ABSTRACT
G-quadruplexes are non-canonical DNA secondary structures involved in several genomic and molecular processes. Here, we summarize the main G-quadruplex features and evidences proving the in vivo role on the transcriptional regulation of genes required for zebrafish embryonic development. We also discuss alternative strategies for specifically interfering G-quadruplex in vivo.
According to the model proposed by James D. Watson and Francis Crick in 1953, the DNA molecule consists of two polynucleotide chains wound around each other to form a clockwise double helix. This structure is known as B form of DNA (B-DNA), and is the predominant conformation adopted by DNA under relaxed conditions. However, non-canonical DNA secondary structures are widespread in all living organisms, where they have profound effects on replication, transcription and genome stability. Guanine quadruplex (G-quadruplex), i-motifs, triplexes, cruciforms and hairpins secondary structures can lead to double strand breaks, induction or inhibition of transcription, and initiation or stalling of replication (). The formation of secondary structures may contribute to the generation of genetic diversity, polymorphism, and genome evolution. Conversely, they may result in a variety of genetic disorders, hereditary diseases, and cancer through chromosomal rearrangements, mutagenesis, dysregulation of gene expression, and changes in the DNA replication process.
Figure 1. Non-canonical DNA secondary structures influencing transcription. (A) Scheme representing the most relevant non-canonical DNA secondary structures and their effects on genome and gene expression. (B) Transcriptional regulation by G-quadruplexes. After transcription onset, transcription bubble generates transiently exposed single-strand segments able to fold as G-quadruplexes. Two putative scenarios are represented (i) G-quadruplexes may form upstream the transcription start site (TSS), causing positive or negative effects on transcription depending on their capability of interfering with RNA Polymerase II or transcription factors binding, recruiting G-quadruplex binding proteins or maintaining an open DNA conformation that facilitates transcription re-initiation. (ii) G-quadruplexes may form downstream the TSS, usually causing positive effects on transcription when located in the coding strand due to favoring transcription re-initiation, or negative effects on transcription when located in the template strand due to stalling the progression of RNA polymerase.
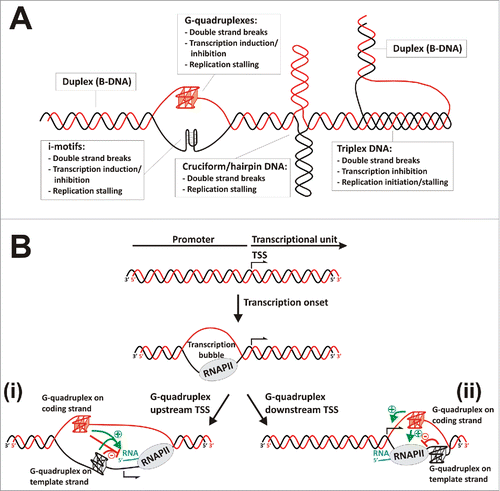
Among various non-canonical DNA structures, G-quadruplexes have attracted enormous research attention as prospective targets for chemical intervention of biological functions. Guanine-rich single-stranded DNA has a strong propensity to fold into G-quadruplex in vitro. The consensus sequence [G3N1–7G3N1–7G3N1–7G3] allows four sets of G-triplets to form three layers of G tetrads stabilized by Hoogsteen hydrogen bonding and K+ chelation.Citation1,2 There are 376,000 putative quadruplex sequences (PQS) in the human genome that have been identified through genome-wide surveys based on quadruplex-folding rules.Citation3,4 PQS are highly frequent in proto-oncogene promotersCitation5,6; however, not all of them may exist in vivo. A prerequisite for the formation of G-quadruplex is the destabilization of the B-DNA double-strand helix, which is highly stable. The current hypothesis states that transcription bubble generates regions of positive and negative supercoiling, which can be propagated along the DNA-helix until reaching sites susceptible of structural transitions. In this condition, transiently exposed single-strand segments become able to fold as G-quadruplexesCitation7 (). G-quadruplexes may affect gene transcriptional activity either by upregulation or downregulation, a function that can be evidenced by stabilizing or disrupting G-quadruplex formation through both interacting small ligands (drugs) and/or specific nucleic acid binding proteins.Citation8,9
The validation of drug-targeted G-quadruplex DNA and the modulation of oncogenes expression intensely increased in the recent past. Despite a few of new anticancer drugs have entered preclinical or clinical trials,Citation10,11 the selectivity of these compounds has yet to be improved. Up to now, there are no drugs that are able to discriminate between G-quadruplexes affecting genome stability from those controlling gene expression. Even more, drugs cannot discern between G-quadruplexes controlling the transcription of oncogenes from those ones controlling the transcription of other essential genes. In this context, a novel strategy consisting of the use of short antisense DNA sequences or oligonucleotides (ASO) blocking the formation of a specific G-quadruplex has been recently reported.Citation12-14
Numerous studies performed in cellulo have demonstrated the influence of different PQS patterns and loop lengths,Citation15,16 the effect of ions,Citation17,18 and the action of specific G-quadruplex ligandsCitation8,9,19 on G-quadruplex stability and the transcriptional process. Mostly, these studies consisted in assessing the effect of specific G-quadruplex ligands on the transcriptional expression of reporter genes governed by promoter elements containing PQS. Several reports suggest that G-quadruplexes act as transcriptional repressors by impeding transcription factor binding to duplex-DNA or stalling the progression of RNA polymerase, mostly when they are located downstream the transcription start site in the template strand.Citation2,20-22 Conversely, other reports showed that G-quadruplex may enhance the transcription of particular genes by favoring the binding of specific transcription factorsCitation2,12,22 or by holding the DNA molecule open thus facilitating the re-initiation of transcriptionCitation2,7,14,22 (). Therefore, the hypothesis about a common behavior of G-quadruplexes on transcriptional control would be erroneous.
The in cellulo existence and potential impact of G-quadruplex on pathological processes is now accepted, but questions regarding their functions and mechanisms of action in vivo remain to be fully addressed. It was reported that microinjection of small G-quadruplex ligands in zebrafish embryos caused G-quadruplex stabilization along with downregulation of Cdh5 transcription and the generation of embryonic phenotypes mimicking Cdh5-morphants.Citation23 However, such study was unable to conclusively demonstrate that phenotypes were due to the tested ligand on the Cdh5 G-quadruplex. In view of the high number of PQS present in zebrafish genome,Citation14 the possibility of non-specific or pleiotropic effects of such ligands could not be ruled out.
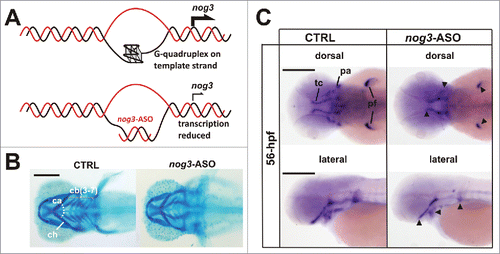
Although in past years the knowledge about the biochemical features and cellular roles of the G-quadruplex has made significant progress, the challenge was to demonstrate the direct role of these structures on a specific biological process carried out by complex multicellular organisms. The embryonic development resulted as an appropriate scenario to accomplish this goal. During embryonic development, gene expression is orchestrated by specific and highly evolutionarily conserved mechanisms that take place accurately, both at spatial and temporal levels.Citation24 An intricate array of cis-regulatory sequences controlling individual genes leads to a fine-tuning of gene expression. The last decades have provided compelling evidence that not only protein-mediated transcriptional control but also chromatin state play essential roles in orchestrating stages of embryonic development.Citation25,26 Although the enrichment of G-quadruplex in promoter regions of developmentally regulated genes had been found in in silico analyses several years ago,Citation27 no experimental evidences proving the role of G-quadruplex in vertebrate embryonic development had been reported. In a recent work, evolutionarily conserved G-quadruplexes located within the proximal promoter region of genes required for proper craniofacial cartilage formation, notochord elongation, and eye development were identified. The disruption of these G-quadruplexes in vivo by microinjection of specific ASOs in developing zebrafish resulted in lower transcription of the targeted genes, as well as in the re-capitulation of the embryonic and larvae phenotypes reported for the respective morphants or mutantsCitation14 (). This pioneer work demonstrated the role in vivo of G-quadruplexes as cis-acting elements contributing to the transcriptional regulation during the embryonic development, one of the most regulated processes of vertebrate's biology.
Figure 2. Noggin 3 (nog3), a gene required for proper craniofacial cartilages development, is regulated in vivo by G-quadruplex. (A) Strategy to specifically block G-quadruplex formation using an antisense oligonucleotide (nog3-ASO) microinjected in zebrafish embryos. (B) Alcian blue staining showing craniofacial cartilages (ca, ceratohyal cartilages angle; cb (3–7): ), ceratobranchial cartilages 3 to 7; ch, ceratohyal cartilage) of 4 days post-fertilization (4-dpf) larvae. Compared to controls (CTRL), nog3-ASO microinjected larvae display reduced head structures and abnormal craniofacial cartilage pattern. (C) Lateral and dorsal views of whole-mount in situ hybridizations showing reduced expression of nog3-mRNA in 56 hours post-fertilization (56-hpf) larvae microinjected with nog3-ASO when compared with controls (CTRL). pa, pharyngeal arches; pf, pectoral fin; tc, trabeculae cranii. Scale bars = 200 μm.
PQS in gene promoters may contain more than four G-stretches or more than three guanines in each stretch, thus resulting in a mixture of conformational isomers in a multiple dynamic equilibrium. Additionally, PQS genetic polymorphisms in G-stretches or even loop-regions may modify G-quadruplex formation and/or stability, thus significantly affecting gene expression among individuals.Citation28 In a particular cellular context, specific proteins could associate with a subpopulation of conformational isomers shifting the equilibrium toward a particular structure or bind differentially to polymorphic PQS. Consequently, both G-quadruplex alternative conformation and PQS polymorphisms might function as molecular switches enabling gene expression modulation by transitions in DNA structure. Of note, several proteins were identified associated with PQS, most of them are highly conserved zinc-finger DNA-binding proteins. Because the versatility of zinc-finger binding pocket is remarkable, it is interesting to consider the implications of G-quadruplex–zinc-finger interactions as a pair.Citation29 In this context, variations in G-quadrupex conformational isomers, PQS genetic polymorphisms, and protein domains may contribute to a fine tuning of transcriptional control.
Conclusions and perspectives
Knowledge about G-quadruplex structure and biochemical features opens up a new field for exploring their biological functions, which might be relevant for understanding the regulation of several cellular and biological processes. G-quadruplexes existence had already been shown both in cellulo and in vivo and their role in trascriptional regulation is now evident. Profound insights into the mechanisms of G-quadruplex folding, polymorphism affecting G-stretches or even loop-regions, drugs and proteins binding affecting G-quadruplex stability should now be the focus of the scientific community.
G-quadruplexes have been linked with diseases such as cancer, neurodegenerative, and genetic disorders, thus providing new clues for the customized design of novel therapeutic strategies. The design of specific molecules capable to discriminate among G-quadruplexes could be helpful at the moment of planning efficient strategies to fight against diseases. The ASO approach allows blocking in vivo the formation of a specific G-quadruplex, thus controlling the expression of defined genes. This and similar approaches would emerge as alternative tools to selectively modulate the G-quadruplex-mediated transcription of precise genes allowing to inhibit either disease development or progression reducing undesired collateral effects.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genetics Dev 2014; 25:22-29; PMID:24584093; http://dx.doi.org/10.1016/j.gde.2013.10.012
- Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 2012; 13:770-780; PMID:23032257; http://dx.doi.org/10.1038/nrg3296
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res 2005; 33:2908-2916; PMID:15914667; http://dx.doi.org/10.1093/nar/gki609
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res 2005; 33:2901-2907; PMID:15914666; http://dx.doi.org/10.1093/nar/gki553
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol 2006; 13:1055-1059; PMID:17146462; http://dx.doi.org/10.1038/nsmb1171
- Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res 2009; 37:4194-4204; PMID:19211664; http://dx.doi.org/10.1093/nar/gkn1076
- Du Z, Zhao Y, Li N. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res 2008; 18:233-241; PMID:18096746; http://dx.doi.org/10.1101/gr.6905408
- Duchler M. G-quadruplexes: targets and tools in anticancer drug design. J Drug Target 2012; 20:389-400; PMID:22424091; http://dx.doi.org/10.3109/1061186X.2012.669384
- Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 2011; 10:261-275; PMID:21455236; http://dx.doi.org/10.1038/nrd3428
- Duan W, Rangan A, Vankayalapati H, Kim MY, Zeng Q, Sun D, Han H, Fedoroff OY, Nishioka D, Rha SY, et al. Design and synthesis of fluoroquinophenoxazines that interact with human telomeric G-quadruplexes and their biological effects. Mol Cancer Ther 2001; 1:103-120; PMID:12467228; http://dx.doi.org/10.1.1.138.390
- Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 2009; 86:151-164; PMID:19454272; http://dx.doi.org/10.1016/j.yexmp.2009.01.004
- Kumar N, Patowary A, Sivasubbu S, Petersen M, Maiti S. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry 2008; 47:13179-13188; PMID:19053274; http://dx.doi.org/10.1021/bi801064j
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015; 525:56-61; PMID:26308891; http://dx.doi.org/10.1038/nature14973
- David AP, Margarit E, Domizi P, Banchio C, Armas P, Calcaterra NB. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res 2016; 44:4163-4173; PMID:26773060; http://dx.doi.org/10.1093/nar/gkw011
- Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. J Phys Chem B 2009; 113:8784-8792; PMID:19480441; http://dx.doi.org/10.1021/jp809486g
- Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res 2010; 38:7858-7868; PMID:20660477; http://dx.doi.org/10.1093/nar/gkq639
- Marathias VM, Bolton PH. Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry 1999; 38:4355-4364; PMID:10194354; http://dx.doi.org/10.1021/bi982604+
- Schultze P, Hud NV, Smith FW, Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)). Nucleic Acids Res 1999; 27:3018-3028; PMID:10454595; http://dx.doi.org/10.1093/nar/27.15.3018
- Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex ligands. Org Biomol Chem 2008; 6:627-636; PMID:18264563; http://dx.doi.org/10.1039/B714772B
- Agarwal T, Roy S, Kumar S, Chakraborty TK, Maiti S. In the sense of transcription regulation by G-quadruplexes: asymmetric effects in sense and antisense strands. Biochemistry 2014; 53:3711-3718; PMID:24850370; http://dx.doi.org/10.1021/bi401451q
- Maizels N. G4-associated human diseases. EMBO Rep 2015; 16:910-922; PMID:26150098; http://dx.doi.org/10.15252/embr.201540607
- Smestad JA, Maher LJ, 3rd. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Medical Genetics 2015; 16:91; PMID:26449372; http://dx.doi.org/10.1186/s12881-015-0236-4
- Agarwal T, Lalwani MK, Kumar S, Roy S, Chakraborty TK, Sivasubbu S, Maiti S. Morphological effects of G-quadruplex stabilization using a small molecule in zebrafish. Biochemistry 2014; 53:1117-1124; PMID:24476096; http://dx.doi.org/10.1021/bi4009352
- Wolpert L. Do we understand development? Science 1994; 266:571-572; PMID:7939707; http://dx.doi.org/10.1126/science.7939707
- Misteli T. Self-organization in the genome. Proc Natl Acad Sci U S A 2009; 106:6885-6886; PMID:19416923; http://dx.doi.org/10.1073/pnas.0902010106
- Rajapakse I, Perlman MD, Scalzo D, Kooperberg C, Groudine M, Kosak ST. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Natl Acad Sci USA 2009; 106:6679-6684; PMID:19276122; http://dx.doi.org/10.1073/pnas.0900986106
- Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res 2007; 35:406-413; PMID:17169996; http://dx.doi.org/10.1093/nar/gkl1057
- Baral A, Kumar P, Halder R, Mani P, Yadav VK, Singh A, Das SK, Chowdhury S. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res 2012; 40:3800-3811; PMID:22238381; http://dx.doi.org/10.1093/nar/gkr1258
- Kumar P, Yadav VK, Baral A, Saha D, Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res 2011; 39:8005-8016; PMID:21729868; http://dx.doi.org/10.1093/nar/gkr536
