ABSTRACT
Transcription factors (TFs) are among the most frequently detected targets of sumoylation, and effects of the modification have been studied for about 200 individual TFs to date. TF sumoylation is most often associated with reduced target gene expression, which can be mediated by enhanced interactions with corepressors or by interference with protein modifications that promote transcription. However, recent studies show that sumoylation also regulates gene expression by controlling the levels of TFs that are associated with chromatin. SUMO can mediate this by modulating TF DNA-binding activity, promoting clearance of TFs from chromatin, or indirectly, by influencing TF abundance or localization.
Introduction
Sumoylation is a conserved eukaryotic post-translational modification (PTM) that regulates numerous cellular processes, including several that are involved in gene expression (reviewed in Refs. Citation1-3). Gene Ontology (GO) analysis of a recent human SUMO proteomics study indicates that nearly 300 sequence-specific DNA-binding transcription factors (TFs), that function as activators or repressors of transcription by RNA polymerase II (RNAP II), are SUMO conjugates, which represents over 50% of all TFs in this category, suggesting that TFs are particularly subject to regulation by SUMO.Citation4 To date, the effects of sumoylation on ∼200 individual TFs have been examined in over 350 published studies, and, with notable exceptions,Citation2 SUMO modifications are generally associated with reduced or repressed transcription.Citation5,6 SUMO can impair transcription by promoting the recruitment of transcriptional corepressor complexes or by interfering with transcription-promoting PTMs, such as acetylation and phosphorylation. However, as supported by several recent studies, it is becoming evident that a major consequence of TF sumoylation is altered levels of TFs that are associated with their target binding sites on chromatin. This can occur directly, by influencing DNA-binding ability or promoting clearance of TFs from chromatin, or indirectly, by regulating TF abundance or localization. Here, we discuss, with recent examples, these mechanisms by which sumoylation regulates TF function and association with chromatin, and how SUMO frequently imparts its negative effects on gene expression by antagonizing transcriptional activators through diverse mechanisms.
The sumoylation pathway
Sumoylation involves the covalent attachment of a ∼12 kDa SUMO polypeptide to specific Lys residues on target proteins.Citation1-3 There are five SUMO isoforms in mammals, SUMO1, the nearly identical SUMO2 and SUMO3, and the less-studied isoforms SUMO4 and SUMO5, whereas D. melanogaster, C. elegans and lower eukaryotes, including yeasts, express a single SUMO form.Citation7,8 SUMO polypeptides are expressed as precursors that are cleaved to their mature form by members of the SUMO protease family of isopeptidases, which exposes a C-terminal di-glycine motif that is required for conjugation to target proteins. Like ubiquitination, the SUMO conjugation pathway involves a cascade of enzymatic activities.Citation1 First, the mature SUMO polypeptide is activated by the E1 class activating enzyme, composed of SAE1 and SAE2 subunits, in an ATP hydrolysis-dependent manner. SUMO is then transferred to the sole E2 conjugating enzyme Ubc9, which catalyzes the covalent SUMO modification of the target protein. Although not essential for sumoylation in vitro, SUMO E3 ligases provide target specificity and enhanced sumoylation efficiency.
Unlike ubiquitination, the SUMO acceptor site is often situated within a consensus motif whose core sequence is Ψ-K-x-D/E, where Ψ represents a large hydrophobic residue, K is the modified Lys residue, x is any residue, and D/E represents Asp or Glu.Citation1,3 Through bioinformatics analysis of multiple SUMO acceptor sites, extended consensus motifs have also been identified, such as the negatively charged amino acid-dependent sumoylation motif (NDSM), which includes clusters of acidic residues downstream of the core motif.Citation9 Recognition of SUMO acceptor sites within some target proteins requires prior phosphorylation of nearby residues, and these sites are consequently referred to as phosphorylation-dependent sumoylation motifs (PDSM).Citation10 The dependence on phosphorylation for these SUMO targets reflects the frequently observed cross-talk between phosphorylation and sumoylation on TFs, with one modification promoting or inhibiting the other.Citation11 For example, MAPK-mediated phosphorylation of Bcl11b promotes its desumoylation through recruitment of the SENP1 SUMO protease during T-cell development.Citation12 SENP1 is one of multiple mammalian SUMO proteases of the SENP family, whose ubiquitous activity is at least partly responsible for the low level of sumoylation typically seen with most SUMO targets.Citation13
The molecular consequences of SUMO modifications depend on the specific target proteins, but often result in altered subcellular localization, activity, or stability.Citation1-3 The SUMO moiety is thought to mediate these effects by modulating protein–protein interactions, in some cases enabling and in other cases precluding the interactions. Proteins that associate with SUMO-modified partners contain one or more SUMO-interacting motifs (SIMs), typically consisting of several hydrophobic residues situated adjacent to an acidic patch of amino acid residues, which bind directly to a surface on SUMO polypeptides.Citation11 Conjugated SUMO2/3 polypeptides themselves can be sumoylated, forming SUMO chains that are recognized by proteins that contain multiple adjacent SIMs, such as the SUMO-targeted ubiquitin ligases (STUbLs).Citation14 STUbLs trigger the degradation of target polysumoylated proteins by promoting their ubiquitination and subsequent degradation through the 26S proteasome.Citation15 As with sumoylation and phosphorylation, STUbL-mediated ubiquitination is another example of the frequently observed interdependence of SUMO and other protein modifications. The effects of sumoylation on downstream PTMs of target proteins, including phosphorylation, ubiquitination, and acetylation, are discussed below.
Interaction with HDACs and other transcriptional coregulators
In 2004, sumoylation of the ETS domain TF Elk-1 was shown to promote the recruitment of the histone deacetylase complex (HDAC) HDAC-2, a transcriptional corepressor, to promoters of Elk-1 target genes.Citation16 Since then, mutation of SUMO acceptor sites on numerous other TFs has been found to reduce the association of the TFs with HDACs, and/or impair the recruitment of HDACs to their target genes (; reviewed in refs. Citation5,17). Recent studies continue to support HDAC recruitment as a major effect of TF SUMO modification. For example, a sumoylation-deficient form of GFI1, a repressor of haematopoietic stem cell genes, fails to associate efficiently with components of the CoREST HDAC and is defective in transcriptional repression,Citation18 while a form of glucocorticoid receptor (GR) that cannot be sumoylated is unable to recruit the NCoR1/SMRT HDAC to a subset of GR-repressed target genes.Citation19 HDACs are the most commonly implicated, but recruitment of other types of corepressors is also promoted by TF sumoylation. For example, G9a, a histone H3 Lys 9 (H3K9) methyltransferase, is targeted to muscle-specific promoters by sumoylation of the repressor Sharp-1 to inhibit skeletal muscle differentiation.Citation20 The frequently observed connection between TF sumoylation and corepressor recruitment is likely explained by the observation that multiple components of corepressor complexes, including CoREST, NCoR1, and SMRT, are capable of interacting directly with SUMO isoforms.Citation17,18,21 This suggests a straightforward mechanism of SUMO-mediated transcriptional repression through recruitment of corepressor complex subunits by sumoylated, DNA-bound TFs ().
Figure 1. SUMO controls TF function through multiple mechanisms. (A) Sumoylation of many repressor TFs enhances transcription inhibition by recruiting HDACs. SUMO interacts directly with components of HDACs, which repress transcription. (B) Sumoylation of some TFs results in recruitment of coactivators CBP/p300, which bind directly to SUMO1, thereby stimulating transcription. (C) SUMO competes with acetylation for target Lys residues on multiple TFs. As TF acetylation frequently promotes gene activation, sumoylation has an inhibitory effect on transcription in these cases. (D) SUMO interferes with phosphorylation at nearby residues on transcription factors that require phosphorylation for full activation of target genes. Examples for each scenario, as described in the text, are listed below each panel. Mechanisms of SUMO action are represented by red arrows; repressive effects of sumoylation on transcription are indicated by red X marks, and stimulation of transcription is indicated by a green arrow. Encircled S represents SUMO; encircled Ac, acetyl; encircled P, phosphate.
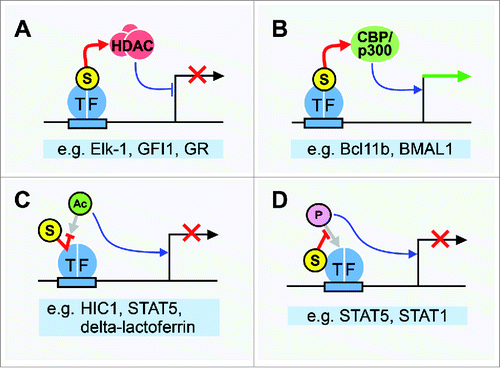
Reflecting SUMO's less frequent role in promoting transcriptional activation, in only a few studies have SUMO modifications been shown to promote recruitment or association with coactivators, particularly the homologous histone acetyltransferases CBP and p300 ().Citation12,22,23 Sumoylation of the T cell TF Bcl11b enables its interaction with p300, which is necessary for derepression of genes that it normally represses.Citation12 Similarly, association of CBP with the CLOCK1-BMAL1 TF complex is dependent on BMAL1 sumoylation, allowing transcription of the circadian rhythm-regulated gene Per1.Citation22 CBP/p300 contains a type of ZZ zinc finger domain that mediates a direct interaction with SUMO1, suggesting that additional TFs might recruit CBP/p300 through their sumoylation.Citation24 In other cases, TF sumoylation promotes transcription not by recruiting coactivators, but by disrupting the recruitment or assembly of corepressor complexes. For example, HDAC3 can suppress gene activation through TFII-I, which binds the initiator element of several promoters, but sumoylation of TFII-I impairs its interaction with HDAC3, and, consequently, a sumoylation-deficient form of TFII-I showed reduced induction of transcription from the c-fos promoter.Citation25 Further work is needed to understand how SUMO modifications selectively regulate the recruitment of either corepressors or coactivators in a context-dependent manner.
Inhibiting acetylation and phosphorylation
In addition to directly modulating the recruitment of transcriptional coregulator complexes to DNA-bound TFs, SUMO modifications often indirectly influence TF activity by inhibiting other types of PTMs on the TF ( and ). Since conjugation of SUMO to Lys precludes other modifications on the residue, sumoylation can compete with other Lys-based modifications to regulate TF function. Acetylation of the tumor suppressor gene product HIC1 impairs its association with the NuRD HDAC component MTA1, thereby reducing its potential as a repressor of target genes. However, sumoylation restores the repressive functions of HIC1 by competing with acetylation for the same Lys residue and promoting the interaction with MTA1.Citation26 Similarly, one of the five SUMO acceptor sites on the intracellular TF form of lactoferrin, delta-lactoferrin, is also its main site of acetylation, K13. Acetylation at this residue was shown to preclude sumoylation and thereby promote transcriptional activation on a delta-lactoferrin responsive reporter gene.Citation27 Competition between SUMO and Lys-based ubiquitin modifications has also been observed, usually influencing TF stability (discussed below), but the competition between sumoylation and acetylation often represents a switch between active and inactive forms of modified TFs, with SUMO typically acting as the “off” switch.Citation26-28
Although sumoylation and phosphorylation occur on different amino acid residues, SUMO modifications can influence the phosphorylation status of nearby residues. STAT5, which drives transcription of genes required for development and immune cell function, is sumoylated at two neighboring residues, K696 and 700. Sumoylation of STAT5 inhibits phosphorylation at nearby Y694 and competes with acetylation at K696, thereby impeding two transcription-promoting modifications.Citation29 During cytokine-induced activation of another member of the STAT family, STAT1, phosphorylation at Y701 promotes the formation of polymers called paracrystals, which are associated with the active form of the TF. Sumoylation at nearby K703 obstructs phosphorylation at this position, thereby interfering with polymer formation and limiting gene activation.Citation30 These examples highlight how the transcriptional repressive effects of SUMO can be mediated by antagonizing other PTMs that promote transcription.
Association with and availability to bind chromatin
In addition to regulating TF activity through interactions with transcriptional coregulators and other PTMs, sumoylation controls the association of TFs with their target sites on chromatin through multiple mechanisms (). This has become evident through several recent studies in which the effects of sumoylation on the TF-chromatin interaction were assayed by chromatin immunoprecipitation (ChIP). In all cases, altering TF sumoylation levels resulted in altered TF chromatin occupancy (see ). Considering also, as discussed below, the frequently-observed effects of TF sumoylation on the subcellular localization and stability of TFs, it is becoming increasingly apparent that a major direct or indirect consequence of TF sumoylation is altered levels of chromatin-bound TFs. Indirectly, sumoylation can control the abundance or subcellular distribution of TFs, thereby influencing the number of TFs that are available for binding to their chromatin target sites, whereas, more directly, sumoylation can influence DNA binding or promote the clearance of DNA-bound TFs from chromatin. These mechanisms are discussed in the four sections below.
Figure 2. SUMO controls levels of chromatin-associated TFs through diverse mechanisms. (A) Some TFs are sumoylated specifically when bound to DNA during active transcription of target genes, and the modification acts to promote their clearance from DNA, thereby restricting gene expression. (B) STUbLs associate with polysumoylated targets, which results in their ubiquitination and degradation through the 26S proteasome (upper panel). In other cases, TF sumoylation interferes with subsequent ubiquitination, often through competition for target Lys residues, thereby preventing degradation of the target (lower panel). Encircled Ub represents ubiquitin. (C) For many TFs, SUMO has a more direct role in regulating the association with chromatin by inhibiting (upper panel) or promoting (lower panel) their DNA-binding activities. (D) Sumoylation regulates the subcellular localization of TFs, either inhibiting (upper panel) or promoting (lower panel) nuclear retention, or targeting TFs to specific nuclear regions (not depicted). Examples for each scenario, as described in the text, are listed below each panel. The consequences of sumoylation on the association of TFs with chromatin is shown on the right of each panel, with the empty DNA symbol indicating that sumoylation reduces the association, and DNA depicted with a TF indicating that sumoylation promotes the association. Bent arrows represent transcriptional start sites situated downstream of TF binding sites.
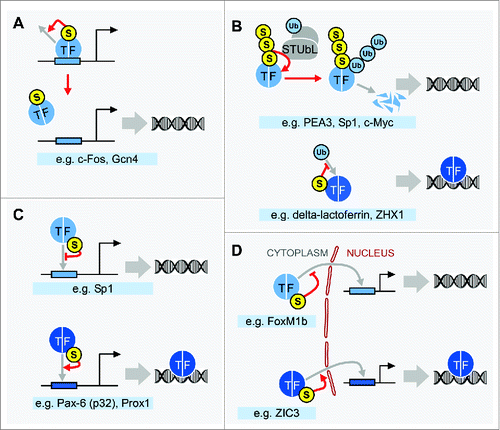
Table 1. Effects of sumoylation on TF chromatin occupancy, determined by ChIP, and target gene expression.
summarizes the results of studies in which ChIP was used to examine the effects of TF sumoylation on chromatin occupancy. In each study, the level of TF sumoylation was modulated by at least one of three methods: blocked or reduced sumoylation by mutation of TF SUMO acceptor sites or nearby residues, elevated sumoylation by expression of a SUMO-TF fusion protein, or by altering cellular levels of SUMO by overexpression or siRNA-mediated silencing. Although informative, each method carries potential caveats. For example, point mutations can interrupt other types of PTMs, fusion to SUMO does not accurately recapitulate natural Lys sumoylation, and altered expression of sumoylation enzymes influences numerous substrates besides the specific target TF. Predominantly, however, elevated sumoylation levels correlate with reduced TF occupancy, and decreased sumoylation correlates with increased occupancy, suggesting that sumoylation acts in restricting TF association with chromatin. GATA-1 and Prox1, are exceptions, however, as both showed somewhat reduced levels of chromatin occupancy when their SUMO acceptor sites were mutated, and SUMO fusion to p53 resulted in higher occupancy on some target genes and reduced occupancy on others.Citation31-34 Whether sumoylation has a widespread role in reducing TF chromatin occupancy will be determined as future studies continue to explore the consequences of TF sumoylation on target site occupancy. However, such a role might be limited to situations where the effects of sumoylation are not dependent on TFs remaining associated with DNA, such as in the recruitment of HDACs.
In all the studies listed in , there is a correlation between the change in TF occupancy and the transcriptional consequences that result from altered sumoylation. For example, SUMO acceptor site mutation of FOXA1, a pioneer TF involved in prostate development, resulted in higher occupancy of the TF on three target loci, and consequently, higher induction of these genes.Citation35 For Scm, a member of the Drosophila Polycomb group of transcriptional repressors, reduced global sumoylation and SUMO acceptor site mutation also correlated with higher target site occupancy, but consistent with its role as a repressor of homeotic genes, this resulted in decreased expression of its target gene Ubx.Citation36 A link between TF sumoylation and human disease was made in an insightful pair of studies in which a polymorphism in the TF MITF that predisposes affected individuals to melanoma and renal cell carcinoma was discovered to severely reduce its sumoylation.Citation37,38 The polymorphism results in a missense substitution, E318R, that impairs sumoylation at adjacent K317, and reduced sumoylation was found to correlate with increased occupancy of MITF on its target gene HIF1A, with a consequential increase in HIF1A expression.Citation38 These studies and the others summarized in point to a strong relationship between the transcriptional consequences of TF sumoylation and the effects of sumoylation on chromatin occupancy of those TFs. It will therefore be critical in future studies to determine whether the transcriptional effects that are attributed to TF sumoylation are at least partly due to SUMO-dependent changes in the association of TFs with their chromatin targets.
(i) SUMO promotes TF clearance from chromatin
Multiple lines of evidence suggest that at least some TFs are modified by SUMO specifically when they are associated with chromatin. For example, in so-called re-ChIP experiments, in which sequential TF and SUMO immunoprecipitations were performed, sumoylated forms of Elk-1, FXR, and Oct4 were found to be bound to their target gene promoters.Citation39-41 Sumoylation of the hematopoiesis-associated TF Ikaros was shown to depend on its intact DNA-binding domain, suggesting that SUMO regulates Ikaros function specifically after it binds to its chromatin targets.Citation42 A number of groups have used ChIP in yeast or mammalian cells to show that Ubc9 and sumoylated proteins are detected specifically near promoters or transcriptional start sites of actively transcribed RNAP II-dependent genes, which implies that sumoylation plays a general role in regulating TFs during active transcription.Citation43-47 For the most part, the specific function of SUMO at these transcriptionally active chromatin sites is unknown, and whether sumoylation affects occupancy of TFs while bound to DNA during active transcription has not been examined.
In a series of recent studies, however, sumoylation was found to promote the clearance of two TFs from DNA, suggesting that this might be a common mechanism by which SUMO modifications can regulate the occupancy of TFs on target DNA (). Using one of the only antibodies reported to date that recognizes a SUMO-modified target specifically, Tempe and colleagues used ChIP to demonstrate that c-Fos is sumoylated when it is bound to its target DNA elements on the promoter of a transcriptionally active reporter gene, thereby correlating c-Fos sumoylation with active transcription. Nonetheless, blocking c-Fos sumoylation resulted in increased activity of the reporter gene and higher levels of c-Fos on its promoter, suggesting that SUMO modifies DNA-bound c-Fos during active transcription to limit its association with promoter DNA.Citation48 Similarly, we recently showed that sumoylation of the yeast activator Gcn4, which is triggered by DNA binding itself, reduces its association with chromatin and thereby dampens transcription levels of its target genes.Citation49,50 In this case, sumoylation of DNA-bound Gcn4 was found to stimulate its phosphorylation by the RNAP II Mediator-associated kinase Cdk8, which then signals for its ubiquitination and proteolysis, thereby clearing Gcn4 from target sites on chromatin. For both c-Fos and Gcn4, sumoylation of the TF on active promoters is thought to restrict its occupancy by enabling its clearance once it has functioned in the recruitment of RNAP II, thereby preventing uncontrolled expression of target genes. Particularly, if a similar mechanism is found to be at play with other TFs in future analyses, these studies might at least partly explain why an apparently repressive mark, SUMO, is so strongly associated with transcriptionally active genes.
(ii) SUMO influences TF stability
Regardless of whether or not sumoylation occurs specifically when TFs are bound to chromatin, SUMO modifications frequently control TF stability by interfering with or promoting subsequent ubiquitination (). Evidence for widespread dependence of ubiquitination on prior sumoylation was provided in a recent study in which overexpression of SUMO3 in HEK293T cells resulted in increased global levels of protein ubiquitination, while overexpression of the SUMO protease SENP3 had the opposite effect.Citation51 Much of this is likely due to the activity of STUbLs, which recognize specific sumoylated targets and mark them with ubiquitin chains for degradation. For example, PEA3, a TF involved in neuronal pathfinding, and the ubiquitous TFs Sp1 and c-Myc are recognized in their sumoylated form by the STUbL RNF4 which promotes their ubiquitination and degradation.Citation52-54 Despite the correlation between global levels of sumoylation and ubiquitination, in many cases, sumoylation acts to increase target protein stability by inhibiting ubiquitination. This was first observed with IκBα, the inhibitor of the TF NF-κB, for which sumoylation antagonizes its ubiquitination, leaving it resistant to degradation.Citation55 For delta-lactoferrin and the homeodomain TF ZHX1, sumoylation and ubiquitination are also thought to compete for modification of the same Lys residues, with sumoylation favoring stability of the TFs by blocking ubiquitin-mediated proteolysis.Citation27,56 In other cases, TF sumoylation inhibits ubiquitination with a consequential increase in TF stability, but whether the modifications compete for the same Lys residues is not yet known (e.g., refs. Citation57-59). The multiple examples of cross-talk between sumoylation and ubiquitination suggest that SUMO frequently controls TF abundance, and consequently regulates the levels of TFs that are available for binding to chromatin.
(iii) SUMO controls TF DNA binding
For many TFs, SUMO has a more direct role in controlling access to target sites on chromatin by regulating their ability to bind DNA (). Multiple studies have examined the effects of sumoylation on DNA binding through in vitro assays. For Sp1, a zinc finger TF with multiple target genes, sumoylation by SUMO2, but not SUMO1, leads to reduced binding to the βB1-crystallin gene promoter in electrophoretic mobility shift analysis and, in agreement, co-expression of SUMO2 with Sp1 in transfected human lens epithelial cells resulted in reduced activation of β-crystallin genes.Citation60 In contrast, the p32 isoform of Pax-6, a homeodomain TF that regulates brain and eye development, does not efficiently bind a DNA sequence containing its target P3 element in vitro unless it is already sumoylated with SUMO1.Citation61 Another homeodomain TF, Prox1, also shows dependence on SUMO1 modification for binding to its DNA targets. In a DNA affinity precipitation assay, wild-type Prox1 derived from a human endothelial cell line was capable of binding an oligonucleotide containing the VEGF promoter sequence, whereas a SUMO acceptor site mutant form of Prox1 (K556R) bound only weakly.Citation34 Supporting the notion that these in vitro observations reflect authentic effects of SUMO on DNA binding in living cells, this effect was corroborated in vivo by ChIP analysis, which showed that wild-type Prox1, but not the K556R mutant, occupies the natural VEGF promoter.Citation34 Influencing the ability of TFs to bind DNA is a direct mechanism by which SUMO can influence the recruitment of TFs to chromatin, but, in most cases, further studies are needed to determine the structural consequences of SUMO modification that lead to changes in DNA binding.
(iv) SUMO controls TF subcellular localization
Sumoylation regulates the subcellular localization of many TFs, and as a consequence, the transcriptional effects of sumoylation can often be at least partly explained by restricted or enhanced access of modified TFs to their chromatin targets (). This is the case with FoxM1b, a TF involved in mitotic entry and progression, whose transcriptional activity is suppressed by sumoylation. SUMO acceptor site mutation of FoxM1b causes increased nuclear localization, whereas a SUMO-FoxM1b fusion protein showed increased levels in the cytoplasm, indicating that sumoylation promotes cytoplasmic retention of this TF.Citation62 On the other hand, sumoylation is associated with nuclear retention for ZIC3, an X-linked transcriptional repressor required for normal heart development, likely explaining why the intact SUMO acceptor site on ZIC3 is required for efficient repression of the cardiac α-actin gene promoter.Citation63 Consistent with the multiple observations that SUMO conjugates frequently accumulate in nuclear foci, specifically PML bodies in mammals,Citation64 sumoylation can regulate TFs by targeting them to specific subnuclear regions. For example, in human CD4+ T-cells, sumoylation is required for the recruitment of NFATc1 to PML bodies where it interacts with histone deacetylases, resulting in transcriptionally inactive chromatin and repression of its target gene interleukin-2.Citation65 Controlling nuclear availability and subnuclear localization, therefore, appear to be important mechanisms by which SUMO regulates TF function and the access of TFs to their chromatin targets.
SUMO: A general antagonist of transcription?
The link between SUMO and transcriptional repression first emerged from early studies on TFs, including Sp3, c-Jun, and c-Myc, all of which showed enhanced ability to activate target genes when their SUMO acceptor sites were mutated.Citation66–69 As more sumoylated TFs were identified and studied, the link was upheld. Indeed, approximately 70% of all published studies that examined transcriptional effects of TF sumoylation report a negative effect for the modification on expression of target genes. As described above, for TFs that act as repressors, SUMO generally enhances the repressive effect by directly recruiting HDACs or other corepressors to DNA-bound, sumoylated TFs. Many sumoylated TFs, however, are transcriptional activators whose ability to induce transcription is impaired by SUMO modification through one or multiple mechanisms. For example, sumoylation of FoxM1b promotes its cytosolic translocation and facilitates its ubiquitin-mediated degradation, which results in reduced expression of its target genes.Citation62 Similarly, the ubiquitous transcriptional activator ATF7 is impaired by SUMO1 modification that inhibits its nuclear translocation, impedes its interaction with the general transcription machinery, and prevents it from binding to target DNA sequences.Citation70 In some cases, the antagonistic effects of SUMO on transcriptional activators are dramatic and result in a full reversal of function, with activators becoming repressors when sumoylated.Citation71,72 It would seem, then, that SUMO frequently functions as a general antagonist of transcriptional activators, working at many levels, potentially as a means of restricting their activity and preventing uncontrolled expression of their target genes.Citation48,49
Considering that numerous TFs are modified by SUMO and that the modification almost always affects transcription levels of their target genes, is it possible that cells can coordinately regulate global transcription patterns by modulating cellular sumoylation levels? For example, one might expect that increasing overall sumoylation levels would have a general repressive effect on global transcription, while promoting expression of the small fraction of genes that are positively regulated by TF sumoylation. Such increases in steady-state sumoylation levels are observed under several stress conditions, including temperature, genotoxic, oxidative, and osmotic stresses, and indeed, several TFs become hypersumoylated in stress conditions, including heat shock.Citation3,73-75 Moreover, stress-induced sumoylation has been shown to influence the function of some TFs. For example, a variety of stress conditions increases sumoylation levels of c-Myb, which impairs its function as a transcriptional activator, while heat shock-induced sumoylation of the heat shock TF HSF2 increases expression of its targets.Citation23,76,77 Whereas the numerous studies on sumoylation of individual TFs point to roles in restricting or fine-tuning expression of their target genes under normal conditions, it remains to be determined, on a global scale, what role coordinated changes to TF sumoylation levels might play in shifting transcription patterns during stress and changing environmental conditions. Learning how sumoylation affects their individual functions and whether sumoylation of multiple TFs can occur in a coordinated manner in response to changing conditions will be key toward understanding how cells use SUMO modifications to regulate transcription of specific genes and genome-wide.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors
Acknowledgments
We thank Patricia Richard (Columbia University) for comments and suggestions.
Funding
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC; grant number RGPIN-04208–2014) and the Canadian Institutes of Health Research (CIHR; grant number MOP-142282) to E.R.
References
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013; 82:357-385; PMID:23746258; https://doi.org/10.1146/annurev-biochem-061909-093311
- Chymkowitch P, Nguéa PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. Bioessays 2015; 37:1095-1105; PMID:26354225; https://doi.org/10.1002/bies.201500065
- Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 2016; 17:581-595; PMID:27435506; https://doi.org/10.1038/nrm.2016.81
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 2017; 24:325-336; PMID:28112733; https://doi.org/10.1038/nsmb.3366
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev 2005; 15:536-541; PMID:16095902; https://doi.org/10.1016/j.gde.2005.07.004
- Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans 2007; 35:1389-1392; PMID:18031228; https://doi.org/10.1042/BST0351389
- Flotho A, Melchior F. Sumoylation: A regulatory protein modification in health and disease. Annu Rev Biochem 2013; 82:357-385; PMID:23746258; https://doi.org/10.1146/annurev-biochem-061909-093311
- Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML, Yang WM. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci Rep 2016; 6:26509; PMID:27211601; https://doi.org/10.1038/srep26509
- Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J 2006; 25:5083-5093; PMID:17036045; https://doi.org/10.1038/sj.emboj.7601383
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 2006; 103:45-50; PMID:16371476; https://doi.org/10.1073/pnas.0503698102
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861-871; PMID:21102611; https://doi.org/10.1038/nrm3011
- Zhang L, Vogel WK, Liu X, Topark-Ngarm A, Arbogast BL, Maier CS, Filtz TM, Leid M. Coordinated regulation of transcription factor Bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation. J Biol Chem 2012; 287:26971-26988; PMID:22700985; https://doi.org/10.1074/jbc.M112.344176
- Nayak A, Müller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 2014; 15:422; PMID:25315341; https://doi.org/10.1186/s13059-014-0422-2
- Keusekotten K, Bade VN, Meyer-Teschendorf K, Sriramachandran AM, Fischer-Schrader K, Krause A, Horst C, Schwarz G, Hofmann K, Dohmen RJ et al. Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem J 2014; 457:207-214; PMID:24151981; https://doi.org/10.1042/BJ20130753
- Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta 2014; 1843:75-85; PMID:24018209; https://doi.org/10.1016/j.bbamcr.2013.08.022
- Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 2004; 13:611-617; PMID:14992729; https://doi.org/10.1016/S1097-2765(04)00060-7
- Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics 2009; 4:440-444; PMID:19829068; https://doi.org/10.4161/epi.4.7.9807
- Andrade D, Velinder M, Singer J, Maese L, Bareyan D, Nguyen H, Chandrasekharan MB, Lucente H, McClellan D, Jones D et al. SUMOylation regulates growth factor independence 1 in transcriptional control and hematopoiesis. Mol Cell Biol 2016; 36:1438-1450; PMID:26951200; https://doi.org/10.1128/MCB.01001-15
- Hua G, Paulen L, Chambon P. GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc Natl Acad Sci 2016; 113:E626-634; PMID:26712002; https://doi.org/10.1073/pnas.1522821113
- Wang Y, Shankar SR, Kher D, Ling BMT, Taneja R. Sumoylation of the basic helix-loop-helix transcription factor sharp-1 regulates recruitment of the histone methyltransferase G9a and function in myogenesis. J Biol Chem 2013; 288:17654-17662; PMID:23637228; https://doi.org/10.1074/jbc.M113.463257
- Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 2009; 34:145-154; PMID:19394292; https://doi.org/10.1016/j.molcel.2009.03.013
- Lee Y, Chun SK, Kim K. Sumoylation controls CLOCK-BMAL1-mediated clock resetting via CBP recruitment in nuclear transcriptional foci. Biochim Biophys Acta 2015; 1853:2697-2708; PMID:26164627; https://doi.org/10.1016/j.bbamcr.2015.07.005
- Ledsaak M, Bengtsen M, Molværsmyr A-K, Fuglerud BM, Matre V, Eskeland R, Gabrielsen OS. PIAS1 binds p300 and behaves as a coactivator or corepressor of the transcription factor c-Myb dependent on SUMO-status. Biochim Biophys Acta 2016; 1859:705-718; PMID:27032383; https://doi.org/10.1016/j.bbagrm.2016.03.011
- Diehl C, Akke M, Bekker-Jensen S, Mailand N, Streicher W, Wikström M. Structural analysis of a complex between small ubiquitin-like modifier 1 (SUMO1) and the ZZ domain of CREB-binding protein (CBP/p300) reveals a new interaction surface on SUMO. J Biol Chem 2016; 291:12658-12672; PMID:27129204; https://doi.org/10.1074/jbc.M115.711325
- Tu J, Chen Y, Cai L, Xu C, Zhang Y, Chen Y, Zhang C, Zhao J, Cheng J, Xie H et al. Functional proteomics study reveals SUMOylation of TFII-I is involved in liver cancer cell proliferation. J Proteome Res 2015; 14:2385-2397; PMID:25869096; https://doi.org/10.1021/acs.jproteome.5b00062
- Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guérardel C, Leprince D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol 2010; 30:4045-4059; PMID:20547755; https://doi.org/10.1128/MCB.00582-09
- Escobar-Ramirez A, Vercoutter-Edouart AS, Mortuaire M, Huvent I, Hardivillé S, Hoedt E, Lefebvre T, Pierce A. Modification by SUMOylation controls both the transcriptional activity and the stability of delta-lactoferrin. PLoS One 2015; 10:e0129965; PMID:26090800; https://doi.org/10.1371/journal.pone.0129965
- Kim D-H, Xiao Z, Kwon S, Sun X, Ryerson D, Tkac D, Ma P, Wu SY, Chiang CM, Zhou E et al. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J 2015; 34:184-199; PMID:25425577; https://doi.org/10.15252/embj.201489527
- Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, Chin YE, Dong C, Yeh ET. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell 2012; 45:210-221; PMID:22284677; https://doi.org/10.1016/j.molcel.2011.12.026
- Droescher M, Begitt A, Marg A, Zacharias M, Vinkemeier U. Cytokine-induced paracrystals prolong the activity of signal transducers and activators of transcription (STAT) and provide a model for the regulation of protein solubility by small ubiquitin-like modifier (SUMO). J Biol Chem 2011; 286:18731-18746; PMID:21460228; https://doi.org/10.1074/jbc.M111.235978
- Cheema A, Knights CD, Rao M, Catania J, Perez R, Simons B, Dakshanamurthy S, Kolukula VK, Tilli M, Furth PA et al. Functional mimicry of the acetylated C-terminal tail of p53 by a SUMO-1 acetylated domain, SAD. J Cell Physiol 2010; 225:371-384; PMID:20458745; https://doi.org/10.1002/jcp.22224
- Wu SY, Chiang CM, Anckar J, Hietakangas V, Denessiouk K, Thiele D, Johnson M, Sistonen L, Appella E, Anderson C et al. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J 2009; 28:1246-1259; PMID:19339993; https://doi.org/10.1038/emboj.2009.83
- Lee HY, Johnson KD, Fujiwara T, Boyer ME, Kim SI, Bresnick EH. Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol Cell 2009; 36:984-995; PMID:20064464; https://doi.org/10.1016/j.molcel.2009.11.005
- Pan MR, Chang TM, Chang HC, Su JL, Wang HW, Hung WC. Sumoylation of Prox1 controls its ability to induce VEGFR3 expression and lymphatic phenotypes in endothelial cells. J Cell Sci 2009; 122:3358-3364; PMID:19706680; https://doi.org/10.1242/jcs.050005
- Sutinen P, Rahkama V, Rytinki M, Palvimo JJ. Nuclear mobility and activity of FOXA1 with androgen receptor are regulated by SUMOylation. Mol Endocrinol 2014; 28:1719-1728; PMID:25127374; https://doi.org/10.1210/me.2014-1035
- Smith M, Mallin DR, Simon JA, Courey AJ. Small ubiquitin-like modifier (SUMO) conjugation impedes transcriptional silencing by the polycomb group repressor Sex Comb on Midleg. J Biol Chem 2011; 286:11391-11400; PMID:21278366; https://doi.org/10.1074/jbc.M110.214569
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011; 480:99-103; PMID:22080950; https://doi.org/10.1038/nature10630
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, D'Hayer B, Mohamdi H, Remenieras A et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011; 480:94-98; PMID:22012259; https://doi.org/10.1038/nature10539
- Cheng J, Huang M, Zhu Y, Xin YJ, Zhao YK, Huang J, Yu JX, Zhou WH, Qiu Z. SUMOylation of MeCP2 is essential for transcriptional repression and hippocampal synapse development. J Neurochem 2014; 128:798-806; PMID:24188180; https://doi.org/10.1111/jnc.12523
- Balasubramaniyan N, Luo Y, Sun AQ, Suchy FJ. SUMOylation of the farnesoid X receptor (FXR) regulates the expression of FXR target genes. J Biol Chem 2013; 288:13850-13862; PMID:23546875; https://doi.org/10.1074/jbc.M112.443937
- Wei F, Schöler HR, Atchison ML. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J Biol Chem 2007; 282:21551-21560; PMID:17525163; https://doi.org/10.1074/jbc.M611041200
- Apostolov A, Litim-Mecheri I, Oravecz A, Goepp M, Kirstetter P, Marchal P, Ittel A, Mauvieux L, Chan S, Kastner P. Sumoylation inhibits the growth suppressive properties of ikaros. PLoS One 2016; 11:e0157767; PMID:27315244; https://doi.org/10.1371/journal.pone.0157767
- Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev 2010; 24:1242-1252; PMID:20504900; https://doi.org/10.1101/gad.1917910
- Chymkowitch P, Nguéa AP, Aanes H, Koehler CJ, Thiede B, Lorenz S, Meza-Zepeda LA, Klungland A, Enserink JM. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res 2015; 25:897-906; PMID:25800674; https://doi.org/10.1101/gr.185793.114
- Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res 2013; 23:1563-1579; PMID:23893515; https://doi.org/10.1101/gr.154872.113
- Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res 2012; 40:10172-10186; PMID:22941651; https://doi.org/10.1093/nar/gks819
- Seifert A, Schofield P, Barton GJ, Hay RT. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci Signal 2015; 8:rs7; PMID:26152697; https://doi.org/10.1126/scisignal.aaa2213
- Tempe D, Vives E, Brockly F, Brooks H, De Rossi S, Piechaczyk M, Bossis G. SUMOylation of the inducible (c-Fos:c-Jun)/AP-1 transcription complex occurs on target promoters to limit transcriptional activation. Oncogene 2014; 33:921-927; PMID:23396363; https://doi.org/10.1038/onc.2013.4
- Akhter A, Rosonina E. Chromatin association of Gcn4 is limited by post-translational modifications triggered by its DNA-binding in saccharomyces cerevisiae. Genetics 2016; 204:1433-1445; PMID:27992414; https://doi.org/10.1534/genetics.116.194134
- Rosonina E, Duncan SM, Manley JL. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev 2012; 26:350-355; PMID:22345516; https://doi.org/10.1101/gad.184689.111
- Wang M, Sang J, Ren Y, Liu K, Liu X, Zhang J, Wang H, Wang J, Orian A, Yang J et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation. Protein Cell 2016; 7:63-77; PMID:26511642; https://doi.org/10.1007/s13238-015-0216-7
- Wang M, Sang J, Ren Y, Liu K, Liu X, Zhang J, Wang H, Wang J, Orian A, Yang J et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation. Protein Cell 2016; 7:63-77; PMID:26511642; https://doi.org/10.1007/s13238-015-0216-7
- González-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 2015; 14:1859-1872; PMID:25895136; https://doi.org/10.1080/15384101.2015.1040965
- Guo B, Sharrocks AD. Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol Cell Biol 2009; 29:3204-3218; PMID:19307308; https://doi.org/10.1128/MCB.01128-08
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 1998; 2:233-239; PMID:9734360; https://doi.org/10.1016/S1097-2765(00)80133-1
- Chen S, Yu X, Lei Q, Ma L, Guo D. The sumoylation of zinc-fingers and homeoboxes 1 (ZHX1) by ubc9 regulates its stability and transcriptional repression activity. J Cell Biochem 2013; 114:2323-2333; PMID:23686912; https://doi.org/10.1002/jcb.24579
- Xing X, Bi H, Chang AK, Zang MX, Wang M, Ao X, Li S, Pan H, Guo Q, Wu H. SUMOylation of AhR modulates its activity and stability through inhibiting its ubiquitination. J Cell Physiol 2012; 227:3812-3819; PMID:22495806; https://doi.org/10.1002/jcp.24092
- Zhang ZB, Ruan CC, Chen DR, Zhang K, Yan C, Gao PJ. Activating transcription factor 3 SUMOylation is involved in angiotensin II-induced endothelial cell inflammation and dysfunction. J Mol Cell Cardiol 2016; 92:149-157; PMID:26850942; https://doi.org/10.1016/j.yjmcc.2016.02.001
- Ramachandran H, Herfurth K, Grosschedl R, Schäfer T, Walz G. SUMOylation Blocks the ubiquitin-mediated degradation of the nephronophthisis gene product Glis2/NPHP7. PLoS One 2015; 10:e0130275; PMID:26083374; https://doi.org/10.1371/journal.pone.0130275
- Gong L, Ji WK, Hu XH, Hu WF, Tang XC, Huang ZX, Li L, Liu M, Xiang SH, Wu E et al. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc Natl Acad Sci USA 2014; 111:5574-5579; PMID:24706897; https://doi.org/10.1073/pnas.1315034111
- Yan Q, Gong L, Deng M, Zhang L, Sun S, Liu JJ, Ma H, Yuan D, Chen PC, Hu X et al. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc Natl Acad Sci U S A 2010; 107:21034-21039; PMID:21084637; https://doi.org/10.1073/pnas.1007866107
- Zhang J, Yuan C, Wu J, Elsayed Z, Fu Z. Polo-like kinase 1-mediated phosphorylation of Forkhead box protein M1b antagonizes its SUMOylation and facilitates its mitotic function. J Biol Chem 2015; 290:3708-3719; PMID:25533473; https://doi.org/10.1074/jbc.M114.634386
- Chen L, Ma Y, Qian L, Wang J. Sumoylation regulates nuclear localization and function of zinc finger transcription factor ZIC3. Biochim Biophys Acta 2013; 1833:2725-2733; PMID:23872418; https://doi.org/10.1016/j.bbamcr.2013.07.009
- Sahin U, de Thé H, Lallemand-Breitenbach V. PML nuclear bodies: Assembly and oxidative stress-sensitive sumoylation. Nucleus 2014; 5:499-507; PMID:25482067; https://doi.org/10.4161/19491034.2014.970104
- Nayak A, Glöckner-Pagel J, Vaeth M, Schumann JE, Buttmann M, Bopp T, Schmitt E, Serfling E, Berberich-Siebelt F. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J Biol Chem 2009; 284:10935-10946; PMID:19218564; https://doi.org/10.1074/jbc.M900465200
- Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 2002; 21:5206-5215; PMID:12356736; https://doi.org/10.1093/emboj/cdf510
- Ross S, Best JL, Zon LI, Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 2002; 10:831-842; PMID:12419227; https://doi.org/10.1016/S1097-2765(02)00682-2
- Bies J, Markus J, Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem 2002; 277:8999-9009; PMID:11779867; https://doi.org/10.1074/jbc.M110453200
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 2000; 275:13321-13329; PMID:10788439; https://doi.org/10.1074/jbc.275.18.13321
- Hamard PJ, Boyer-Guittaut M, Camuzeaux B, Dujardin D, Hauss C, Oelgeschläger T, Vigneron M, Kedinger C, Chatton B. Sumoylation delays the ATF7 transcription factor subcellular localization and inhibits its transcriptional activity. Nucleic Acids Res 2007; 35:1134-1144; PMID:17264123; https://doi.org/10.1093/nar/gkl1168
- Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J Cell Biol 2012; 198:799-813; PMID:22927467; https://doi.org/10.1083/jcb.201204161
- van den Burg HA, Takken FL. SUMO-, MAPK-, and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signal Behav 2010; 5:1597-1601; PMID:21150289; https://doi.org/10.4161/psb.5.12.13913
- Lewicki MC, Srikumar T, Johnson E, Raught B. The S. cerevisiae SUMO stress response is a conjugation–deconjugation cycle that targets the transcription machinery. J Proteomics 2015; 118:39-48; PMID:25434491; https://doi.org/10.1016/j.jprot.2014.11.012
- Enserink JM. SUMO and the cellular stress response. Cell Div 2015; 10:4; PMID:26101541; https://doi.org/10.1186/s13008-015-0010-1
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2009; 2:ra24; PMID:19471022; https://doi.org/10.1126/scisignal.2000282
- Sramko M, Markus J, Kabat J, Wolff L, Bies J. Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J Biol Chem 2006; 281:40065-40075; PMID:17077080; https://doi.org/10.1074/jbc.M609404200
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD, Goodson M. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 2001; 276:40263-40267; PMID:11514557; https://doi.org/10.1074/jbc.M104714200
- Singh S, Pradhan AK, Chakraborty S. SUMO1 negatively regulates the transcriptional activity of EVI1 and significantly increases its co-localization with EVI1 after treatment with arsenic trioxide. Biochim Biophys Acta 2013; 1833:2357-2368; PMID:23770046; https://doi.org/10.1016/j.bbamcr.2013.06.003
- Lin BS, Tsai PY, Hsieh WY, Tsao HW, Liu MW, Grenningloh R, Wang LF, Ho IC, Miaw SC. SUMOylation attenuates c-Maf-dependent IL-4 expression. Eur J Immunol 2010; 40:1174-1184; PMID:20127678; https://doi.org/10.1002/eji.200939788
- Suico MA, Nakamura H, Lu Z, Saitoh H, Shuto T, Nakao M, Kai H. SUMO down-regulates the activity of Elf4/Myeloid Elf-1-like factor. Biochem Biophys Res Commun 2006; 348:880-888; PMID:16904644; https://doi.org/10.1016/j.bbrc.2006.07.151
