ABSTRACT
We recently identified PHF13 as an H3K4me2/3 chromatin reader and transcriptional co-regulator. We found that PHF13 interacts with RNAPIIS5P and PRC2 stabilizing their association with active and bivalent promoters. Furthermore, mass spectrometry analysis identified ∼50 spliceosomal proteins in PHF13s interactome. Here, we will discuss the potential role of PHF13 in RNAPII pausing and co-transcriptional splicing.
Introduction
The role of histone binding and chromatin modulating proteins in various transcriptional regulatory processes has been well documented. We recently identified PHF13 as a novel H3K4me2/3 chromatin reader and transcriptional co-regulator.Citation1 We found that PHF13 interacts with and tethers PRC2 (a H3K27 methyltransferase) to H3K4me3/H3K27me3 bivalent promoters and that its depletion led to increased expression at these targets and reduced PRC2 binding. In addition to bivalent targets, PHF13 was also found at a greater number of active promoters (H3K4me3-only) genome wide and its depletion led to reduced expression from these targets. Together these findings imply a general role for PHF13 in transcription regulation, an assumption that is supported by the finding that PHF13 also co-localized genome-wide with promoter-associated RNA polymerase II (RNAPII) S5P and S7P. More conclusively, we could demonstrate that PHF13 interacts with RNAPII and that its depletion reduced the interaction of RNAPII S5P with H3K4me3 and H3K27me3, indicating a role for PHF13 in stabilizing RNAPII S5P at H3K4me3 active and H3K4me3/H3K27me3 bivalent chromatin. Last, while we did not explore these interactions in any detail, mass spectrometry-based analysis of PHF13 interacting proteins identified more than 50 different spliceosome proteins in its interactome, arguing for an additional role of PHF13 in co-transcriptional splicing. Here, we will discuss the implications of PHF13 affiliation with RNAPII S5P at bivalent genes, with RNAPII S5P/S7P at promoter proximal pausing and with RNAPII S2P/S5P at splicing junctions.
RNAPII, CTD modification and the transcription cycle
RNA polymerase II (RNAPII) is a highly conserved multi-subunit enzymatic complex, consisting of 12 polypeptides that are required to transcribe DNA sequences into RNA, namely mRNAs and ncRNAs.Citation2 The largest subunit is Rbp1 (Polr2a) that is conserved from bacteria to mammals and harbors the enzymatic activity of the RNAPII complex.Citation2 Eukaryotic Rbp1 contains an exposed C-terminal domain (CTD) composed of tandem heptad repeats (Y1S2P3T4S5P6S7), ranging from 26 in budding yeast to 52 in vertebrates.Citation2 Some conserved substitutions exist in specific heptads, with only 21 of 52 heptads perfectly matching the consensus sequence.Citation2 The high conservation of the 52 heptad repeats in vertebrates indicates that polymorphisms in this sequence are not tolerated. It is supposed that the CTD of Rbp1 acts as RNAPII code, which depending on the combinatorial modification pattern, recruits different histone and chromatin modifiers to impact transcription and RNA processing. To this end, we have recently identified the chromatin reader and effector protein PHF13, as a novel RNAPII S5 interacting protein.Citation1
Modifications of all CTD amino acids have been described (namely; phosphorylation, glycosylation, methylation, acetylation and isomerization) and their timely appearance is required for coordination of the transcription cell cycle.Citation3 The CTD of RNAPII can be phosphorylated at Y1, S2, T4, S5 and S7 and the phosphorylation of each of these residues is affiliated with distinct functions in the transcription cycle.Citation4 All of these residues are important for cell viability and their substitution to either phenylalanine or alanine, non-phosphorylatable residues, is lethal.Citation3,Citation4 Similarly mutation of S2, T4, S5 or S7 with glutamate, an acidic phospho-mimic is lethal,Citation5,Citation6 indicating that RNAPII's ability to return to a hypo-phosphorylated state is as important as the sequential phosphorylation of its CTD. Hypo-phosphorylated RNAPII is recruited to promoters with the help of the general transcription factors (TFIIA-TFIIH) and mediator complex which assemble to form the pre-initiation complex (PIC).Citation7 The PIC is phosphorylated by Cdk7, a component of the general transcription factor TFIIH, at position S5 and S7 of RNAPII CTD, causing its release from the mediator complex and leading to its translocation to the TSS or just downstream of the TSS.Citation4,Citation8,Citation9 RNAPII S5P/S7P is transcriptionally competent but is paused before elongation, a delay that is stabilized by the recruitment of two negative regulators of transcription, namely negative elongation factor (NELF) and DRB sensitivity inducing factor (DSIF).Citation10 In order for productive transcriptional elongation to commence, Cdk9 phosphorylates both NELF and DSIF converting them into positive regulators of transcription and concomitantly phosphorylates S2.Citation11,Citation12 Early in transcription elongating RNAPII is phosphorylated at S2 and S5, and as elongation proceeds S5P is successively removed whereas S2P gradually increases toward the 3′ end.Citation4 Therefore, the phosphorylation status of the CTD domain and key regulatory interacting factors is mediated by specific Cdk kinases, which in turn regulates the transcription cycle. Interestingly, PHF13 also contains several putative SP/TP Cdk phosphorylation sites and a Cdk consensus sequence S/T RX K/R,Citation13 raising the possibility that it may also be a substrate of Cdk7 and/or Cdk9.
The diverse functions of RNAPII S5P
Serine 5 phosphorylation of RNAPII has been demonstrated to be functionally important for bivalent poising, 5′capping, RNAPII pausing and co-transcriptional splicing.Citation4,Citation8,Citation14-Citation16 Thus, RNAPII S5P localizes to different chromatin regions, including repressed bivalent genes,Citation14 active promoters/promoter proximal regionsCitation3 and at splice site junctions.Citation3,Citation15 Furthermore, most of these RNAPII S5P contexts are affiliated with a pausing of RNAPII,Citation14,Citation17,Citation18 indicating that S5P is additionally important for co-transcriptional processing of pre-mRNA. Similarly, PHF13 localizes to each of these regions and interacts with RNAPII phosphorylated at serine 5 and serine 7, H3K4me2/3, PRC2 and several splicing factorsCitation1 linking PHF13 to the various described functions of RNAPII S5P.
Polycomb poising
Polycomb complexes (PRC1/PRC2) repress many developmental genes in embryonic stem cells (ESCs) which are characterized by bivalent histone marks (H3K4me2/3 and H3K27me3) and are associated with RNAPII containing only S5P.Citation14 ERK1/2 is responsible for S5 phosphorylation of RNAPII at bivalent repressed genes and not Cdk719, potentially explaining the absence of S7 and S2 phosphorylation on RNAPII at bivalent loci. RNAPII-mediated transcription is severely impaired in bivalent chromatin landscapes due to reduced histone acetylationCitation18 and the absence of the elongating S2P.Citation14 Coincidentally, PHF13 is highest expressed in stem cells,Citation20 where PRC2 repression of developmental promoters is important for pluripotency and likewise where PRC2-mediated repression of pluripotency factors is required for differentiation. Knockout (KO) of either PHF13 or core PRC2 factors results in defects in stem cell differentiation and maintenance arguing for their importance in these state transitions.Citation20,Citation21 Furthermore, PHF13 forms a common complex with H3K4me2/3, PRC2 and RNAPII S5P in mouse ESCs as was demonstrated by their co-elution and co-precipitation in column chromatography.Citation1 Consistently, PHF13 co-localized at a subset of PRC2 bound genes and its depletion led to increased expression from these bivalent targets, arguing that it acts as a transcriptional co-regulator at a subset of polycomb regulated genes.Citation1 Furthermore, PHF13 depletion led to the reduction of both PRC2 and RNAPII S5P at H3K4me3/H3K27me3 demarcated chromatin, arguing for a specific role of PHF13 in the recruitment or stabilization of these complexes.Citation1 Taken together, these findings strongly suggest a mutual relationship between PHF13, PRC2 and RNAPIIS5P in the transcriptional co-regulation of specific targets.
Promoter/promoter proximal pausing
In contrast to bivalent-repressed chromatin, RNAPII which is localized to H3K4me3 active promoters and promoter proximal regions is phosphorylated at both S5 and S7. RNAPII S5P/S7P is associated with promoter/promoter proximal pausing that allows for 5′ capping of the nascent RNA transcript,Citation8 which can be visualized in ChIP sequencing as a bimodal accumulation of reads near the TSS and at the +1 nucleosome.Citation22 Interestingly, the mean exon length in mouse and humans is 147 nt and corresponds to the length of DNA wrapped around a nucleosome.Citation23 For a detailed reviews on RNAPII pausing, we refer the reader to Liu et al. (2015) and Jonkers et al. (2015)Citation24,Citation25. RNAPII S5P/S7P and RNAPII that is hypo-phosphorylated at S2, highly correlate genome wide with H3K4me3 and PHF13 enrichment, at or just downstream of the TSSs.Citation1 Consistently, PHF13 co-elutes with H3K4me3, RNAPII S2P, S5P and S7P in column chromatography, however, it preferentially co-precipitates with H3K4me3, RNAPII S5P and S7P from overlapping fractions, in line with their stronger correlation genome wide.Citation1 These findings indicate an intimate relationship of PHF13 with promoter and promoter proximal-associated RNAPII, which is lost in the elongating form of RNAPII, as S5P becomes depleted. Furthermore, PHF13 depletion specifically reduced the interaction of H3K4me3 with RNAPII S5P and not with RNAPII S2P with a concomitant decreased in gene expression from several of these targets. This suggests that PHF13 tethers promoter affiliated RNAPII to H3K4me3 and has less of an impact on elongating RNAPII which is enriched in gene bodies devoid of H3K4me3 and marked by H3K36me3. Together, these findings implicate a role for PHF13 in stabilizing promoter-associated RNAPII and in the co-transcriptional regulation of active genes. It remains, however, to be determined which activating transcriptional complexes PHF13 is involved with other than RNA polymerase II itself and whether or not PHF13 can recruit H3K4 methyltransferases to active promoters, in a manner similar to its ability to recruit PRC2 to bivalent promoters. Interestingly, PHF13 has been previously shown to play a role in DNA damage response (DDR) and DNA repair and likewise to interact with several proteins important for DNA repair and surveillance.Citation26 One such factor, TRIM28 (also known as TIF1β and KAP1), has been recently shown to globally regulate RNAPII promoter proximal pausing and release, which hinged on whether or not it was phosphorylated by DNA-PK and ATM, two important DDR kinases.Citation27,Citation28 PHF13 interacts with all of these proteins to modulate DDR, and we have recently shown that in the absence of induced DNA damage (albeit DNA damage and recovery is a continuous and naturally occurring process, even in the absence of DNA damage inducing agents) PHF13 interacts with TRIM28 and ATM.Citation1 This raises the possibility that PHF13 may cooperate with these proteins in RNAPII promoter proximal pausing and release.
Splicing checkpoint
At splicing site junctions, RNAPII is phosphorylated at S2P/S5P. S5P is required for efficient co-transcriptional splicing and has been reported to be involved in spliceosome assembly at the 5′ and 3′ splice sites triggering a splicing checkpoint and RNAPII pausing at intron–exon junctions.Citation15-Citation17,Citation29-Citation31 Mammalian NET-seq demonstrated RNAPII S5P enrichment at 5′ and 3′ splice sitesCitation15 and phospho-specific RNAPII immunoprecipitations have revealed that RNAPII S5P interacts with key proteins involved spliceosomal assembly.Citation15,Citation16,Citation29 For a detailed reviews on co-transcriptional splicing, we refer the reader to Saldi et al. (2016) and Jonkers et al. (2015)Citation25,Citation32. Spliceosome assembly is a multi-step process that occurs on the pre-mRNA transcript, starting with recruitment of U1 snRNP at the 5′ splice siteCitation33 and U2 snRNP to the exonic nucleosome at the 3SSCitation34 and then at the branch point to form the pre-spliceosome (complex A). U4/U6 and U5 snRNPs are then recruited to form complex B, which is then reorganized to displace U1 and U4 and form the activated spliceosome (complex C). Mass spectrometry of PHF13s interactome revealed that it co-precipitated with ∼50 spliceosomal related proteins that were enriched in complex A, B and C specific factors and U2 snRNPs (). PHF13 co-localized genome wide with H3K4me3 and RNAPII S5P at 5′ splice sites after the first exon of expressed genes (). A functional interdependence between H3K4me3, RNAPII S5P, spliceosome assembly and co-transcriptional pre-mRNA processing has been previously shown.Citation15,Citation30,Citation35 Considering that PHF13 depletion reduces binding of RNAPII S5P to H3K4me3, this suggests a possible role for PHF13 in coupling transcription and co-transcriptional splicing at H3K4me3. Interestingly, U2 snRNPs also bind to H3K4me3 at the exonic 3SSCitation35 and may suggest a looping mechanism between the 5SS and 3SS exons during splicing and the formation of complex A.
Figure 1. Schematic depiction of splicing and splicing-related PHF13 interacting proteins. These PHF13 interacting proteins were identified by mass spectrometry of co-precipitating proteins from chromatin lysate in mouse embryonic stem cells. The interacting proteins have grouped according to their function in splicing and splicing-associated complexes.
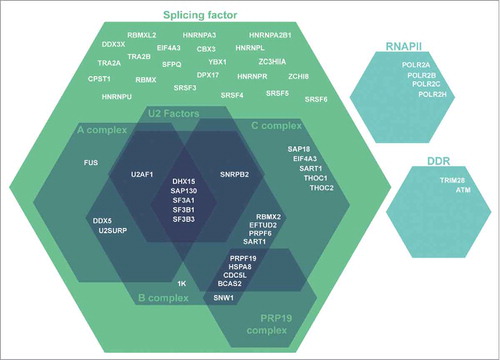
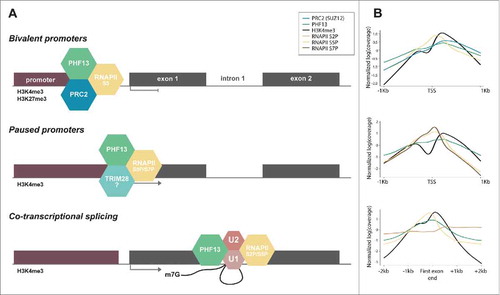
Figure 2. Schematic depiction and ChIP-seq profiles representing the relationship of PHF13 with RNAPII S5P. (A) Schematic depiction of PHF13 localization and interactions at bivalent promoters (top panel), at active paused promoters (middle panel) and at 5′ splice site junctions of the first exon (lower panel). (B) Normalized ChIP-seq read densities were plotted at PHF13 positive bivalent (H3K4me3/H3K27me3) TSS's (top panel), at PHF13 paused TSS's (middle panel) and at all PHF13 positive active (H3K4me3) genes at the end of the first exon (lower panel). ChIP-seq data accession numbers from the short read archive (http://www.ncbi.nlm.nih.gov/sra) PHF13 (E-MTAB-2636), H3K4me3 (SRX026272, SRX149184, SRX149188, SRX185845, SRX186795), H3K27me3 (SRX006976, SRX026276), SUZ12 (SRX206420), RNAPII 8WG16 (SRR391036, SRR391037), RNAPII S2P (SRR391034, SRR391038, SRR391039), S5P (SRR391032, SRR391033, SRR391050) and S7P (SRR391035, SRR391040). The ChIP-seq reads for each experiment were individually mapped against the mouse genome (genome version mm10) using Bowtie2 with default parameters. ChIP-seq enrichment calling was done using normR R package (Johannes Helmuth and Ho-Ryun Chung (2016). normr: Normalization and difference calling in ChIP-seq data. R package version 0.99.7. https://github.com/your-highness/normR) for PHF13, RNAPII (8WG16 antibody), H3K4me3 and H3K27me3 against individual input controls (FDR ≤ 0.05). For further analysis only PHF13 positive promoters were considered. Promoters enriched for H3K4me3 were regarded as active, while bivalent promoters were defined as H3K4me3 and H3K27me3 positive. Pausing index was calculated with custom script using RNAPII ChIP-seq data, genes with pausing index >10 were considered as paused. For data visualization, the signals were normalized and plotted in R.
H3K4me3 and co-transcriptional splicing
Several arguments support a role for H3K4me3 in co-transcriptional splicing: (1) H3K4me3 is found at active promoters predominantly overlapping with hypomethylated CpG islands, localizing from the TSS to the 5′ splice site of the first exon.Citation36 (2) A recent study exploring alternative splicing, which used large data sets from ENCODE, has demonstrated the presence of promoter like signatures (H3K4me3, H3K27Ac and H3K9Ac) in exons which marked them for inclusion.Citation37 (3) In vitro transcription assays have demonstrated that H3K4me3 is not essential for transcription,Citation38 indicating that its presence either requires other effector proteins to impact transcription and/or that it is important for co-transcriptional processes and not transcription per se. (4) The deposition of H3K4me3 by H3K4-specific histone methyltransferases (HMTs) requires active transcription,Citation38 arguing that its positive correlation with transcription is rather a consequence than a cause and suggests that H3K4me3 may be more important for co-transcriptional processes. (5) H3K4me3 and pre-mRNA splicing are interdependent as attenuation of one negatively impacts the other.Citation35,Citation36,Citation39 Together, these findings support that H3K4me3 is more relevant for co-transcriptional splicing than transcription itself. Intriguingly, CHD1, an H3K4me3 reader and chromatin remodeler was demonstrated to bridge H3K4me3 to the U2 snRNP and depletion of either CHD1 or H3K4me3, perturbed pre-mRNA splicing.Citation35 This indicates that H3K4me3 found at splice sites of the first exon and at exons marked for alternative splicing, act as a molecular cue to direct the co-transcriptional splicing machinery via H3K4me3 molecular readers. PHF13 is another good candidate for such a role, making it tempting to speculate that PHF13 acts at the interface between RNAPII mediated transcription and co-transcriptional splicing.
Tying it all together with PHF13
Serine 5 phosphorylation of RNAPII is associated with different forms of transcriptional pausing: at bivalent promoters, at promoter proximal regions and at 5′/3′ splice sites. This correlation with paused transcription similarly implies that PHF13 which is associated with RNAPII S5P in each of these contexts may also contribute to RNAPII pausing or retention. We have schematically depicted the different roles and relationship of PHF13 with RNAPII S5P in and have used data obtained from ChIP sequencing experiments to support these models (for accession codes see figure legend). We suggest that PHF13 in conjunction with TRIM28 may act to regulate RNAPII S5P promoter proximal pausing, with PRC2 to enforce transcriptional pausing at bivalent loci and with spliceosomal proteins at splice site junctions to exert a transient splicing checkpoint. In conclusion, we speculate that PHF13 tethers and stabilizes RNAPII S5P to H3K4me3 that culminate in RNAPII pausing to allow for specific and essential co-transcriptional regulatory processes to occur.
In the future, it will be important to see if PHF13 can interact with and recruit specific H3K4 histone methyltransferases (HMTs) to chromatin, similar to its described function with PRC2 (a H3K27 HMT) and to explore the impact of PHF13 depletion on H3K4me3 deposition and splicing as both processes are transcriptionally coupled. Furthermore, it will also be clearly important to experimentally and functionally characterize the specific impact of PHF13 on splicing and/or alternative splicing and it will be interesting to explore the consequence of PHF13 depletion on RNAPII pausing and the downstream processes that are associated with it. Answers to these questions will provide substantial insight into the detailed mechanisms of PHF13 co-transcriptional regulatory functions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Xinyi Yang for her support with the bioinformatic analysis.
Funding
This work was supported by the Max Planck Society.
References
- Chung HR, Xu C, Fuchs A, Mund A, Lange M, Staege H, Schubert T, Bian C, Dunkel I, Eberharter A et al. PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3. Elife 2016; 5:e10607; PMID: 27223324; http://dx.doi.org/10.7554/eLife.10607
- Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 2013; 113(11):8456-8490; PMID:23952966; http://dx.doi.org/10.1021/cr400071f
- Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 2013; 1829(1):55-62; PMID:22982363; http://dx.doi.org/10.1016/j.bbagrm.2012.08.013
- Srivastava R, Ahn SH. Modifications of RNA polymerase II CTD: Connections to the histone code and cellular function. Biotechnol Adv 2015; 33(6 Pt 1):856-872; PMID: 26241863; http://dx.doi.org/10.1016/j.biotechadv.2015.07.008
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 1995; 140(4):1223-1233; PMID: 7498765
- Zhang DW, Mosley AL, Ramisetty SR, Rodríguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 2012; 287(11):8541-8551; PMID: 22235117; http://dx.doi.org/10.1074/jbc.M111.335687
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 2009; 21(3):344-351; PMID: 19411170; http://dx.doi.org/10.1016/j.ceb.2009.03.006
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 2009; 29(20):5455-5464; PMID:19667075; http://dx.doi.org/10.1128/MCB.00637-09
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 1998; 2(1):43-53; PMID: 9702190; http://dx.doi.org/10.1016/S1097-2765(00)80112-4
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999; 97(1):41-51; PMID: 10199401; http://dx.doi.org/10.1016/S0092-8674(00)80713-8
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 2004; 24(2):787-795; PMID: 14701750; http://dx.doi.org/10.1128/MCB.24.2.787-795.2004
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 1996; 271(43):27176-27183; PMID: 8900211; http://dx.doi.org/10.1074/jbc.271.43.27176
- Kinkley S, Staege H, Mohrmann G, Rohaly G, Schaub T, Kremmer E, Winterpacht A, Will H. SPOC1: a novel PHD-containing protein modulating chromatin structure and mitotic chromosome condensation. J Cell Sci 2009; 122(Pt 16):2946-2956; PMID: 19638409; http://dx.doi.org/10.1242/jcs.047365
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 2012; 10(2):157-170; PMID: 22305566; http://dx.doi.org/10.1016/j.stem.2011.12.017
- Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 2015; 161(3):526-540; PMID: 25910207; http://dx.doi.org/10.1016/j.cell.2015.03.027
- Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, Churchman LS. Comprehensive RNA Polymerase II Interactomes Reveal Distinct and Varied Roles for Each Phospho-CTD Residue. Cell Rep 2016; 15(10):2147-2158; PMID: 27239037; http://dx.doi.org/10.1016/j.celrep.2016.05.010
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 2010; 40(4):582-593; PMID: 21095588; http://dx.doi.org/10.1016/j.molcel.2010.11.005
- Tie F, Banerjee R, Fu C, Stratton CA, Fang M, Harte PJ. Polycomb inhibits histone acetylation by CBP by binding directly to its catalytic domain. Proc Natl Acad Sci U S A 2016; 113(6):E744-E753; PMID: 26802126; http://dx.doi.org/10.1073/pnas.1515465113
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 2014; 156(4):678-690; PMID: 24529373; http://dx.doi.org/10.1016/j.cell.2014.01.009
- Bordlein A, Scherthan H, Nelkenbrecher C, Molter T, Bösl MR, Dippold C, Birke K, Kinkley S, Staege H, Will H et al. SPOC1 (PHF13) is required for spermatogonial stem cell differentiation and sustained spermatogenesis. J Cell Sci 2011; 124(Pt 18):3137-3148; PMID: 21852425; http://dx.doi.org/10.1242/jcs.085936
- Obier N, Lin Q, Cauchy P, Hornich V, Zenke M, Becker M, Müller AM. Polycomb protein EED is required for silencing of pluripotency genes upon ESC differentiation. Stem Cell Rev 2015; 11(1):50-61; PMID: 25134795; http://dx.doi.org/10.1007/s12015-014-9550-z
- Quinodoz M, Gobet C, Naef F, Gustafson KB. Characteristic bimodal profiles of RNA polymerase II at thousands of active mammalian promoters. Genome Biol 2014; 15(6):R85; PMID: 24972996; http://dx.doi.org/10.1186/gb-2014-15-6-r85
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G. Changes in exon-intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 2012; 22(1):35-50; PMID: 21974994; http://dx.doi.org/10.1101/gr.119834.110
- Liu X, Kraus WL, Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem Sci 2015; 40(9):516-525; PMID: 26254229; http://dx.doi.org/10.1016/j.tibs.2015.07.003
- Jonkers I, JT Lis. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16(3):167-177; PMID:25693130; http://dx.doi.org/10.1038/nrm3953
- Mund A, Schubert T, Staege H, Kinkley S, Reumann K, Kriegs M, Fritsch L, Battisti V, Ait-Si-Ali S, Hoffbeck AS et al. SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response. Nucleic Acids Res 2012; 40(22):11363-11379; PMID:23034801; http://dx.doi.org/10.1093/nar/gks868
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G et al. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol 2014; 21(10):876-883; PMID: 25173174; http://dx.doi.org/10.1038/nsmb.2878
- McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D'Orso I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol Cell 2016; 61(1):39-53; PMID:26725010; http://dx.doi.org/10.1016/j.molcel.2015.11.004
- Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 2014; 53(5):779-790; PMID: 24560925; http://dx.doi.org/10.1016/j.molcel.2014.01.017
- Park D, Shivram H, Iyer VR. Chd1 co-localizes with early transcription elongation factors independently of H3K36 methylation and releases stalled RNA polymerase II at introns. Epigenetics Chromatin 2014; 7(1):32; PMID: 25395991; http://dx.doi.org/10.1186/1756-8935-7-32
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 2015; 161(3):541-554; PMID: 25910208; http://dx.doi.org/10.1016/j.cell.2015.03.010
- Saldi T, Cortazar MA, Sheridan RM, Bentley DL. Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing. J Mol Biol 2016; 428(12):2623-2635; PMID: 27107644; http://dx.doi.org/10.1016/j.jmb.2016.04.017
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 1994; 368(6467):119-124; PMID: 8139654; http://dx.doi.org/10.1038/368119a0
- Kfir N, Lev-Maor G, Glaich O, Alajem A, Datta A, Sze SK, Meshorer E, Ast G. SF3B1 association with chromatin determines splicing outcomes. Cell Rep 2015; 11(4):618-629; PMID: 25892229; http://dx.doi.org/10.1016/j.celrep.2015.03.048
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 2007; 28(4):665-676; PMID: 18042460; http://dx.doi.org/10.1016/j.molcel.2007.11.010
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep 2012; 2(1):62-68; PMID: 22840397; http://dx.doi.org/10.1016/j.celrep.2012.05.019
- Curado J, Iannone C, Tilgner H, Valcárcel J, Guigó R. Promoter-like epigenetic signatures in exons displaying cell type-specific splicing. Genome Biol 2015; 16:236; PMID: 26498677; http://dx.doi.org/10.1186/s13059-015-0797-8
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006; 125(4):703-717; PMID:16713563; http://dx.doi.org/10.1016/j.cell.2006.04.029
- Davie JR, Xu W, Delcuve GP. Histone H3K4 trimethylation: dynamic interplay with pre-mRNA splicing. Biochem Cell Biol 2016; 94(1):1-11; PMID: 26352678; http://dx.doi.org/10.1139/bcb-2015-0065
