mammary cells. However, several factors hinder this study: (i) normal cells in culture tend to rapidly loose hormone receptors, (ii) the amount of cells is limiting, and (iii) normal cells require the three-dimensional (3D) structure of the mammary gland to maintain their identity.
Several groups reported the recruitment of PR to progesterone-target genes as RANKL, β-casein, and ID4 in HC-11 mouse mammary epithelial cells,Citation3,5,14 but the genomic distribution of PR in normal human mammary epithelial cells is still unknown.
Finally, given the relevance of the downregulated genes for breast cancer cell proliferation and apoptosis, the factors involved in the repression mechanism could be the potential targets for the management of hormone-dependent cancers.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The experimental work was supported by grants from the Departament d’Innovació Universitat i Empresa (DIUiE). We acknowledge support of the Spanish Ministry of Economy and Competitiveness (SAF2013-42497-P), “Centro de Excelencia Severo Ochoa 2013–2017,” SEV-2012-0208, and ERC Synergy Grant “4DGenome” nr 609989.
References
- Acevedo ML, Kraus WL. Transcriptional activation by nuclear receptors. Essays Biochem 2004; 40:73-88; PMID:15242340; http://dx.doi.org/10.1042/bse0400073
- Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 2013; 49:67-79; PMID:23177737; http://dx.doi.org/10.1016/j.molcel.2012.10.019
- Buser AC, Obr AE, Kabotyanski EB, Grimm SL, Rosen JM, Edwards DP. Progesterone receptor directly inhibits beta-casein gene transcription in mammary epithelial cells through promoting promoter and enhancer repressive chromatin modifications. Mol Endocrinol 2011; 25:955-968; PMID:21527503; http://dx.doi.org/10.1210/me.2011-0064
- Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol 2001; 21:7933-7943; PMID:11689686; http://dx.doi.org/10.1128/MCB.21.23.7933-7943.2001
- Fernandez-Valdivia R, Mukherjee A, Creighton CJ, Buser AC, DeMayo FJ, Edwards DP, Lydon JP. Transcriptional response of the murine mammary gland to acute progesterone exposure. Endocrinology 2008; 149:6236-6250; PMID:18687774; http://dx.doi.org/10.1210/en.2008-0768
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007; 128:505-518; PMID:17289570; http://dx.doi.org/10.1016/j.cell.2006.12.038
- Giangrande PH, McDonnell DP. The A and B isoforms of the human progesterone receptor: two functionally different transcription factors encoded by a single gene. Recent Progress Hormone Res 1999; 54:291-313; discussion 313-294; PMID:10548881
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007; 8:35-46; PMID:17173056; http://dx.doi.org/10.1038/nrg2008
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011; 43:27-33; PMID:21151129; http://dx.doi.org/10.1038/ng.730
- Kariagina A, Aupperlee MD, Haslam SZ. Progesterone receptor isoform functions in normal breast development and breast cancer. Crit Rev Eukaryot Gene Expr 2008; 18:11-33; PMID:18197783; http://dx.doi.org/10.1615/CritRevEukarGeneExpr.v18.i1.20
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 2012; 151:1608-1616; PMID:23260146; http://dx.doi.org/10.1016/j.cell.2012.11.018
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437:436-439; PMID:16079795; http://dx.doi.org/10.1038/nature04020
- Nacht AS, Pohl A, Zaurin R, Soronellas D, Quilez J, Sharma P, Wright RH, Beato M, Vicent GP. Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J 2016; 35:1822-1843; PMID:27390128; http://dx.doi.org/10.15252/embj.201593260
- Obr AE, Edwards DP. The biology of progesterone receptor in the normal mammary gland and in breast cancer. Mol Cell Endocrinol 2012; 357:4-17; PMID:22193050; http://dx.doi.org/10.1016/j.mce.2011.10.030
- Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet 2007; 8:544-554; PMID:17572692; http://dx.doi.org/10.1038/nrg2100
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 2010; 11:109-123; PMID:20084085; http://dx.doi.org/10.1038/nrg2736
- Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem 2001; 276:47733-47741; PMID:11602581; http://dx.doi.org/10.1074/jbc.M105590200
- Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell 2006; 22:669-679; PMID:16762839; http://dx.doi.org/10.1016/j.molcel.2006.04.019
- Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem 2002; 277:5209-5218; PMID:11717311; http://dx.doi.org/10.1074/jbc.M110090200
- Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet 2008; 4:e1000227; PMID:18927631; http://dx.doi.org/10.1371/journal.pgen.1000227
- Santos GM, Fairall L, Schwabe JW. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol Metab 2011; 22:87-93; PMID:21196123; http://dx.doi.org/10.1016/j.tem.2010.11.004
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 2011; 145:224-241; PMID:21496643; http://dx.doi.org/10.1016/j.cell.2011.03.027
- Tsai SC, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem 2002; 277:31826-31833; PMID:12082111; http://dx.doi.org/10.1074/jbc.M204149200
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 2006; 24:367-381; PMID:17081988; http://dx.doi.org/10.1016/j.molcel.2006.10.011
- Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev 2013; 27:1179-1197; PMID:23699411; http://dx.doi.org/10.1101/gad.215293.113
- Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, Vermeulen M, Mann M, Beato M. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet 2009; 5:e1000567; PMID:19609353; http://dx.doi.org/10.1371/journal.pgen.1000567
- Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics 2007; 6:72-87; PMID:17043054; http://dx.doi.org/10.1074/mcp.M600255-MCP200
- Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, Nacht AS, Samino S, Font-Mateu J, Vicent GP et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 2016; 352:1221-1225; PMID:27257257; http://dx.doi.org/10.1126/science.aad9335
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 1992; 258:1598-1604; PMID:1360703; http://dx.doi.org/10.1126/science.1360703
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011; 25:2227-2241; PMID:22056668; http://dx.doi.org/10.1101/gad.176826.111
- Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Zhang Y, Shen YF. A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res 2010; 20:1345-1360; PMID:21079652; http://dx.doi.org/10.1038/cr.2010.155
