ABSTRACT
TFIID is a large protein complex required for the recognition and binding of eukaryotic gene core promoter sequences and for the recruitment of the rest of the general transcription factors involved in initiation of eukaryotic protein gene transcription. Cryo-electron microscopy studies have demonstrated the conformational complexity of human TFIID, where one-third of the mass of the complex can shift its position by well over 100 Å. This conformational plasticity appears to be linked to the capacity of TFIID to bind DNA, and suggests how it would allow both the recognition of different core promoter elements and the tuning of its binding affinity by regulatory factors.
Introduction
Eukaryotic transcription initiation requires the assembly of a large transcription preinitiation complex (PIC) at the core promoter of each protein gene. The PIC includes, in addition to RNA Pol II, a number of Pol II-associated general transcription factors (GTFs) whose role is to bring the polymerase to the core promoter, in some cases positioning it very precisely with respect to the transcription start site, as well as helping Pol II with the opening of the duplex DNA and the stabilization of the initial transcription bubble. Among the GTFs are TFIIA, TFIIB, TFIID, TFIIF, TFIIE and TFIIH.Citation22,9,28 TFIID, the largest GTF (∼1.2 MDa), is composed of the TATA-box binding protein (TBP) and 13 TBP-associated factors (TAFs).Citation6,1 The roles of TFIID include the recognition and binding of all gene core promoter sequences.Citation34,9,28,5,15,32,31 Core promoter sequences vary widely throughout the genome, although a set of core promoter motifs seems to be present, in different combinations, in a significant number of gene core promoters.Citation14 These often include a combination of the TATA box, the Initiator motif (Inr), the downstream promoter element (DPE) and/or the Motif Ten Element (MTE). The Kadonaga lab designed a super core promoter (SCP) by combining optimal sequences and spacing for TATA, Inr, DPE and MTE. This SCP has a significantly higher affinity for TFIID and leads to improved transcriptional output via increased promoter usage.Citation13 The SCP has now been used to facilitate biophysical studies of the process of TFIID binding and transcription initiation, such as single molecule experimentsCitation37 or cryo-electron microscopy (cryo-EM) studies.Citation7,20
The size of TFIID, its poor biochemical stability and the complexity of its subunit stoichiometry, are among the factors that have so far precluded the development of expression systems for purification of large amounts of this complex. Thus, in vitro studies still rely on purification from endogenous sources, which are very limited. This limitation has severely hampered the structural characterization of TFIID by X-ray crystallography, and even challenged other techniques with simpler sample requirements, like cryo-EM.Citation24 Human TFIID is typically produced by immunopurification from HeLa cells, with yields of around 5–10 μg per 10 Ls of cells.
Initial structural studies of TFIID
The first structural glimpses of TFIID came from early EM studies of both human and budding yeast TFIID using negatively stained samples. At resolutions of 30–40 Å, these studies showed TFIID to be composed on three main lobes, termed A, B and C, surrounding a central cavity.Citation2,4 Antibody labeling studies led to a proposal of subunit distribution within those lobes that included two copies of some of the TAFs in different regions of the complex.Citation16,17,26 More functional studies followed, investigating the structure of different TFIID isoforms,Citation18 its interaction with activatorsCitation19 and/or its binding to DNA.Citation25 Biochemical efforts lead to the reconstitution of TFIID subcomplexes, including a symmetrical complex containing two copies each of TAF-4, −5, −6, −9, and −12, and an asymmetrical one after addition of TAF8–TAF10, both of which were characterized by cryo-EM.Citation3
An important realization was that TFIID is a very flexible complex,Citation10,26 but how this flexibility related to the mechanism of action of TFIID was not initially clear. Recent cryo-EM studies have shed new light onto the complex conformational landscape of TFIID and its functional relevance in the binding of core promoters.
Conformational states of TFIID and DNA binding
Through careful EM image analysis of both negatively stained and frozen hydrated samples, it became possible to determine that the extreme conformational heterogeneity of human TFIID was due to changes in the position of lobe A with respect to a more stable BC core.Citation7 TFIID transitions in a continuous fashion between two broadly defined states, referred to as “canonical” and “rearranged.” While in the former, lobe A is in contact with lobe C, in the rearranged state it has moved by more than 100 Å to contact lobe B (). Given that lobe A is always present in our TFIID images, it is clear that it never detaches completely from the BC core, but needs to remain covalently attached. The fine details of this connection are not yet known.
Figure 1. Conformational rearrangement of TFIID. 3D cryo-EM reconstructions of apo TFIID in the canonical state (left) and of the TFIID–IIA–DNA complex in the rearranged state (right) revealed that TFIID binds to core promoter DNA in the rearranged state.Citation7 The density for the stable BC-core, outlined on the bottom for either structure (dotted black line), stays relatively consistent between the two states, while the flexible lobe A (yellow) transits from one side of the BC-core to the other between the two states. In the rearranged state, lobe A can be further divided into lobe A1 (orange), which contains TBP and TFIIA and binds the TATA-containing upstream promoter DNA (see ), and lobe A2 (yellow).
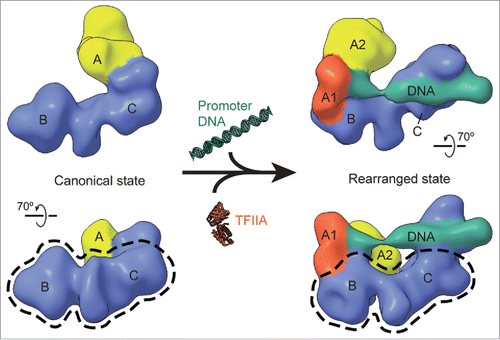
What could be the possible biological relevance of such dramatic structural reorganization? A clue came from the quantitative comparison of lobe A positions obtained from cryo-EM images of apo TFIID versus samples also containing TFIIA and SCP. Such comparison showed that the percentage of complexes in the rearranged state increased significantly in the presence of DNA and TFIIA. Indeed, 3D reconstruction later showed that the DNA-bound complexes corresponded to the rearranged state (), thus defining such conformation as the one capable of core promoter engagement.Citation7 The position of the density in the 3D reconstruction ascribed to DNA explained the discrimination by the core promoter DNA of the conformational state of TFIID. Contacts with the upstream and downstream core promoter elements involved, respectively, the simultaneous interaction of the relocated lobe A and lobe C, which, therefore, need to be at a significant and fixed distance from one another. Furthermore, the position of lobe A in the canonical state which is very close, if not overlapping, with the surface of lobe C interacting with the downstream segments – seems incompatible with a simultaneous engagement with DNA by lobe C.
The dramatic conformational plasticity of TFIID makes a lot of functional sense for a molecular hub involved in the integration of signals from cofactors, gene-specific activators and inhibitors, and epigenetic marks.Citation8 All of those signals need to be read by TFIID and then translated into a gene expression outcome related to the capacity of the complex to bind core promoter sequences and nucleate PIC assembly. The structural plasticity of TFIID allows for its tuning by additional factors, and the relationship between conformational state and core promoter binding would link the allosteric effect of factor binding to TFIID to its capacity to engage the promoter, and perhaps even select certain core promoters with different promoter elements architecture. To dissect how such potential allosteric mechanisms act will require visualization of TFIID binding to different promoters in the presence and absence of regulatory factors.
Human TFIID subunit architecture
Our cryo-EM studies described above enabled the detection of the complex conformational mixtures present in human TFIID samples, and, through initial image classification strategies, allowed the partitioning of the images into two main states for 3D reconstruction. However, the resolution of those cryo-EM reconstructions was very limited due to the fact that in reality there is a continuum of states that were still being mixed. We recently obtained significantly better resolution for the cryo-EM structure of TFIID engaged with DNA thanks to an improved experimental strategy, and higher quality cryo-EM images that then allowed for more sophisticated image analysis.
In order to reduce the conformational complexity of the TFIID samples, we purified DNA-bound complexes using a strategy we previously developed in our studies of the human TBP-based transcription PIC.Citation11 We biotinylated the SCP and bound it to streptavidin-coated magnetic beads. This allowed us to incubate with excess TFIID and TFIIA, then wash the excess protein and purify the DNA-bound complexes via release from the beads using a restriction enzyme. This strategy eliminated canonical states. The sample was mildly cross-linked and imaged using a direct detector. The latter improved both contrast and resolution in our images. Data processing using maximum likelihood principles within the RELION softwareCitation30 allowed us to remove images of damaged complexes and to systematically deal with regions that were more flexible. As a result, it was ultimately possible to identify all the protein components involved in DNA binding within TFIID's A and C lobes.Citation20
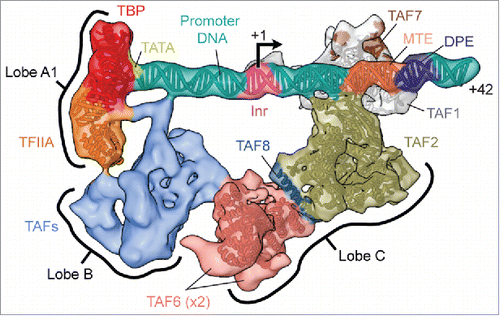
While most of lobe A remains very flexible, we could assign a subregion – lobe A1 – at one end of the DNA density, to the TBP–TFIIA–TATA subcomplex by fitting the available crystal structure () (notice that the rest of lobe A, referred to as lobe A2 ( left, yellow region), remains very flexible, even in the DNA-bound state, and had to be “masked out” for any further analysisCitation20). Accurate positioning of the TATA box within lobe A1 then permitted placement of the other core promoter elements in the SCP (). The subnanometer resolution we obtained for lobe C allowed us to assign the protein regions contacting the DPE and MTE elements by fitting of the TAF1–TAF7 crystal structureCitation35 and a homology model of the aminopeptidase-like domain of TAF2. Most of the remaining lobe C corresponds to a tandem of two HEAT repeats that were assigned to this region of TAF6 by docking of a corresponding crystal structureCitation29 (). More tentatively, based on biochemical evidence and the docking of a large α-helix, the region connecting one of the TAF6 copies with TAF2 was assigned to a segment of TAF8 important for the assembly of TFIID.Citation33
Figure 2. Structure of the promoter-bound core of TFIID. The cryo-EM reconstruction of the promoter-bound core of TFIID comprising lobes A1, B and C, is shown here colored according to current subunit assignments and with available atomic models docked (EMD-3305Citation20). The promoter DNA and the locations of the sequence motifs present in the super core promoter used in the experiment are also modeled. Note that the positions and identities of the TAFs making up lobe B (light blue) are yet unknown, owing to the flexibility and thus lower resolution of this region. For clarity, the flexible part of lobe A (lobe A2) is not shown (see ).
The recent cryo-EM results just described have localized over one-third of the TFIID structure expected to be structured (regions predicted to be disordered abound within TFIID), but a large number of TAFs are still unaccounted for and must correspond to lobe B and the remaining lobe A. Future studies with improved resolution are needed in order to identify these remaining components and describe in more detail the structural rearrangements of TFIID.
An emerging model of the TFIID-based PIC and larger assemblies
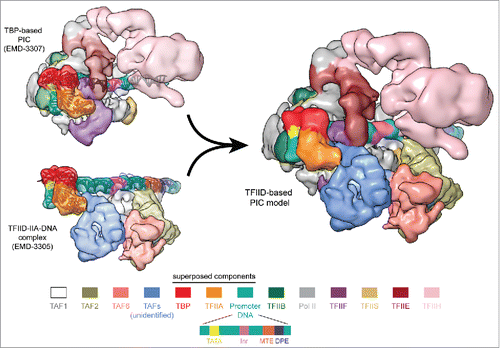
Parallel cryo-EM efforts have recently aimed to describe the structure of the human and budding yeast PIC,Citation11,21,23,12,27 where TFIID had been substituted by TBP in an effort to simplify a very molecularly complex assembly. For the human system, we have been able to combine our structure of TFIID–TFIIA–DNA with that of the TBP-based PIC to arrive at a synthetic model of a full, TFIID-based PIC.Citation20 The two cryo-EM structures show remarkable shape complementarity (). The DNA, as it is being held by TFIID, is presented to the Pol II cleft, while lobe B is positioned next to TFIIF and TFIIE, and lobe C is proximal to TFIIH and the Pol II jaws. Small regions of overlap are indicative of a required conformational change in TFIID upon Pol II recruitment, a process that has previously been detected using time-course footprinting analysis.Citation36 Release of the downstream elements by the lobe C of TFIID while TBP remains engaged with the rest of the PIC is likely to be part of that change. Once again, the plasticity of TFIID may play an essential role in this stage of the initiation process.
Figure 3. Model of the TFIID-based PIC. Superposition of the common elements in the TBP-based PIC (upper left) and the TFIID–IIA–DNA complex (lower left). Superposition of TBP, TFIIA and promoter DNA (shown in ribbon representation) result in a synthetic structural model for the TFIID-based PIC (right,Citation20). The coloring scheme for the PIC components is shown on the bottom, with the TFIID subunits colored similarly as in .
While efficient complex generation will be challenging, cryo-EM may allow us in the future to directly visualize the complete transcriptional prenitiation complexes containing full TFIID, as well as regulatory factors that affect its assembly, recruitment and function.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a grant from NIGMS (GM63072 to E.N.). E.N. is a Howard Hughes Medical Institute Investigator.
References
- Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene 2000; 242:1-13; PMID:10721692; http://dx.doi.org/10.1016/S0378-1119(99)00495-3
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 1999; 286:2153-2156; PMID:10591646; http://dx.doi.org/10.1126/science.286.5447.2153
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. The architecture of human general transcription factor TFIID core complex. Nature 2013; 493:699-702; PMID:23292512; http://dx.doi.org/10.1038/nature11791
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 1999; 286:2151-2153; PMID:10591645; http://dx.doi.org/10.1126/science.286.5447.2151
- Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev 1997; 11:3020-3031; PMID:9367984; http://dx.doi.org/10.1101/gad.11.22.3020
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID). Ann. Rev. Biochem. 1996; 65:769-799; PMID:8811195;[AQ7] http://dx.doi.org/10.1146/annurev.bi.65.070196.004005
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 2013; 152:120-131; PMID:23332750; http://dx.doi.org/10.1016/j.cell.2012.12.005
- Cianfrocco MA, Nogales E. Regulatory interplay between TFIID's conformational transitions and its modular interaction with core promoter DNA. Transcription 2013; 4:120-126; PMID:23863784; http://dx.doi.org/10.4161/trns.25291
- Goodrich JA, Cutler G, Tjian R. Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell 1996; 84:825-830; PMID:8601306; http://dx.doi.org/10.1016/S0092-8674(00)81061-2
- Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: Conformational breathing in the integration of gene regulatory cues. Structure 2006; 14:511-520; PMID:16531235; http://dx.doi.org/10.1016/j.str.2005.11.020
- He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature 2013; 495:481-486; PMID:23446344; http://dx.doi.org/10.1038/nature11991
- He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E. Near-atomic resolution visualization of human transcription promoter opening. Nature 2016; 533:359-365; PMID:27193682; http://dx.doi.org/10.1038/nature17970
- Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods 2006; 3:917-922; PMID:17124735; http://dx.doi.org/10.1038/nmeth937
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter – the gateway to transcription. Curr Opin Cell Biol 2008; 20:253-259; PMID:18436437; http://dx.doi.org/10.1016/j.ceb.2008.03.003
- Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: The downstream core element is recognized by TAF1. Mol Cell Biol 2005; 25:9674-9686; PMID:16227614; http://dx.doi.org/10.1128/MCB.25.21.9674-9686.2005
- Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. Embo J 2002; 21:3424-3433; PMID:12093743; http://dx.doi.org/10.1093/emboj/cdf342
- Leurent C, Sanders SL, Demeny MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Mapping key functional sites within yeast TFIID. Embo J 2004; 23:719-727; PMID:14765106; http://dx.doi.org/10.1038/sj.emboj.7600111
- Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell 2008; 29:81-91; PMID:18206971; http://dx.doi.org/10.1016/j.molcel.2007.11.003
- Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, Dailey G, Nogales E, Tjian R. Structures of three distinct activator-TFIID complexes. Genes Dev 2009; 23:1510-1521; PMID:19571180; http://dx.doi.org/10.1101/gad.1790709
- Louder RK, He Y, Lopez-Blanco JR, Fang J, Chacon P, Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016; 531:604-609; PMID:27007846; http://dx.doi.org/10.1038/nature17394
- Luo J, Cimermancic P, Viswanath S, Ebmeier CC, Kim B, Dehecq M, Raman V, Greenberg CH, Pellarin R, Sali A et al. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell 2015; 59:794-806; PMID:26340423; http://dx.doi.org/10.1016/j.molcel.2015.07.016
- Matsui T, Segall J, Weil PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem 1980; 255:11992-11996; PMID:7440580
- Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci USA 2015; 112:13543-13548; PMID:26483468; http://dx.doi.org/10.1073/pnas.1518255112
- Nogales E, Louder RK, He Y. Cryo-EM in the study of challenging systems: The human transcription pre-initiation complex. Curr Opin Struct Biol 2016; 40:120-127; PMID:27689812; http://dx.doi.org/10.1016/j.sbi.2016.09.009
- Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 2010; 465:956-960; PMID:20559389; http://dx.doi.org/10.1038/nature09080
- Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, Schultz P. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 2009; 17:363-373; PMID:19278651; http://dx.doi.org/10.1016/j.str.2009.01.006
- Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P. Transcription initiation complex structures elucidate DNA opening. Nature 2016; 533:353-358; PMID:27193681; http://dx.doi.org/10.1038/nature17990
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 1996; 21:327-335; PMID:8870495; http://dx.doi.org/10.1016/S0968-0004(96)10050-5
- Scheer E, Delbac F, Tora L, Moras D, Romier C. TFIID TAF6-TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. J Biol Chem 2012; 287:27580-27592; PMID:22696218; http://dx.doi.org/10.1074/jbc.M112.379206
- Scheres SH. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol 2016; 579:125-157; PMID:27572726; http://dx.doi.org/10.1016/bs.mie.2016.04.012
- Theisen JW, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol Cell Biol 2010; 30:3471-3479; PMID:20457814; http://dx.doi.org/10.1128/MCB.00053-10
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 2006; 41:105-178; PMID:16858867; http://dx.doi.org/10.1080/10409230600648736
- Trowitzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong IO, Schaffitzel C, Zou J et al. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun 2015; 6:6011; PMID:25586196; http://dx.doi.org/10.1038/ncomms7011
- Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 1995; 81:1115-1125; PMID:7600579; http://dx.doi.org/10.1016/S0092-8674(05)80016-9
- Wang H, Curran EC, Hinds TR, Wang E H, Zheng N. Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res 2014; 24:1433-1444; PMID:25412659; http://dx.doi.org/10.1038/cr.2014.148
- Yakovchuk P, Gilman B, Goodrich JA, Kugel JF. RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID. J Mol Biol 2010; 397:57-68; PMID:20083121; http://dx.doi.org/10.1016/j.jmb.2010.01.025
- Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, Kim HJ, Abole AK, Doud MK, Lewis TA, Koehler AN et al. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife 2015; 4; PMCID:PMC4582147; http://dx.doi.org/10.7554/eLife.07777
